Abstract
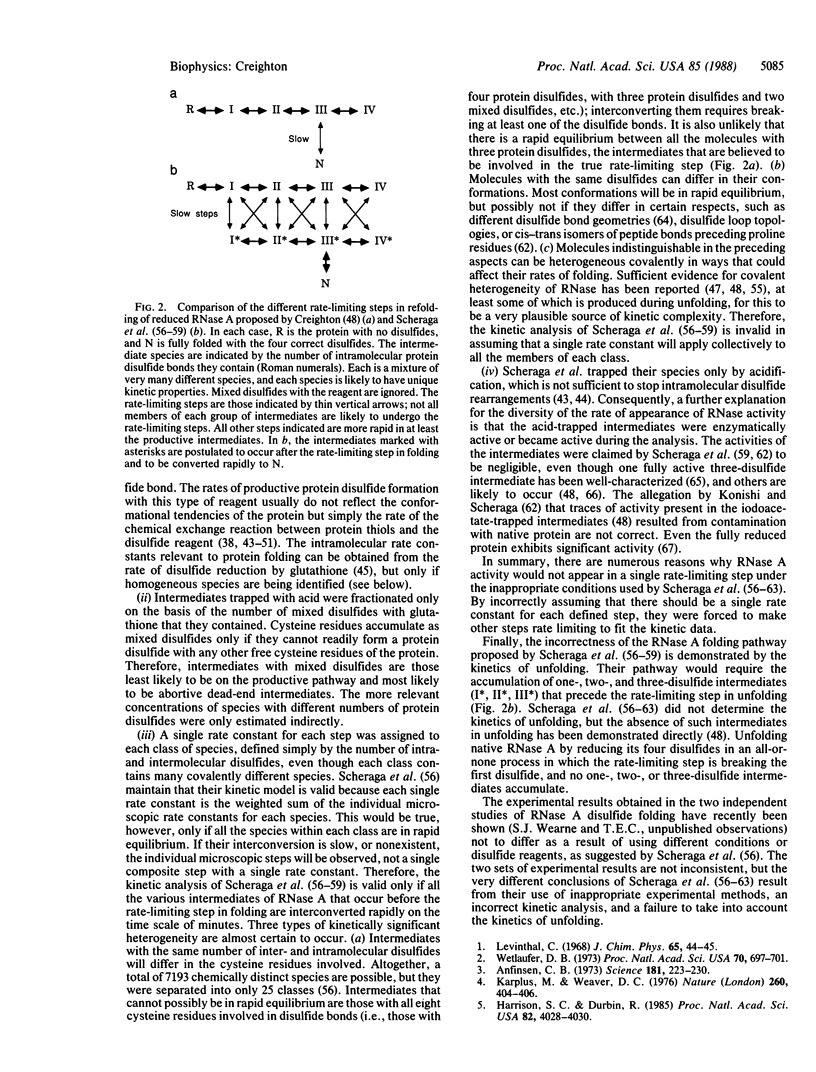
Experimental observations of how unfolded proteins refold to their native three-dimensional structures contrast with many popular theories of protein folding mechanisms. The available experimental evidence (ignoring slow cis-trans peptide bond isomerization) is largely consistent with the following general scheme: under folding conditions, unfolded protein molecules rapidly equilibrate between different conformations prior to complete refolding. This rapid prefolding equilibrium favors certain compact conformations that have somewhat lower free energies than the other unfolded conformations. Some of the favored conformations are important for productive folding. The rate-limiting step occurs late in the pathway and involves a high-energy, distorted form of the native conformation; there appears to be a single transition state through which essentially all molecules refold. Consequently, proteins are not assembled via a large number of independent pathways, nor is folding initiated by a nucleation event in the unfolded protein followed by rapid growth of the folded structure. The known folding pathways involving disulfide bond formation follow the same general principles. An exceptional folding mechanism for reduced ribonuclease A proposed by Scheraga et al. (Scheraga, H.A., Konishi, Y., Rothwarf, D.M. & Mui, P.W. (1987) Proc. Natl. Acad. Sci. USA 84, 5740-5744) is shown to result from experimental shortcomings, an incorrect kinetic analysis, and a failure to consider the kinetics of unfolding.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Brennan M., Lung-Nan Lin Unfolding and refolding occur much faster for a proline-free proteins than for most proline-containing proteins. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4178–4181. doi: 10.1073/pnas.74.10.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts J. F., Halvorson H. R., Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975 Nov 4;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. A three-disulphide intermediate in refolding of reduced ribonuclease A with a folded conformation. FEBS Lett. 1980 Sep 8;118(2):283–288. doi: 10.1016/0014-5793(80)80239-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. An empirical approach to protein conformation stability and flexibility. Biopolymers. 1983 Jan;22(1):49–58. doi: 10.1002/bip.360220110. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Conformational restrictions on the pathway of folding and unfolding of the pancreatic trypsin inhibitor. J Mol Biol. 1977 Jun 25;113(2):275–293. doi: 10.1016/0022-2836(77)90142-5. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bond formation in proteins. Methods Enzymol. 1984;107:305–329. doi: 10.1016/0076-6879(84)07021-x. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Energetics of folding and unfolding of pancreatic trypsin inhibitor. J Mol Biol. 1977 Jun 25;113(2):295–312. doi: 10.1016/0022-2836(77)90143-7. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Goldenberg D. P. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J Mol Biol. 1984 Nov 5;179(3):497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Intermediates in the refolding of reduced ribonuclease A. J Mol Biol. 1979 Apr 15;129(3):411–431. doi: 10.1016/0022-2836(79)90504-7. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Kinetic study of protein unfolding and refolding using urea gradient electrophoresis. J Mol Biol. 1980 Feb 15;137(1):61–80. doi: 10.1016/0022-2836(80)90157-6. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Kinetics of refolding of reduced ribonuclease. J Mol Biol. 1977 Jun 25;113(2):329–341. doi: 10.1016/0022-2836(77)90145-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Pain R. H. Unfolding and refolding of Staphylococcus aureus penicillinase by urea-gradient electrophoresis. J Mol Biol. 1980 Mar 15;137(4):431–436. doi: 10.1016/0022-2836(80)90167-9. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Renaturation of the reduced bovine pancreatic trypsin inhibitor. J Mol Biol. 1974 Aug 15;87(3):563–577. doi: 10.1016/0022-2836(74)90104-1. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Role of the environment in the refolding of reduced pancreatic trypsin inhibitor. J Mol Biol. 1980 Dec 25;144(4):521–550. doi: 10.1016/0022-2836(80)90335-6. [DOI] [PubMed] [Google Scholar]
- Denton J. B., Konishi Y., Scheraga H. A. Folding of ribonuclease A from a partially disordered conformation. Kinetic study under folding conditions. Biochemistry. 1982 Oct 12;21(21):5155–5163. doi: 10.1021/bi00264a008. [DOI] [PubMed] [Google Scholar]
- Garel A. T., Garel J. R. Partially oxidized active intermediates in refolding of reduced ribonuclease. J Biol Chem. 1982 Apr 25;257(8):4031–4033. [PubMed] [Google Scholar]
- Garel J. R., Baldwin R. L. Both the fast and slow refolding reactions of ribonuclease A yield native enzyme. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3347–3351. doi: 10.1073/pnas.70.12.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel J. R. Early steps in the refolding reaction of reduced ribonuclease A. J Mol Biol. 1978 Jan 25;118(3):331–345. doi: 10.1016/0022-2836(78)90232-2. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Nall B. T., Baldwin R. L. Guanidine-unfolded state of ribonuclease A contains both fast- and slow-refolding species. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1853–1857. doi: 10.1073/pnas.73.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmanshin R. I., Ptitsyn O. B. An early intermediate of refolding alpha-lactalbumin forms within 20 ms. FEBS Lett. 1987 Nov 2;223(2):327–329. doi: 10.1016/0014-5793(87)80313-7. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P., Creighton T. E. Energetics of protein structure and folding. Biopolymers. 1985 Jan;24(1):167–182. doi: 10.1002/bip.360240114. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P. Kinetic analysis of the folding and unfolding of a mutant form of bovine pancreatic trypsin inhibitor lacking the cysteine-14 and -38 thiols. Biochemistry. 1988 Apr 5;27(7):2481–2489. doi: 10.1021/bi00407a034. [DOI] [PubMed] [Google Scholar]
- Goto Y., Hamaguchi K. Role of amino-terminal residues in the folding of the constant fragment of the immunoglobulin light chain. Biochemistry. 1987 Apr 7;26(7):1879–1884. doi: 10.1021/bi00381a014. [DOI] [PubMed] [Google Scholar]
- Goto Y., Hamaguchi K. Unfolding and refolding of the reduced constant fragment of the immunoglobulin light chain. Kinetic role of the intrachain disulfide bond. J Mol Biol. 1982 Apr 25;156(4):911–926. doi: 10.1016/0022-2836(82)90147-4. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc Natl Acad Sci U S A. 1985 Jun;82(12):4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollecker M., Creighton T. E. Evolutionary conservation and variation of protein folding pathways. Two protease inhibitor homologues from black mamba venom. J Mol Biol. 1983 Aug 5;168(2):409–437. doi: 10.1016/s0022-2836(83)80026-6. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M., Kuwajima K., Mitani M., Sugai S. Evidence for identity between the equilibrium unfolding intermediate and a transient folding intermediate: a comparative study of the folding reactions of alpha-lactalbumin and lysozyme. Biochemistry. 1986 Nov 4;25(22):6965–6972. doi: 10.1021/bi00370a034. [DOI] [PubMed] [Google Scholar]
- Karplus M., Weaver D. L. Protein-folding dynamics. Nature. 1976 Apr 1;260(5550):404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- Kato S., Okamura M., Shimamoto N., Utiyama H. Spectral evidence for a rapidly formed structural intermediate in the refolding kinetics of hen egg-white lysozyme. Biochemistry. 1981 Mar 3;20(5):1080–1085. doi: 10.1021/bi00508a006. [DOI] [PubMed] [Google Scholar]
- Kato S., Shimamoto N., Utiyama H. Identification and characterization of the direct folding process of hen egg-white lysozyme. Biochemistry. 1982 Jan 5;21(1):38–43. doi: 10.1021/bi00530a007. [DOI] [PubMed] [Google Scholar]
- Kelley R. F., Richards F. M. Replacement of proline-76 with alanine eliminates the slowest kinetic phase in thioredoxin folding. Biochemistry. 1987 Oct 20;26(21):6765–6774. doi: 10.1021/bi00395a028. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Ooi T., Scheraga H. A. Regeneration of RNase A from the reduced protein: models of regeneration pathways. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5734–5738. doi: 10.1073/pnas.79.18.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y., Ooi T., Scheraga H. A. Regeneration of ribonuclease A from the reduced protein. Energetic analysis. Biochemistry. 1982 Sep 14;21(19):4741–4748. doi: 10.1021/bi00262a034. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Ooi T., Scheraga H. A. Regeneration of ribonuclease A from the reduced protein. Isolation and identification of intermediates, and equilibrium treatment. Biochemistry. 1981 Jul 7;20(14):3945–3955. doi: 10.1021/bi00517a001. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Ooi T., Scheraga H. A. Regeneration of ribonuclease A from the reduced protein. Rate-limiting steps. Biochemistry. 1982 Sep 14;21(19):4734–4740. doi: 10.1021/bi00262a033. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Scheraga H. A. Regeneration of ribonuclease A from the reduced protein. 1. Conformational analysis of the intermediates by measurements of enzymatic activity, optical density, and optical rotation. Biochemistry. 1980 Apr 1;19(7):1308–1316. doi: 10.1021/bi00548a008. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Scheraga H. A. Regeneration of ribonuclease A from the reduced protein. 2. Conformational analysis of the intermediates by nuclear magnetic resonance spectroscopy. Biochemistry. 1980 Apr 1;19(7):1316–1322. doi: 10.1021/bi00548a009. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Nitta K., Yoneyama M., Sugai S. Three-state denaturation of alpha-lactalbumin by guanidine hydrochloride. J Mol Biol. 1976 Sep 15;106(2):359–373. doi: 10.1016/0022-2836(76)90091-7. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Yamaya H., Miwa S., Sugai S., Nagamura T. Rapid formation of secondary structure framework in protein folding studied by stopped-flow circular dichroism. FEBS Lett. 1987 Aug 31;221(1):115–118. doi: 10.1016/0014-5793(87)80363-0. [DOI] [PubMed] [Google Scholar]
- Lin L. N., Brandts J. F. Further evidence suggesting that the slow phase in protein unfolding and refolding is due to proline isomerization: a kinetic study of carp parvalbumins. Biochemistry. 1978 Sep 19;17(19):4102–4110. doi: 10.1021/bi00612a036. [DOI] [PubMed] [Google Scholar]
- Lin L. N., Brandts J. F. Involvement of prolines-114 and -117 in the slow refolding phase of ribonuclease A as determined by isomer-specific proteolysis. Biochemistry. 1984 Nov 20;23(24):5713–5723. doi: 10.1021/bi00319a009. [DOI] [PubMed] [Google Scholar]
- Lin S. H., Konishi Y., Nall B. T., Scheraga H. A. Influence of an extrinsic cross-link on the folding pathway of ribonuclease A. Kinetics of folding-unfolding. Biochemistry. 1985 May 21;24(11):2680–2686. doi: 10.1021/bi00332a013. [DOI] [PubMed] [Google Scholar]
- Lynn R. M., Konishi Y., Scheraga H. A. Folding of ribonuclease A from a partially disordered conformation. Kinetic study under transition conditions. Biochemistry. 1984 May 22;23(11):2470–2477. doi: 10.1021/bi00306a023. [DOI] [PubMed] [Google Scholar]
- Marks C. B., Naderi H., Kosen P. A., Kuntz I. D., Anderson S. Mutants of bovine pancreatic trypsin inhibitor lacking cysteines 14 and 38 can fold properly. Science. 1987 Mar 13;235(4794):1370–1373. doi: 10.1126/science.2435002. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Creighton T. E. The disulphide folding pathway of ribonuclease T1. J Mol Biol. 1986 Apr 5;188(3):477–486. doi: 10.1016/0022-2836(86)90169-5. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A., Konishi Y., Ooi T. Multiple pathways for regenerating ribonuclease A. Adv Biophys. 1984;18:21–41. doi: 10.1016/0065-227x(84)90005-4. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A., Konishi Y., Rothwarf D. M., Mui P. W. Toward an understanding of the folding of ribonuclease A. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5740–5744. doi: 10.1073/pnas.84.16.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X., Baldwin R. L. Detection of an early intermediate in the folding of ribonuclease A by protection of amide protons against exchange. J Mol Biol. 1979 Nov 25;135(1):199–215. doi: 10.1016/0022-2836(79)90347-4. [DOI] [PubMed] [Google Scholar]
- Schmid F., Blaschek H. An early intermediate in the folding of ribonuclease A is protected against cleavage by pepsin. Biochemistry. 1984 May 8;23(10):2128–2133. doi: 10.1021/bi00305a004. [DOI] [PubMed] [Google Scholar]
- Segawa S., Husimi Y., Wada A. Kinetics of thermal unfolding of lysozyme. Biopolymers. 1973 Nov;12(11):2521–2537. doi: 10.1002/bip.1973.360121107. [DOI] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. I. Effect of protein-solvent interactions on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2473–2488. doi: 10.1002/bip.360231122. [DOI] [PubMed] [Google Scholar]
- Segawa S., Sugihara M. Characterization of the transition state of lysozyme unfolding. II. Effects of the intrachain crosslinking and the inhibitor binding on the transition state. Biopolymers. 1984 Nov;23(11 Pt 2):2489–2498. doi: 10.1002/bip.360231123. [DOI] [PubMed] [Google Scholar]
- Semisotnov G. V., Rodionova N. A., Kutyshenko V. P., Ebert B., Blanck J., Ptitsyn O. B. Sequential mechanism of refolding of carbonic anhydrase B. FEBS Lett. 1987 Nov 16;224(1):9–13. doi: 10.1016/0014-5793(87)80412-x. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Thannhauser T. W., Scheraga H. A. Reversible blocking of half-cystine residues of proteins and an irreversible specific deamidation of asparagine-67 of S-sulforibonuclease under mild conditions. Biochemistry. 1985 Dec 17;24(26):7681–7688. doi: 10.1021/bi00347a027. [DOI] [PubMed] [Google Scholar]
- Touchette N. A., Perry K. M., Matthews C. R. Folding of dihydrofolate reductase from Escherichia coli. Biochemistry. 1986 Sep 23;25(19):5445–5452. doi: 10.1021/bi00367a015. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B., Branca P. A., Chen G. X. The oxidative folding of proteins by disulfide plus thiol does not correlate with redox potential. Protein Eng. 1987 Feb-Mar;1(2):141–146. doi: 10.1093/protein/1.2.141. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973 Mar;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]



