Abstract
We present a comprehensive phylogeny derived from 5 genes, nucSSU, nucLSU rDNA, TEF1, RPB1 and RPB2, for 356 isolates and 41 families (six newly described in this volume) in Dothideomycetes. All currently accepted orders in the class are represented for the first time in addition to numerous previously unplaced lineages. Subclass Pleosporomycetidae is expanded to include the aquatic order Jahnulales. An ancestral reconstruction of basic nutritional modes supports numerous transitions from saprobic life histories to plant associated and lichenised modes and a transition from terrestrial to aquatic habitats are confirmed. Finally, a genomic comparison of 6 dothideomycete genomes with other fungi finds a high level of unique protein associated with the class, supporting its delineation as a separate taxon.
Keywords: Ascomycota, Pezizomycotina, Dothideomyceta, fungal evolution, lichens, multigene phylogeny, phylogenomics, plant pathogens, saprobes, Tree of Life
INTRODUCTION
Multi laboratory collaborative research in various biological disciplines is providing a high level of interaction amongst researchers with diverse interests and backgrounds. For the mycological community, the “Assembling the Fungal Tree of Life” project (AFTOL) provided the first DNA-based comprehensive multigene phylogenetic view of the fungal Kingdom (Lutzoni et al. 2004, James et al. 2006). This has also made it possible to revise the classification of the fungi above the ordinal level (Hibbett et al. 2007). Subsequent work is focused on elucidating poorly resolved nodes that were highlighted in the initial DNA-based phylogeny (McLaughlin et al. 2009).
At the other end of the scale from the tree of life projects, taxon sampling with relatively small numbers of sequence characters are also progressing in various barcoding projects (Seifert et al. 2007, Chase et al. 2009, Seifert 2009). It remains important to link these two ends of the spectrum by also sampling intensively at foci of interest between barcoding and the tree of life. With this in mind it is the aim of this paper and subsequent ones in this volume to provide a broadly sampled phylogeny at class level and below for Dothideomycetes. This result is combined efforts and data from a diverse group of researchers to focus on systematic sampling, therefore developing a more robust fungal class wide phylogeny of Dothideomycetes. This is especially important as a framework for comprehending how fungi have evolved as they shift ecological habitats and adapt to new environments and nutritional modes.
It is apparent that the assemblage of fungi, now defined as Dothideomycetes, exemplifies a dynamic evolutionary history. This is by far the largest and arguably most phylogenetically diverse class within the largest fungal phylum, Ascomycota (Kirk et al. 2008). It contains a heterogeneous group of fungi that subsist in the majority of the niches where fungi can be found. The best-known members of the group are plant pathogens that cause serious crop losses. Species in the genera Cochliobolus, Didymella, Phaeosphaeria, Pyrenophora, Venturia, Mycosphaerella and Leptosphaeria, or their anamorphs, are major pathogens of corn, melons, wheat, barley, apples, bananas and brassicas respectively, in most areas of the world where they are cultivated. Other species are important pathogens in forestry e.g. species in the genera Botryosphaeria and Mycosphaerella and their anamorphs that attack economically important tree species.
Despite a large body of work containing taxonomic, phytopathological, genetic and genomic research, the majority of fungi hypothesised to be members of Dothideomycetes remain under-sampled within a systematic framework. Several studies performed during the course of the last four years have advanced our understanding of these fungi, but phylogenetic relationships of the saprobes, aquatic, asexual and lichenised species remain particularly poorly studied. Indeed, their conspicuous absence in phylogenetic analyses frustrates a broader understanding of dothideomycete evolution.
Dothideomycetes share a number of morphological characters with other fungal classes. It was recently formally described (Eriksson & Winka 1997) replacing in part the long-recognised loculoascomycetes (Luttrell 1955). This redefinition of the loculoascomycetes was mainly prompted by DNA sequencing comparisons of ribosomal RNA genes (Berbee & Taylor 1992, Spatafora et al. 1995) that was subsequently expanded and confirmed (Berbee 1996, Silva-Hanlin & Hanlin 1999, Lindemuth et al. 2001, Lumbsch & Lindemuth 2001). These early phylogenetic studies demonstrated that loculoascomycetes, as it was defined, is not monophyletic, although contrary views exist (Liu & Hall 2004). Nevertheless the majority of analyses have shown that some loculoascomycete taxa, such as the “black yeasts” in Chaetothyriales as well as the lichenised Verrucariales, reside within Eurotiomycetes as subclass Chaetothyriomycetidae (Spatafora et al. 1995, Winka et al. 1998, Geiser et al. 2006, Gueidan et al. 2008). The majority of the remaining loculoascomycete species are now placed in Dothideomycetes. Although finer morphological distinctions between the distantly related members of loculoascomycetes can be made, their synapomorphies remain elusive (Lumbsch & Huhndorf 2007). These findings all point to the fact that a number of loculoascomycete morphological characters are either retained ancestral traits or that they exhibit convergence due to similar selection pressures.
Traditionally the most important morphological characters used to define major groups in Ascomycota were the type of ascus, septation of ascospores, the morphology and development of the ascoma, as well as the structure and organisation of the centrum. Dothideomycetes (and previously, loculoascomycetes) have fissitunicate (or functionally bitunicate) asci, that emerge from ascolocular development in preformed locules within vegetative tissue, that represents the ascoma. The reproductive structures in ascolocular development are derived from cells before fusion of opposing mating types occurs and can contain one or several locules. This form of ascolocular development is in contrast to the ascohymenial development found in most other fungal classes. During ascohymenial development asci are generated in a hymenium and the reproductive structure is derived from cells after fusion of opposing mating types. The fissitunicate ascus has been described for more than a century, but the importance of ascolocular development was first emphasised in 1932 (Nannfeldt 1932). Importantly Nannfeldt's concepts were also the basis for the Santesson's integration of lichens into the fungal classification (Santesson 1952). In fissitunicate asci, generally, the ascospores are dispersed by the rupture of the thick outer layers (ectotunica) at its apex, allowing the thinner inner layer (endotunica) to elongate similar to a “jack in a box”. The elongated endotunica ruptures apically and releases the ascospores forcefully through the ascoma opening. The spores are then released in the air, or in aquatic species, under water. Building on this work and that of others (Miller 1949), Luttrell proposed Loculoascomycetes, synonymous to Nannfeldt's “Ascoloculares” (Luttrell 1955). Importantly, he proposed a correlation between fissitunicate asci and ascolocular development, also emphasising the importance of ascus morphology and dehiscence as well as the development of surrounding elements within the ascoma.
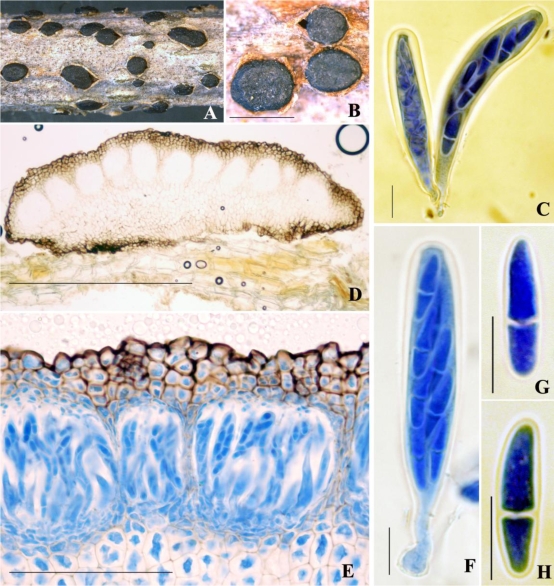
Although the concept of a group of fungi (including the Dothideomycetes) with fissitunicate asci and ascolocular development has been accepted by several authors, much less agreement could be found on ordinal definitions in the era before molecular characters. This ranged from proposing a single order (von Arx & Müller 1975) to three (Müller & von Arx 1962), five (Luttrell 1951, 1955) six (Barr 1979), or seven (Barr 1987). Luttrell initially described a number of important development types centered on descriptions of all tissues inside the ascoma (the centrum concept) and combined this with ascoma structure to define his five orders (Luttrell 1951, 1955). Of Luttrell's initial centrum concepts three are applicable to the Dothideomycetes as they are presently defined. Thus, the Pleospora type, the Dothidea type and the Elsinoë type centra correspond to the dothideomycete orders Pleosporales, Dothideales and Myriangiales, respectively. An important refinement to Luttrell's ideas was introduced with the concept of the hamathecium by Eriksson (Eriksson 1981). This is defined as a neutral term for sterile hyphae or other tissues between the asci in the ascoma (Kirk et al. 2008). For example, hamathecial types can include the presence or absence of pseudoparaphyses, which are sterile cells that extend down from the upper portion of the ascomatal cavity. They become attached at both ends, although the upper part may become free at maturity. Other important concepts introduced by Müller and von Arx (Müller & von Arx 1962) focused on the morphology of the ascoma opening and ascus shape. The Dothidea type centrum in the type species of Dothidea, D. sambuci illustrates several typical dothideomycete morphologies (Fig. 1). These include the thick-walled fissitunicate asci produced within a multilocular stroma.
Fig. 1.
Dothidea sambuci. A–B. Appearance of ascomata on the host surface. C, F. Asci in cotton blue reagent. D. Vertical section through ascomata illustrating the mutilocule at the upper layer. E. Vertical section through ascomata in cotton blue reagent illustrating the locule. G–H. Ascospores in cotton blue reagent. Scale bars: B = 1000 μm; C = 500 μm; E = 100 μm; F–H = 10 μm.
The most recent dothideomycete class-wide morphological assessments were carried out by Barr (Barr 1979, 1987). Her subclasses were determined based on characters in the centrum, including the absence, presence and types of hamathecial tissues. Consistent with several earlier authors, Barr's ordinal classifications were based on ascomatal shape (perithecioid or apothecioid) and manner in which nutrients are obtained by the fungus (Barr 1987). In addition to these characters she emphasised the importance of finer distinctions in the hamathecium such as the shape and structure of the pseudoparaphyses (Barr 1979, 1987).
The introduction of molecular phylogenies for Dothideomycetes (Berbee 1996) provided an opportunity to verify the significance of various morphological characters used in the aforementioned classifications. The clearest correlation with a DNA sequence-based phylogeny was for the presence or absence of pseudoparaphyses, largely agreeing with the first orders proposed by Luttrell (Liew et al. 2000, Lumbsch & Lindemuth 2001). Barr's concept of applying the shape of the pseudoparaphyses to define orders was rejected by molecular phylogenies (Liew et al. 2000). This set the stage for more comprehensive analyses incorporating protein data, and resulted in the definition of two subclasses, Pleosporomycetidae (pseudoparaphyses present) and the Dothideomycetidae (pseudoparaphyses absent; Schoch et al. 2006). Numerous orders and other taxa remained unresolved outside of these two subclasses.
The most recent class level phylogenetic analyses combining sequences from protein coding genes with ribosomal RNA sequences fortified the view that Dothideomycetes is a monophyletic group (Schoch et al. 2009a, b). Furthermore, strong support was found for a sister relationship between Dothideomycetes and the lichenised class Arthoniomycetes (Lumbsch et al. 2005, Spatafora et al. 2006, Schoch et al. 2009a). This clade was recently defined as a rankless taxon “Dothideomyceta” (Schoch et al. 2009a, b). The Arthoniomycetes consists of a single order (Arthoniales) of lichens and lichenicolous fungi (Ertz et al. 2009) that produce bitunicate asci in ascohymenial apothecia and was proposed as an intermediate group or “Zwischengruppe” (Henssen & Thor 1994). This placement raises intriguing questions regarding the origins of ascolocular development and further illustrates the importance of including lichen-forming fungi in dothideomycete phylogenies.
While considerable progress has been made in defining these fungi the placement of Dothideomycetes in relation to the majority of other Ascomycota classes remains unresolved. Here, greater clarity would likely require a huge increase of characters from genome projects. In this regard, the first phylogenomic studies have shown low resolution for this relationship (Fitzpatrick et al. 2006, Kuramae et al. 2006, Robbertse et al. 2006). This could indicate a rapid radiation event, but more likely suggests taxon sampling bias. This latter view is supported by the fact that none of these studies has included lichenised species that represent about 25 % of the number of species in Ascomycota.
The authors of this volume have focused on two primary goals. These are to considerably expand the taxon sampling of existing orders by including saprobes, asexual species and other poorly sampled groups. Secondly we aim to sample widely within specific environmental niches and present a multigene phylogeny that exposes the highly diverse nature of Dothideomycetes.
MATERIAL AND METHODS
DNA extraction, amplification and sequencing
The majority of fungal cultures were obtained from the CBS culture collection and additional sources mentioned in other papers of this volume. DNA was also provided by authors of several papers presented in this volume and the reader is referred to Boehm et al. (2009a), Crous et al. (2009a), Suetrong et al. (2009) and Zhang et al. (2009). For additional details see Table 1 - see online Supplementary Information. Fungal genomic DNA was obtained by scraping mycelium from PDA plates. Samples were subsequently pulverised and the DNA was extracted using the FastDNA® kit and the FastPrep® instrument from MPI Biochemicals (Irvine, CA, U.S.A.). DNA amplifications were completed using Taq polymerase (GenScript, Piscataway, NJ, U.S.A.), with FailSafe™ PCR 2× PreMix E (Epicentre, San Diego, CA, U.S.A.). Primers were used as noted in the Assembling the Fungal Tree of Life project (AFTOL; Schoch et al. 2009a). This resulted in DNA sequence data obtained from the small and large subunits of the nuclear ribosomal RNA genes (SSU, LSU) and three protein coding genes, namely the translation elongation factor-1 alpha (TEF1) and the largest and second largest subunits of RNA polymerase II (RPB1, RPB2). Primer sets used for these genes were as follows: SSU: NS1/NS4; LSU: LR0R/LR5; TEF1 983/2218R (initially obtained from S. Rehner: ocid.nacse.org/research/deephyphae/EF1primer.pdf); RPB2: fRPB2-SF/fRPB2-7cR; RPB1: RPB1-Ac/RPB1-Cr (obtained from V. Hofstetter). Primer sequences are available at the WASABI database at the AFTOL website (aftol.org). PCRs for these genes were performed in various laboratories of the coauthors mentioned but the majority of reactions were run under conditions described previously (Lutzoni et al. 2004, Schoch et al. 2009a). Two duplicate sets of sequences were inadvertently included in the analysis (indicated in Table 1).
Table 1.
Isolates of Dothideomycetes included in this study. Newly deposited sequences are shown in bold.
| Taxon | voucher/culture1 | SSU | LSU | RPB1 | RPB2 | TEF1 |
|---|---|---|---|---|---|---|
| Acanthostigma perpusillum | UAMH | AY856937 | AY856892 | |||
| Aglaospora profusa | CBS 123109 | GU296130 | GU301792 | GU349062 | ||
| Aigialus grandis 1 | 2Q | GU296132 | GU301794 | GU349063 | ||
| Aigialus grandis 2 | JK 5244A | GU296131 | GU301793 | GU371762 | ||
| Aigialus parvus | A6 | GU296133 | GU301795 | GU371771 | GU349064 | |
| Aliquandostipite khaoyaiensis | CBS 118232 | AF201453 | GU301796 | FJ238360 | GU349048 | |
| Alternaria alternata | CBS 916.96 | DQ678031 | DQ678082 | DQ677980 | DQ677927 | |
| Amniculicola parva | CBS 123092 | GU296134 | FJ795497 | GU349065 | ||
| Anteaglonium abbreviatum 1 | ANM 925.1 | GQ221877 | GQ221924 | |||
| Anteaglonium abbreviatum 2 | GKM 1029 | GQ221878 | GQ221915 | |||
| Anteaglonium globosum 1 | SMH 5283 | GQ221911 | GQ221919 | |||
| Anteaglonium globosum 2 | ANM 925.2 | GQ221879 | GQ221925 | |||
| Anteaglonium latirostrum | L100N 2 | GQ221876 | GQ221938 | |||
| Anteaglonium parvulum | SMH 5210 | GQ221907 | GQ221917 | |||
| Apiosporina collinsii | CBS 118973 | GU296135 | GU301798 | GU357778 | GU349057 | |
| Apiosporina morbosa | dimosp | EF114694 | ||||
| Arthopyrenia salicis 1 | 1994 Coppins | AY607730 | AY607742 | |||
| Arthopyrenia salicis 2 | CBS 368.94 | AY538333 | AY538339 | GU371814 | ||
| Ascochyta pisi | CBS 126.54 | DQ678018 | DQ678070 | DQ677967 | DQ677913 | |
| Ascocratera manglicola | JK 5262C | GU296136 | GU301799 | GU371763 | ||
| Asteromassaria pulchra | CBS 124082 | GU296137 | GU301800 | GU371772 | GU349066 | |
| Astrosphaeriella aggregata | MAFF 239486 | AB524450 | AB524591 | AB539105 | AB539092 | |
| Astrosphaeriella bakeriana | CBS 115556 | GU301801 | GU357752 | GU349015 | ||
| Astrothelium cinnamomeum | DUKE 0000007 | AY584652 | DQ782896 | |||
| Aulographina pinorum 1 | CBS 302.71 | GU371766 | ||||
| Aulographina pinorum 2 | CBS 174.90 | GU296138 | GU301802 | GU357763 | GU371737 | GU349046 |
| Aureobasidium pullulans | CBS 584.75 | DQ471004 | DQ470956 | DQ471148 | DQ470906 | DQ471075 |
| Bagnisiella examinans | CBS 551.66 | GU296139 | GU301803 | GU357776 | GU371746 | GU349056 |
| Batcheloromyces proteae | CBS 110696 | AY251102 | EU019247 | |||
| Beverwykella pulmonaria | CBS 283.53 | GU301804 | GU371768 | |||
| Bimuria novae-zelandiae | CBS 107.79 | AY016338 | AY016356 | DQ471159 | DQ470917 | DQ471087 |
| Botryosphaeria dothidea | CBS 115476 | DQ677998 | DQ678051 | GU357802 | DQ677944 | DQ767637 |
| Botryosphaeria tsugae | CBS 418.64 | AF271127 | DQ767655 | DQ767644 | DQ677914 | |
| Byssolophis sphaerioides | IFRDCC2053 | GU296140 | GU301805 | GU456348 | GU456263 | |
| Byssothecium circinans | CBS 675.92 | AY016339 | AY016357 | DQ767646 | GU349061 | |
| Camarosporium quaternatum | CBS 483.95 | GU296141 | GU301806 | GU357761 | GU349044 | |
| Capnobotryella renispora | CBS 215.90 | AY220613 | GQ852582 | |||
| Capnodium coffeae | CBS 147.52 | DQ247808 | DQ247800 | DQ471162 | DQ247788 | DQ471089 |
| Capnodium salicinum | CBS 131.34 | DQ677997 | DQ678050 | DQ677889 | ||
| Catenulostroma abietis (as Trimmatostroma abietis) | CBS 459.93 | DQ678040 | DQ678092 | GU357797 | DQ677933 | |
| Catenulostroma elginense | CBS 111030 | GU214517 | EU019252 | |||
| Catinella olivacea | UAMH 10679 | DQ915484 | EF622212 | |||
| Cenococcum geophilum 1 | HUNT A1 | L76616 | ||||
| Cenococcum geophilum 2 | CGMONT | L76617 | ||||
| Cenococcum geophilum 3 | 10 | L76618 | ||||
| Cercospora beticola | CBS 116456 | DQ678039 | DQ678091 | DQ677932 | ||
| Chaetosphaeronema hispidulum | CBS 216.75 | EU754045 | EU754144 | GU357808 | GU371777 | |
| Cladosporium cladosporioides | CBS 170.54 | DQ678004 | DQ678057 | GU357790 | DQ677952 | DQ677898 |
| Cladosporium iridis (teleomorph Davidiella macrospora) | CBS 138.40 | DQ008148 | ||||
| Clathrospora elynae | CBS 196.54 | GU296142 | GU323214 | |||
| Cochliobolus heterostrophus | CBS 134.39 | AY544727 | AY544645 | DQ247790 | DQ497603 | |
| Cochliobolus sativus | DAOM 226212 | DQ677995 | DQ678045 | DQ677939 | ||
| Columnosphaeria fagi | CBS 171.93 | AY016342 | AY016359 | DQ677966 | ||
| Comminutispora agavaciensis | CBS 619 95 | Y18699 | EU981286 | |||
| Conidioxyphium gardeniorum | CPC 14327 | GU296143 | GU301807 | GU357774 | GU371743 | GU349054 |
| Coniothyrium palmarum | CBS 400.71 | DQ678008 | DQ767653 | DQ677956 | DQ677903 | |
| Corynespora cassiicola 1 | CBS 100822 | GU296144 | GU301808 | GU357772 | GU371742 | GU349052 |
| Corynespora cassiicola 2 | CCP | GU296145 | ||||
| Corynespora olivacea | CBS 114450 | GU301809 | GU349014 | |||
| Corynespora smithii | CABI 5649b | GU323201 | GU371804 | GU371783 | GU349018 | |
| Cryptothelium amazonum | 47 | GU327713 | GU327731 | |||
| Cryptothelium pulchrum | 63C | GU327714 | ||||
| Cystocoleus ebeneus 1 | L348 | EU048573 | EU048580 | |||
| Cystocoleus ebeneus 2 | L315 | EU048572 | ||||
| Davidiella tassiana | CBS 399.80 | DQ678022 | DQ678074 | GU357793 | DQ677971 | DQ677918 |
| Delitschia cf. chaetomioides 1 | GKM 3253.2 | GU390656 | ||||
| Delitschia cf. chaetomioides 2 | GKM 1283 | GU385172 | ||||
| Delitschia didyma 1 (duplicate) | UME 31411 | DQ384090 | ||||
| Delitschia didyma 2 | UME 31411 | AF242264 | DQ384090 | |||
| Delitschia winteri | CBS 225.62 | DQ678026 | DQ678077 | DQ677975 | DQ677922 | |
| Delphinella strobiligena | CBS 735.71 | DQ470977 | DQ471175 | DQ677951 | DQ471100 | |
| Devriesia staurophora | CBS 375.81 | EF137359 | DQ008151 | |||
| Devriesia strelitziae | CBS 122379 | GU296146 | GU301810 | GU371738 | GU349049 | |
| Didymella bryoniae (as Phoma cucurbitacearum) | CBS 133.96 | GU301863 | GU371767 | |||
| Didymella exigua | CBS 183.55 | GU296147 | GU357800 | GU371764 | ||
| Didymocrea sadasivanii | CBS 438 65 | DQ384066 | DQ384103 | |||
| Diplodia mutila (teleomorph Botryosphaeria stevensii) | CBS 431.82 | DQ678012 | DQ678064 | DQ677960 | DQ677907 | |
| Dissoconium aciculare | CBS 204.89 | GU214523 | GQ852587 | |||
| Dissoconium commune (teleomorph Mycosphaerella communis) | CBS 110747 | GU214525 | GQ852589 | |||
| Dissoconium dekkeri (teleomorph Mycosphaerella lateralis) | CBS 111282 | GU214531 | GU214425 | |||
| Dothidea hippophaës | CBS 188.58 | U42475 | DQ678048 | GU357801 | DQ677942 | DQ677887 |
| Dothidea insculpta | CBS 189.58 | DQ247810 | DQ247802 | DQ471154 | AF107800 | DQ471081 |
| Dothidea sambuci | DAOM 231303 | AY544722 | AY544681 | DQ522854 | DQ497606 | |
| Dothiora cannabinae | CBS 737.71 | DQ479933 | DQ470984 | DQ471182 | DQ470936 | DQ471107 |
| Dothiora elliptica | CBS 736.71 | GU301811 | GU349013 | |||
| Dothistroma septosporum 1 (teleomorph Mycosphaerella pini) | CBS 543 74 | GU301853 | GU371730 | |||
| Dothistroma septosporum 2 | CBS 112498 | GU214533 | GQ852597 | |||
| Elsinoë centrolobi | CBS 222.50 | DQ678041 | DQ678094 | GU357798 | DQ677934 | |
| Elsinoë phaseoli | CBS 165.31 | DQ678042 | DQ678095 | GU357799 | DQ677935 | |
| Elsinoë veneta | CBS 150.27 | DQ767651 | DQ767658 | DQ767641 | ||
| Endosporium aviarium | UAMH 10530 | EU304349 | EU304351 | |||
| Endosporium populi-tremuloidis | UAMH 10529 | EU304346_ | EU304348 | |||
| Entodesmium rude | CBS 650.86 | GU301812 | GU349012 | |||
| Falciformispora lignatilis 1 | BCC 21118 | GU371835 | GU371827 | GU371820 | ||
| Falciformispora lignatilis 2 | BCC 21117 | GU371834 | GU371826 | GU371819 | ||
| Farlowiella carmichaeliana 2 | CBS 179.73 | GU296148 | ||||
| Farlowiella carmichealiana 1 (as anamorph Acrogenospora sphaerocephala) | CBS 164.76 | GU296129 | GU301791 | GU357780 | GU371748 | GU349059 |
| Floricola striata | JK 56781 | GU296149 | GU301813 | GU371758 | ||
| Friedmanniomyces endolithicus | CCFEE 522 | DQ066715 | ||||
| Friedmanniomyces simplex | CBS 116775 | DQ066716 | ||||
| Gibbera conferta | CBS 191.53 | GU296150 | GU301814 | GU357758 | GU349041 | |
| Gloniopsis arciformis | GKM L166A | GU323180 | GU323211 | |||
| Gloniopsis praelonga 1 | CBS 112415 | FJ161134 | FJ161173 | FJ161113 | FJ161090 | |
| Gloniopsis praelonga 2 | CBS 123337 | FJ161154 | FJ161195 | FJ161103 | FJ161103 | |
| Gloniopsis subrugosa | CBS 123346 | FJ161170 | FJ161210 | GU371808 | FJ161131 | |
| Glonium circumserpens 1 | CBS 123342 | FJ161168 | FJ161208 | |||
| Glonium circumserpens 2 | CBS 123343 | FJ161160 | FJ161200 | GU371806 | FJ161126 | FJ161108 |
| Glonium stellatum | CBS 207.34 | FJ161140 | FJ161179 | FJ161095 | ||
| Guignardia bidwellii | CBS 237.48 | DQ678034 | DQ678085 | GU357794 | DQ677983 | |
| Guignardia citricarpa | CBS 102374 | GU296151 | GU301815 | GU357773 | GU349053 | |
| Guignardia gaultheriae | CBS 447.70 | DQ678089 | GU357796 | DQ677987 | ||
| Halomassarina ramunculicola 1 (as Massarina ramunculicola) | BCC 18404 | GQ92538 | GQ925853 | |||
| Halomassarina ramunculicola 2 (as Massarina ramunculicola) | BCC 18405 | GQ925839 | GQ925854 | |||
| Halomassarina thalassiae (as Massarina thalassia) | JK 5262D | GU301816 | GU349011 | |||
| Helicomyces roseus | CBS 283.51 | DQ678032 | DQ678083 | DQ677981 | DQ677928 | |
| Hortaea acidophila | CBS 113389 | GU323202 | GU357768 | |||
| Hortaea werneckii | CBS 708.76 | GU296153 | GU301818 | GU357779 | GU371747 | GU349058 |
| Hortaea werneckii | CBS 100496 | GU296152 | GU301817 | GU371739 | GU349050 | |
| Hysterium angustatum | CBS 123334 | FJ161167 | FJ161207 | FJ161129 | FJ161111 | |
| Hysterium barrianum 1 | ANM 1495 | GU323182 | GQ221885 | |||
| Hysterium barrianum 2 | ANM 1442 | GU323181 | GQ221884 | |||
| Hysterobrevium mori 1 | CBS 123336 | FJ161164 | FJ161204 | |||
| Hysterobrevium mori 2 | SMH 5273 | GU301820 | GQ221936 | |||
| Hysterobrevium mori 3 | GKM 1013 | GU301819 | GU397338 | |||
| Hysterobrevium smilacis 1 | CBS 114601 | FJ161135 | FJ161174 | GU357806 | FJ161114 | FJ161091 |
| Hysterobrevium smilacis 2 | SMH 5280 | GU323183 | GQ221912 | GU371810 | GU371784 | |
| Hysteropatella clavispora | CBS 247.34 | DQ678006 | AY541493 | DQ677955 | DQ677901 | |
| Hysteropatella elliptica | CBS 935.97 | EF495114 | DQ767657 | DQ767647 | DQ767640 | |
| Jahnula aquatica | R68-1 | EF175633 | EF175655 | |||
| Jahnula bipileata | F49-1 | EF175635 | EF175657 | |||
| Jahnula seychellensis | SS2113.1 | EF175644 | EF175665 | |||
| Julella avicenniae 1 | BCC 18422 | GU371831 | GU371823 | GU371787 | GU371816 | |
| Julella avicenniae 2 | BCC 20173 | GU371830 | GU371822 | GU371786 | GU371815 | |
| Kabatiella caulivora | CBS 242.64 | EU167576 | EU167576 | GU357765 | ||
| Kalmusia scabrispora 1 | MAFF 239517 | AB524452 | AB524593 | AB539093 | AB539106 | |
| Kalmusia scabrispora 2 | NBRC 106237 | AB524453 | AB524594 | AB539094 | AB539107 | |
| Karstenula rhodostoma | CBS 690.94 | GU296154 | GU301821 | GU371788 | GU349067 | |
| Katumotoa bambusicola | MAFF 239641 | AB524454 | AB524595 | AB539095 | AB539108 | |
| Keissleriella cladophila | CBS 104.55 | GU296155 | GU301822 | GU371735 | GU349043 | |
| Kirschsteiniothelia elaterascus | A22-5A / HKUCC7769 | AF053727 | AY787934 | |||
| Kirschsteiniothelia maritima | CBS 221.60 | GU323203 | GU349001 | |||
| Laurera megasperma | AFTOL 2094 | FJ267702 | ||||
| Lentithecium aquaticum | CBS 123099 | GU296156 | GU301823 | GU371789 | GU349068 | |
| Lentithecium arundinaceum | CBS 619.86 | GU296157 | GU301824 | FJ795473 | ||
| Lentithecium fluviatile | CBS 122367 | GU296158 | GU301825 | GU349074 | ||
| Lepidosphaeria nicotiae | CBS 101341 | DQ678067 | DQ677963 | DQ677910 | ||
| Leptosphaeria biglobosa | CBS 303.51 | GU301826 | GU349010 | |||
| Leptosphaeria doliolum | CBS 505.75 | GU296159 | GU301827 | GU349069 | ||
| Leptosphaeria dryadis | CBS 643.86 | GU301828 | GU371733 | GU349009 | ||
| Leptosphaerulina argentinensis | CBS 569.94 | GU301829 | GU357759 | GU349008 | ||
| Leptosphaerulina australis | CBS 317.83 | GU296160 | GU301830 | GU371790 | GU349070 | |
| Leptosphearia maculans | DAOM 229267 | DQ470993 | DQ470946 | DQ471136 | DQ470894 | DQ471062 |
| Leptoxyphium fumago | CBS 123.26 | GU296161 | GU301831 | GU357771 | GU371741 | GU349051 |
| Letendraea helminthicola | CBS 884.85 | AY016345 | AY016362 | |||
| Letendraea padouk | CBS 485.70 | GU296162 | AY849951 | |||
| Lindgomyces breviappendiculata | HHUF 28193 | AB521733 | AB521748 | |||
| Lindgomyces ingoldianus | ATCC_200398 | AB521719 | AB521736 | |||
| Lindgomyces rotundatus | HHUF_27999 | AB521723 | AB521740 | |||
| Lophiostoma alpigenum | GKM 1091b | GU385193 | ||||
| Lophiostoma arundinis | CBS 621.86 | DQ782383 | DQ782384 | DQ782386 | DQ782387 | |
| Lophiostoma caulium 1 | CBS 623.86 | GU296163 | GU301833 | GU371791 | ||
| Lophiostoma caulium 2 | CBS 624.86 | GU301832 | GU349007 | |||
| Lophiostoma compressum | IFRD 2014 | GU296164 | GU301834 | FJ795457 | ||
| Lophiostoma crenatum | CBS 629.86 | DQ678017 | DQ678069 | DQ677965 | DQ677912 | |
| Lophiostoma fuckelii | GKM 1063 | GU385192 | ||||
| Lophiotrema brunneosporum | CBS 123095 | GU296165 | GU301835 | GU349071 | ||
| Lophiotrema lignicola | CBS 122364 | GU296166 | GU301836 | GU349072 | ||
| Lophiotrema nucula | CBS 627.86 | GU296167 | GU301837 | GU371792 | GU349073 | |
| Lophium elegans | EB 0366 | GU323184 | GU323210 | |||
| Lophium mytilinum 1 | CBS 114111 | EF596819 | EF596819 | |||
| Lophium mytilinum 2 | CBS 269.34 | DQ678030 | DQ678081 | DQ677979 | DQ677926 | |
| Loratospora aestuarii | JK 5535B | GU296168 | GU301838 | GU371760 | ||
| Macrophomina phaseolina | CBS 227.33 | DQ678037 | DQ678088 | DQ677986 | DQ677929 | |
| Macrovalsaria megalospora 1 | 178150 | FJ215707 | FJ215701 | |||
| Macrovalsaria megalospora 2 | 178149 | FJ215706 | FJ215700 | |||
| Massaria anomia | CBS 591.78 | GU296169 | GU301839 | GU371769 | ||
| Massaria platani | CBS 221.37 | DQ678013 | DQ678065 | DQ677961 | DQ677908 | |
| Massarina arundinariae 1 | MAFF 239461 | AB524455 | AB524596 | AB539096 | AB524817 | |
| Massarina arundinariae 2 | NBRC 106238 | AB524456 | AB524597 | AB539097 | AB524818 | |
| Massarina eburnea | CBS 473.64 | GU296170 | GU301840 | GU357755 | GU371732 | GU349040 |
| Massarina igniaria | CBS 845.96 | GU296171 | GU301841 | GU371793 | ||
| Massariosphaeria grandispora | CBS 613 86 | GU296172 | GU301842 | GU357747 | GU371725 | GU349036 |
| Massariosphaeria phaeospora | CBS 611.86 | GU296173 | GU301843 | GU371794 | ||
| Massariosphaeria typhicola 1 | CBS 123126 | GU296174 | GU301844 | GU371795 | ||
| Massariosphaeria typhicola 2 | KT 797 | AB521730 | AB521747 | |||
| Mauritiana rhizophorae 1 | BCC 28866 | GU371832 | GU371824 | GU371796 | GU371817 | |
| Mauritiana rhizophorae 2 | BCC 28867 | GU371833 | GU371825 | GU371797 | GU371818 | |
| Melanomma pulvis-pyrius 1 | SMH 3291 | GU385197 | ||||
| Melanomma pulvis-pyrius 2 | CBS 371.75 | GU301845 | GU371798 | GU349019 | ||
| Melanomma rhododendri | ANM 73 | GU385198 | ||||
| Microthyrium microscopicum | CBS 115976 | GU296175 | GU301846 | GU371734 | GU349042 | |
| Microxyphium aciculiforme | CBS 892.73 | GU296176 | GU301847 | GU357762 | GU371736 | GU349045 |
| Microxyphium citri | CBS 451.66 | GU296177 | GU301848 | GU357750 | GU371727 | GU349039 |
| Microxyphium theae | CBS 202.30 | GU296178 | GU301849 | GU357781 | GU349060 | |
| Monascostroma innumerosum | CBS 345.50 | GU296179 | GU301850 | GU349033 | ||
| Monotosporella tuberculata | CBS 256.84 | GU301851 | GU349006 | |||
| Montagnula opulenta | CBS 168.34 | AF164370 | DQ678086 | DQ677984 | ||
| Mycosphaerella endophytica | CBS 114662 | GU214538 | DQ246255 | |||
| Mycosphaerella eurypotami | JK 5586J | GU301852 | GU371722 | |||
| Mycosphaerella graminicola 1 | CBS 292.38 | DQ678033 | DQ678084 | DQ677982 | ||
| Mycosphaerella graminicola 2 | CBS 115943 | GU214540 | GU214436 | |||
| Mycosphaerella heimii | CBS 110682 | GU214541 | GQ852604 | |||
| Mycosphaerella latebrosa | CBS 687.94 | DQ848331 | GU214444 | |||
| Mycosphaerella marksii | CBS 110942 | GU214549 | GQ852612 | |||
| Mycosphaerella punctiformis (anamorph Ramularia endophylla) | CBS 113265 | DQ471017 | DQ470968 | DQ471165 | DQ470920 | DQ471092 |
| Myriangium duriaei | CBS 260.36 | AY016347 | DQ678059 | DQ677954 | DQ677900 | |
| Myriangium hispanicum | CBS 247.33 | GU296180 | GU301854 | GU357775 | GU371744 | GU349055 |
| Mytilinidion acicola | EB 0349 | GU323185 | GU323209 | GU371757 | ||
| Mytilinidion andinense | CBS 123562 | FJ161159 | FJ161199 | FJ161125 | FJ161107 | |
| Mytilinidion californicum | EB 0385 | GU323186 | GU323208 | |||
| Mytilinidion mytilinellum | CBS 303.34 | FJ161144 | FJ161184 | GU357810 | FJ161119 | FJ161100 |
| Mytilinidion resinicola | CBS 304.34 | FJ161145 | FJ161185 | FJ161101 | FJ161101 | FJ161120 |
| Mytilinidion rhenanum | EB 0341 | GU323187 | GU323207 | |||
| Mytilinidion scolecosporum | CBS 305.34 | FJ161146 | FJ161186 | GU357811 | FJ161121 | FJ161102 |
| Mytilinidion thujarum | EB 0268 | GU323188 | GU323206 | |||
| Mytilinidion tortile | EB 0377 | GU323189 | GU323205 | |||
| Neofusicoccum ribis (teleomorph Botryosphaeria ribis) | CBS 115475 | DQ678000 | DQ678053 | GU357789 | DQ677947 | DQ677893 |
| Neophaeosphaeria filamentosa | CBS 102202 | GQ387516 | GQ387577 | GU357803 | GU371773 | GU349084 |
| Neottiosporina paspali | CBS 331.37 | EU754073 | EU754172 | GU357812 | GU371779 | GU349079 |
| Oedohysterium insidens 1 | CBS 238.34 | FJ161142 | FJ161182 | FJ161118 | FJ161097 | |
| Oedohysterium insidens 2 | ANM 1443 | GU323190 | GQ221882 | GU371811 | GU371785 | |
| Oedohysterium sinense | CBS 123345 | FJ161169 | FJ161209 | GU371807 | FJ161130 | |
| Opegrapha dolomitica | DUKE 0047528 | DQ883706 | DQ883717 | DQ883714 | DQ883732 | |
| Ophiosphaerella herpotricha | CBS 620.86 | DQ678010 | DQ678062 | DQ677958 | DQ677905 | |
| Ophiosphaerella sasicola | MAFF 239644 | AB524458 | AB524599 | AB539098 | AB539111 | |
| Otthia spiraeae 1 | CBS 114124 | EF204515 | EF204498 | |||
| Otthia spiraeae 2 | CBS 113091 | EF204516 | EF204499 | GU357777 | ||
| Paraconiothyrium minitans | CBS 122788 | EU754074 | EU754173 | GU357807 | GU371776 | GU349083 |
| Patellaria atrata | CBS 958.97 | GU296181 | GU301855 | GU357749 | GU371726 | GU349038 |
| Patellaria cf. atrata 1 | BCC 28876 | GU371836 | GU371828 | |||
| Patellaria cf. atrata 2 | BCC 28877 | GU371837 | GU371829 | |||
| Phacellium paspali | CBS 113093 | GU214669 | GQ852627 | |||
| Phaeocryptopus gaeumannii 1 | CBS 244.38 | GU357766 | GU371740 | |||
| Phaeocryptopus gaeumannii 2 | CBS 267.37 | EF114722 | EF114698 | GU357770 | ||
| Phaeocryptopus nudus | CBS 268.37 | GU296182 | GU301856 | GU357745 | GU349034 | |
| Phaeodothis winteri | CBS 182.58 | GU296183 | GU301857 | DQ677917 | ||
| Phaeosclera dematioides | CBS 157.81 | GU296184 | GU301858 | GU357764 | GU349047 | |
| Phaeosphaeria ammophilae | CBS 114595 | GU296185 | GU301859 | GU357746 | GU371724 | GU349035 |
| Phaeosphaeria avenaria | DAOM 226215 | AY544725 | AY544684 | DQ677941 | DQ677885 | |
| Phaeosphaeria brevispora 1 | NBRC 106240 | AB524460 | AB524601 | AB539100 | AB539113 | |
| Phaeosphaeria brevispora 2 | MAFF 239276 | AB524459 | AB524600 | AB539099 | AB539112 | |
| Phaeosphaeria caricis | CBS 120249 | GU301860 | GU349005 | |||
| Phaeosphaeria eustoma | CBS 573.86 | DQ678011 | DQ678063 | DQ677959 | DQ677906 | |
| Phaeosphaeria juncicola | CBS 595.86 | GU349016 | ||||
| Phaeosphaeria luctuosa | CBS 308.79 | GU301861 | GU349004 | |||
| Phaeosphaeria nodorum | Broad | Genome | Genome | Genome | Genome | Genome |
| Phaeosphaeriopsis musae | CBS 120026 | GU296186 | GU301862 | GU357748 | GU349037 | |
| Phaeotrichum benjaminii | CBS 541.72 | AY016348 | AY004340 | GU357788 | DQ677946 | DQ677892 |
| Phoma betae | CBS 109410 | EU754079 | EU754178 | GU357804 | GU371774 | GU349075 |
| Phoma complanata | CBS 268.92 | EU754081 | EU754180 | GU357809 | GU371778 | GU349078 |
| Phoma exigua | CBS 431.74 | EU754084 | EU754183 | GU357813 | GU371780 | GU349080 |
| Phoma glomerata | CBS 528.66 | EU754085 | EU754184 | GU371781 | GU349081 | |
| Phoma herbarum | CBS 276.37 | DQ678014 | DQ678066 | GU357792 | DQ677962 | DQ677909 |
| Phoma heteromorphospora | CBS 115.96 | EU754089 | EU754188 | GU371775 | GU349077 | |
| Phoma radicina | CBS 111.79 | EU754092 | EU754191 | GU357805 | GU349076 | |
| Phoma zeae-maydis | CBS 588.69 | EU754093 | EU754192 | GU357814 | GU371782 | GU349082 |
| Piedraia hortae | CBS 480.64 | AY016349 | AY016366 | DQ677990 | ||
| Pleomassaria siparia | CBS 279.74 | DQ678027 | DQ678078 | DQ677976 | DQ677923 | |
| Pleospora ambigua | CBS 113979 | AY787937 | GU357760 | |||
| Pleospora herbarum | CBS 191.86 | DQ247812 | DQ247804 | DQ471163 | DQ247794 | DQ471090 |
| Polyplosphaeria fusca | MAFF 239685 | AB524463 | AB524604 | |||
| Polythrincium trifolii (as Cymadothea trifolii) | 133 | EU167612 | EU167612 | |||
| Preussia funiculata | CBS 659.74 | GU296187 | GU301864 | GU371799 | GU349032 | |
| Preussia lignicola (as Sporormia lignincola) | CBS 264.69 | GU296197 | GU301872 | GU371765 | GU349027 | |
| Preussia terricola | DAOM 230091 | AY544726 | AY544686 | DQ471137 | DQ470895 | DQ471063 |
| Pseudocercospora fijiensis (teleomorph Mycosphaerella fijiensis) | OSC 100622 | DQ767652 | DQ678098 | DQ677993 | ||
| Pseudocercospora griseola f. griseola | CPC 10461 | GU323191 | GU348997 | |||
| Pseudocercospora vitis | CPC 11595 | DQ289864 | GU214483 | |||
| Pseudotetraploa curviappendiculata | MAFF 239495 | AB524467 | AB524608 | |||
| Psiloglonium araucanum | CBS 112412 | FJ161133 | FJ161172 | GU357743 | FJ161112 | FJ161089 |
| Psiloglonium clavisporum 1 | CBS 123338 | FJ161156 | FJ161197 | FJ161123 | ||
| Psiloglonium clavisporum 2 | GKM L172A | GU323192 | GU323204 | |||
| Psiloglonium simulans | CBS 206.34 | FJ161139 | FJ161178 | FJ161116 | FJ161094 | |
| Pyrenochaeta nobilis | CBS 407.76 | DQ678096 | DQ677991 | DQ677936 | ||
| Pyrenophora phaeocomes | DAOM 222769 | DQ499595 | DQ499596 | DQ497614 | DQ497607 | |
| Pyrenophora tritici-repentis 1 | OSC 100066 | AY544672 | DQ677882 | |||
| Pyrenophora tritici-repentis 2 | CBS 328.53 | GU349017 | ||||
| Quadricrura septentrionalis | CBS 125429 | AB524474 | AB524615 | |||
| Quintaria lignatilis | CBS 117700 | GU296188 | GU301865 | GU371761 | ||
| Quintaria submersa | CBS 115553 | GU301866 | GU357751 | GU349003 | ||
| Racodium rupestre 1 | L423 | EU048576 | EU048581 | |||
| Racodium rupestre 2 | L424 | EU048577 | EU048582 | |||
| Ramichloridium apiculatum | CBS 156.59 | GU296189 | GU371770 | |||
| Ramichloridium cerophilum | CBS 103.59 | GU296190 | EU041855 | |||
| Rasutoria tsugae | ratstk | EF114730 | EF114705 | GU371809 | ||
| Rhytidhysterium rufulum 2 | CBS 306.38 | GU296191 | FJ469672 | FJ238444 | GU349031 | |
| Rhytidhysteron rufulum 1 | GKM 361A | GU296192 | GU301867 | |||
| Rimora mangrovei | JK 5246A | GU296193 | GU301868 | GU371759 | ||
| rock isolate TRN 111 | CBS 118294 | GU323193 | GU323220 | GU357783 | GU371751 | GU349088 |
| rock isolate TRN 123 | CBS 117932 | GU323194 | GU323219 | GU357784 | GU371753 | |
| rock isolate TRN 137 | CBS 118300 | GU323195 | GU323218 | GU357782 | GU371749 | |
| rock isolate TRN 138 | CBS 118301 | GU323196 | GU323217 | GU371750 | ||
| rock isolate TRN 152 | CBS 118346 | GU323197 | GU323223 | GU371752 | ||
| rock isolate TRN 211 | CBS 117937 | GU323198 | GU323222 | GU357785 | GU371754 | |
| rock isolate TRN 235 | CBS 118605 | GU323199 | GU357787 | GU371756 | GU349087 | |
| rock isolate TRN 43 | CBS 117950 | GU323200 | GU323221 | GU357786 | GU371755 | GU349086 |
| Roussoella hysterioides 1 | MAFF 239636 | AB524480 | AB524621 | AB539101 | AB539114 | |
| Roussoella hysterioides 2 | CBS 125434 | AB524481 | AB524622 | AB539102 | AB539115 | |
| Roussoella pustulans | MAFF 239637 | AB524482 | AB524623 | AB539103 | AB539116 | |
| Roussoellopsis tosaensis | MAFF 239638 | AB524625 | AB539104 | AB539117 | ||
| Saccharata proteae | CBS 115206 | GU296194 | GU301869 | GU357753 | GU371729 | GU349030 |
| Saccothecium sepincola | CBS 278.32 | GU296195 | GU301870 | GU371745 | GU349029 | |
| Schismatomma decolorans | DUKE 0047570 | AY548809 | AY548815 | DQ883715 | DQ883725 | |
| Schizothyrium pomi 1 | CBS 406.61 | EF134949 | EF134949 | |||
| Schizothyrium pomi 2 | CBS 486.50 | EF134948 | EF134948 | |||
| Schizothyrium pomi 3 | CBS 228.57 | EF134947 | EF134947 | |||
| Scorias spongiosa | CBS 325.33 | DQ678024 | DQ678075 | DQ677973 | DQ677920 | |
| Setomelanomma holmii | CBS 110217 | GU296196 | GU301871 | GU371800 | GU349028 | |
| Setosphaeria monoceras | AY016368 | AY016368 | ||||
| Spencermartinsia viticola (teleomorph Botryosphaeria viticola) | CBS 117009 | DQ678036 | DQ678087 | GU357795 | DQ677985 | |
| Sporormiella minima | CBS 524.50 | DQ678003 | DQ678056 | DQ677950 | DQ677897 | |
| Stagonospora macropycnidia | CBS 114202 | GU296198 | GU301873 | GU349026 | ||
| Stylodothis puccinioides | CBS 193.58 | AY004342 | FJ238427 | DQ677886 | ||
| Sydowia polyspora | CBS 116.29 | DQ678005 | DQ678058 | GU357791 | DQ677953 | DQ677899 |
| Teratosphaeria associata (as Teratosphaeria jonkershoekensis) | CBS 112224 | GU296200 | GU301874 | GU357744 | GU371723 | GU349025 |
| Teratosphaeria cryptica (as Mycosphaerella cryptica) | CBS 110975 | GU214602 | GQ852682 | |||
| Teratosphaeria fibrillosa 1 | CBS 121707 | GU296199 | GU323213 | GU357767 | ||
| Teratosphaeria fibrillosa 2 | CPC 1876 | GU214506 | ||||
| Teratosphaeria stellenboschiana (as Colletogloeopsis stellenboschiana) | CBS 116428 | GU214583 | EU019295 | |||
| Teratosphaeria suberosa (as Mycosphaerella suberosa) | CPC 11032 | GU214614 | GQ852718 | |||
| Tetraplosphaeria sasicola | MAFF 239677 | AB524490 | AB524631 | |||
| Thyridaria rubronotata | CBS 419.85 | GU301875 | GU371728 | GU349002 | ||
| Tremateia halophila | JK 5517J | GU296201 | GU371721 | |||
| Trematosphaeria pertusa | CBS 122371 | GU348999 | GU301876 | GU371801 | GU349085 | |
| Trichodelitschia bisporula 1 | CBS 262.69 | GU349000 | GU348996 | GU371812 | GU371802 | GU349020 |
| Trichodelitschia bisporula 2 (duplicate) | CBS 262.69 | GU296202 | ||||
| Trichodelitschia munkii | Kruys201 | DQ384070 | DQ384096 | |||
| Triplosphaeria maxima | MAFF 239682 | AB524496 | AB524637 | |||
| Trypethelium nitidiusculum 1 | 139 | GU327728 | GU327732 | |||
| Trypethelium nitidiusculum 2 | AFTOL 2099 | FJ267701 | ||||
| Trypethelium tropicum | 25 | GU327730 | ||||
| Tubeufia cerea | CBS 254.75 | DQ471034 | DQ470982 | DQ471180 | DQ470934 | DQ471105 |
| Tubeufia paludosa | CBS 120503 | GU296203 | GU301877 | GU357754 | GU371731 | GU349024 |
| Tubeufia paludosa (as anamorph Helicosporium phragmitis) | CBS 245.49 | DQ767649 | DQ767654 | DQ767643 | DQ767638 | |
| Tyrannosorus pinicola | CBS 124.88 | DQ471025 | DQ470974 | DQ471171 | DQ470928 | DQ471098 |
| Ulospora bilgramii | CBS 110020 | DQ678025 | DQ678076 | DQ677974 | DQ677921 | |
| Venturia inaequalis 1 | CBS 594.70 | GU296205 | GU301879 | GU357757 | GU349022 | |
| Venturia inaequalis 2 | CBS 815.69 | GU296204 | GU301878 | GU357756 | GU349023 | |
| Venturia inaequalis 3 (as Spilocaea pomi) | CBS 176.42 | GU348998 | GU349089 | |||
| Venturia populina | CBS 256.38 | GU296206 | GU323212 | GU357769 | ||
| Verrucisporota daviesiae | CBS 116002 | GU296207 | GQ852730 | |||
| Verruculina enalia | JK 5253A | DQ678028 | DQ678079 | DQ677977 | DQ677924 | |
| Westerdykella angulata (as Eremodothis angulata) | CBS 610.74 | DQ384067 | DQ384105 | GU371805 | GU371821 | |
| Westerdykella cylindrica | CBS 454.72 | AY016355 | AY004343 | DQ471168 | DQ470925 | DQ497610 |
| Westerdykella ornata | CBS 379.55 | GU296208 | GU301880 | GU371803 | GU349021 | |
| Wettsteinina lacustris | CBS 618.86 | DQ678023 | DQ677972 | DQ677919 | ||
| Wicklowia aquatica | AF289-1 | GU045446 | ||||
| Wicklowia aquatica | CBS 125634 | GU266232 | GU045445 | GU371813 | ||
| Zasmidium cellare | CBS 146.36 | EF137362 | EU041878 | |||
| Zopfia rhizophila | CBS 207.26 | DQ384086 | DQ384104 |
BCC: Belgian Coordinated Collections of Microorganisms; CABI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, U.K.; CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; DAOM: Plant Research Institute, Department of Agriculture (Mycology), Ottawa, Canada; DUKE: Duke University Herbarium, Durham, North Carolina, U.S.A.; HHUF: Herbarium of Hirosaki University, Japan; IFRDCC: Culture Collection, International Fungal Research & Development Centre, Chinese Academy of Forestry, Kunming, China; MAFF: Ministry of Agriculture, Forestry and Fisheries, Japan; NBRC: NITE Biological Resource Centre, Japan; OSC: Oregon State University Herbarium, U.S.A.; UAMH: University of Alberta Microfungus Collection and Herbarium, Edmonton, Alberta, Canada; UME: Herbarium of the University of Umeå, Umeå, Sweden; Culture and specimen abbreviations: ANM: A.N. Miller; CPC; P.W. Crous; EB: E.W.A. Boehm; EG: E.B.G. Jones; GKM: G.K. Mugambi; JK: J. Kohlmeyer; KT: K. Tanaka; SMH: S.M. Huhndorf.
Sequence alignment and phylogenetic analyses
Sequences were obtained from WASABI (Kauff et al. 2007) as well as from previous publications (e.g. Lutzoni et al. 2004, Schoch et al. 2009a). Introns were removed and an initial core set of 171 taxa were aligned by using default options for a simultaneous method of estimating alignments and tree phylogenies, SATé (Liu et al. 2009). In order to consider codons without the insertion of unwanted gaps, protein coding fragments were translated in BioEdit v. 7.0.1 (Hall 2004) and aligned within SATé as amino acids. These were then realigned with their respective DNA sequences using the RevTrans 1.4 Server (Wernersson & Pedersen 2003). After the removal of intron sequences the alignment was examined manually in BioEdit with a shade threshold of 40 % and regions with high amounts of gap characters were excluded. This resulted in a reduction of 99 columns in the LSU data set, 118 in RPB1 and 162 in RPB2, for a total of 379. Nothing was removed for TEF1. In order to allow for the extension of our alignment as newly generated sequences became available from other studies in this volume, these were subsequently added to this core alignment with MAFFT v. 6.713 (Katoh et al. 2009). The E-INS-i setting, focused on high accuracy with a high percentage of unalignable regions such as introns, was applied and the SATé alignment was used as a seed. This resulted in a supermatrix of five genes (LSU, SSU TEF1, RPB1, RPB2) consisting of 52 % gaps and undetermined characters out of a total of 6 582 characters. GenBank accession numbers are shown in Table 1.
Conflict tests
Conflict tests on the initial core set of 204 taxa were conducted by selecting single gene data sets and doing comparisons on a gene by gene basis. This was done using the “bootstopping” criterion in RAxML v. 7.0.4 (Stamatakis et al. 2008) under the CIPRES v. 2.1 webportal to produce trees of comparative gene sets where all taxa have the gene present. Comparisons between all potential sets of gene trees with no missing taxa were done using a script (Kauff & Lutzoni 2002) obtained through the Lutzoni lab website and to detect present or absent taxa within clades with a cut-off bootstrap value of 70 %. This is described in more detail elsewhere (Miadlikowska et al. 2006, Schoch et al. 2009a).
Phylogeny
A phylogenetic analysis was performed using RAxML v. 7.0.4 (Stamatakis 2006) applying unique model parameters for each gene and codon. The dataset was divided in 11 partitions as previously described in Schoch et al. (2009a). A general time reversible model (GTR) was applied with a discrete gamma distribution and four rate classes following procedures laid out in Schoch et al. (2009). Ten thorough maximum likelihood (ML) tree searches were done in RAxML v. 7.0.4 under the same model, each one starting from a randomised tree. Bootstrap pseudo replicates were performed 2000 times using the fast bootstrapping option and the best scoring tree form 10 separate runs were selected. The resulting trees were printed with TreeDyn v. 198.3 (Chevenet et al. 2006). All alignments are deposited in TreeBASE. Additionally, the data sets were analyzed in GARLI v. 0.96 (Zwickl 2006) using the GTR-gamma-invariant model. In this case 200 bootstraps were run under default conditions.
Ancestral reconstruction
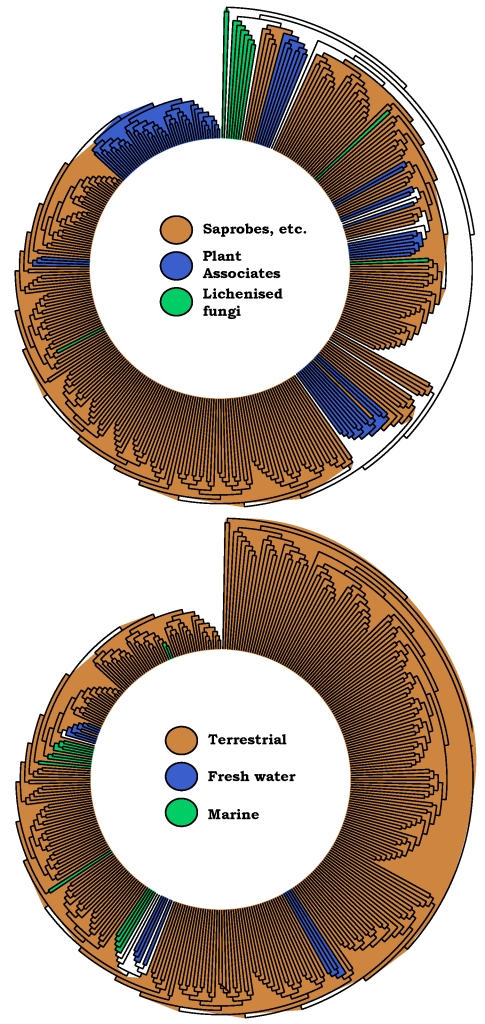
Ancestral reconstructions were performed in Mesquite v. 2.6 with character states traced over 2000 bootstrapped trees obtained with RAxML-MPI v. 7.0.4 (Stamatakis 2006). Following the phylogeny presented (Fig. 2) this reconstruction was performed with a maximum-likelihood criterion using the single parameter Mk1 model. Ancestral states were assigned to a node if the raw likelihood was higher by at least 2 log units than the likelihood value of the other ancestral state(s) according to default settings. Character states were also mapped using TreeDyn v. 198.3 (Chevenet et al. 2006), shown in Fig. 3. This is presented as a clockwise circular tree, starting with outgroup taxa. Only clades with more than two taxa of the same state are shown and bootstrap recovery was not considered in assigning character states. In applying the character states of saprobes (including rock heterotrophs), plant associated fungi (including pathogens, endophytes and mycorrhizae) and lichenised fungi the broad concepts presented were followed as laid out in Schoch et al. (2009a). Some character assessments were taken from Zhang et al. (2009; this volume). Ecological characters of sampling sources, terrestrial, fresh water and marine were assessed based on papers elsewhere in this volume (Suetrong et al. 2009, Shearer et al. 2009).
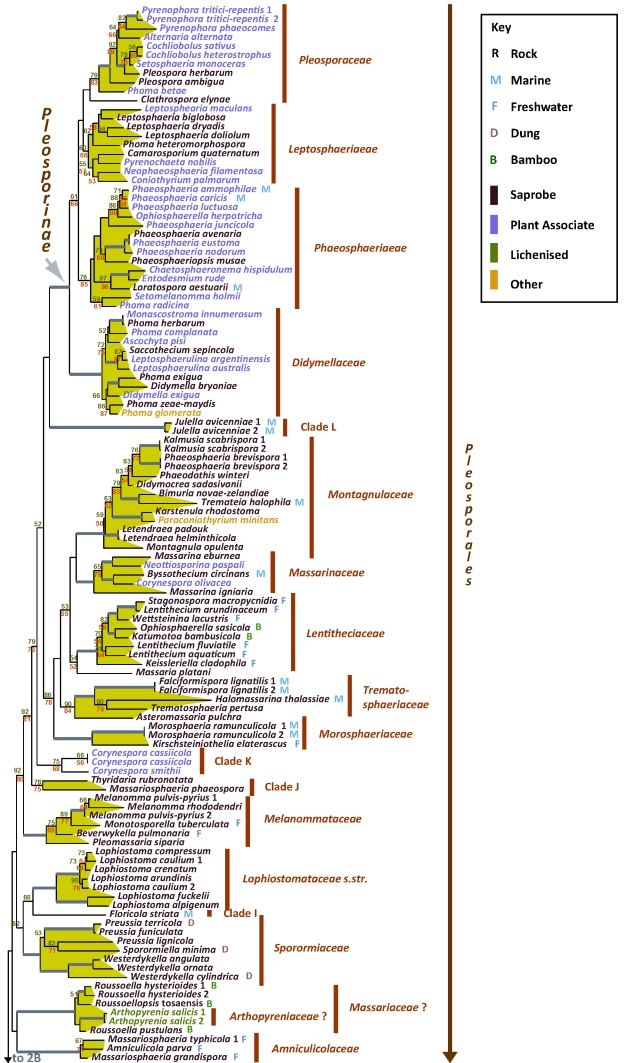
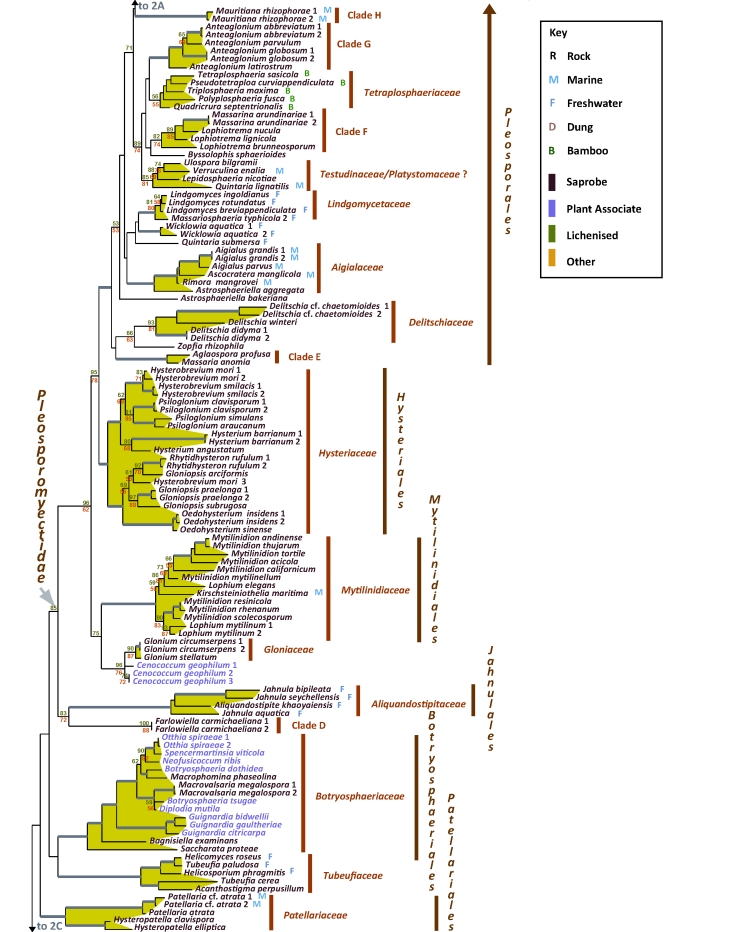
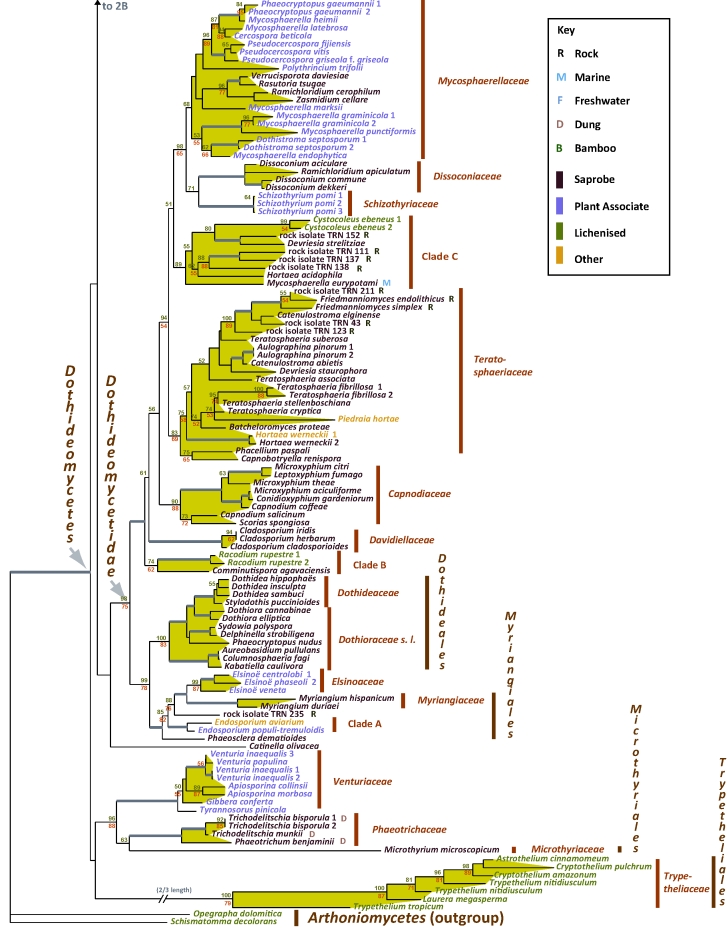
Fig. 2A–C.
Best scoring ML tree with RAxML and GARLI bootstrap values respectively above (green) and below (red) the nodes. Values below 50 % were removed and branches with more than 90 % bootstrap for both methods are thickened without values. Environmental sources relevant to the papers in this volume are indicated in the key (R-Rock; M-Marine; F-Freshwater; D-Dung; B-Bamboo). Nutritional characters are indicated by colour as per the key.
Fig. 3.
Simplified ancestral state reconstructions, showing potential transitions between character states. The same phylogeny as in Fig. 2A–C is shown, with the outgroups positioned at twelve o' clock and subsequent clades arranged in a clockwise manner. Characters were traced over 2 000 bootstrap trees and those that were recovered in the majority are coloured on the nodes. In the case of equivocal construction no colour was used (white). To simplify the figure, only clades with two or more neighbouring character states are shown.
Genome analyses
A MCL (Markov Cluster Algorithm) protein analysis of 52 fungi and one metazoan (Drosophila melanogaster) (Table 2 - see online Supplementary Information) and the phylogenetic placement of these species was used to characterise the phylogenetic profile of each cluster. Chytridiomycota and Mucoromycotina each were represented by one and two species, respectively. In Dikarya, Basidiomycota and Ascomycota were represented by 8 and 40 species respectively. The Pezizomycotina (filamentous ascomycetes) was presented by 26 species in four classes [Sordariomycetes (12), Leotiomycetes (2), Dothideomycetes (6) and Eurotiomycetes (6)].
Table 2.
Genomes used for phylogenetic profile. All are opisthokonts; remaining classifications used in Fig. 4 are indicated in columns: Do – Dothideomycetes, ED - Eurotiomycetes & Dothideomycetes, S – Saccharomyceta, A – Ascomycota, Di — Dikarya, MD - Mucoromycotina & Dikarya, CMD - Chytridiomycota, F - Fungi.
| Genomes | Classifications | |||||||
|---|---|---|---|---|---|---|---|---|
| Alternaria brassicicola | Do | ED | S | A | Di | MD | CMD | F |
| Cochliobolus heterostrophus | Do | ED | S | A | Di | MD | CMD | F |
| Mycosphaerella fijiensis | Do | ED | S | A | Di | MD | CMD | F |
| Mycosphaerella graminicola | Do | ED | S | A | Di | MD | CMD | F |
| Pyrenophora tritici-repentis | Do | ED | S | A | Di | MD | CMD | F |
| Stagonospora nodorum | Do | ED | S | A | Di | MD | CMD | F |
| Aspergillus fumigatus | ED | S | A | Di | MD | CMD | F | |
| Aspergillus nidulans | ED | S | A | Di | MD | CMD | F | |
| Aspergillus terreus | ED | S | A | Di | MD | CMD | F | |
| Coccidioides immitis | ED | S | A | Di | MD | CMD | F | |
| Histoplasma capsulatum | ED | S | A | Di | MD | CMD | F | |
| Uncinocarpus reesii | ED | S | A | Di | MD | CMD | F | |
| Ashbya gossypii | S | A | Di | MD | CMD | F | ||
| Botrytis cinerea | S | A | Di | MD | CMD | F | ||
| Candida albicans | S | A | Di | MD | CMD | F | ||
| Candida glabrata | S | A | Di | MD | CMD | F | ||
| Candida guilliermondii | S | A | Di | MD | CMD | F | ||
| Candida lusitaniae | S | A | Di | MD | CMD | F | ||
| Chaetomium globosum | S | A | Di | MD | CMD | F | ||
| Debaryomyces hansenii | S | A | Di | MD | CMD | F | ||
| Fusarium graminearum | S | A | Di | MD | CMD | F | ||
| Fusarium oxysporum | S | A | Di | MD | CMD | F | ||
| Fusarium verticillioides | S | A | Di | MD | CMD | F | ||
| Kluyveromyces lactis | S | A | Di | MD | CMD | F | ||
| Laccaria bicolor | S | A | Di | MD | CMD | F | ||
| Lodderomyces elongisporus | S | A | Di | MD | CMD | F | ||
| Magnaporthe grisea | S | A | Di | MD | CMD | F | ||
| Nectria haematococca | S | A | Di | MD | CMD | F | ||
| Neurospora crassa | S | A | Di | MD | CMD | F | ||
| Pichia stipitis | S | A | Di | MD | CMD | F | ||
| Podospora anserina | S | A | Di | MD | CMD | F | ||
| Saccharomyces cerevisiae | S | A | Di | MD | CMD | F | ||
| Sclerotinia sclerotiorum | S | A | Di | MD | CMD | F | ||
| Sporobolomyces roseus | S | A | Di | MD | CMD | F | ||
| Trichoderma atroviride | S | A | Di | MD | CMD | F | ||
| Trichoderma reseei | S | A | Di | MD | CMD | F | ||
| Trichoderma virens | S | A | Di | MD | CMD | F | ||
| Verticillium dahliae | S | A | Di | MD | CMD | F | ||
| Yarrowia lipolytica | S | A | Di | MD | CMD | F | ||
| Schizosaccharomyces japonicus | A | Di | MD | CMD | F | |||
| Schizosaccharomyces octosporus | A | Di | MD | CMD | F | |||
| Schizosaccharomyces pombe | A | Di | MD | CMD | F | |||
| Coprinus cinereus | Di | MD | CMD | F | ||||
| Cryptococcus neoformans | Di | MD | CMD | F | ||||
| Phanerochaete chrysosporium | Di | MD | CMD | F | ||||
| Postia placenta | Di | MD | CMD | F | ||||
| Puccinia graminis f. sp. tritici | Di | MD | CMD | F | ||||
| Ustilago maydis | Di | MD | CMD | F | ||||
| Phycomyces blakesleeanus | MD | CMD | F | |||||
| Rhizopus oryzae | MD | CMD | F | |||||
| Batrachochytrium dendrobatidis | CMD | F | ||||||
| Encephalitozoon cuniculi | F | |||||||
| Drosophila melanogaster | ||||||||
RESULTS AND DISCUSSION
Taxon sampling
The phylogram presented in Fig. 2 represents the largest ever phylogenetic assessment of Dothideomycetes to date. Here the focus has been on expanding taxon diversity in the class while specifically avoiding a small number of taxa that other analyses suggest reside on long unstable branches. This still allowed for an extensive sweep of dothideomycete taxon diversity; in doing so we followed the premise of allowing for missing data in our supermatrix (Wiens 2006). An effort was made to intersperse taxa with poor character sampling amongst those having better sampling throughout the tree, but the inclusion of missing characters could still have unanticipated effects on phylogenetic assessments (Lemmon et al. 2009). While recognising this caveat, a recent expansive data set covering all of Ascomycota noted very little changes in major nodes even after the removal of taxa with high proportions of missing characters (Schoch et al. 2009a). The phylogeny presented here agrees well with broad phylogenies in this volume and elsewhere (Schoch et al. 2006, Crous et al. 2007a, Zhang et al. 2008, Crous et al. 2009b). After all introns and 379 ambiguous character positions were removed, the matrix consisted of 52 % missing and indeterminate characters. This maximum-likelihood analysis had 5 069 distinct alignment patterns and produced a best known likely tree with a log likelihood of -207247.761117.
Evolution of nutritional modes
The ancestral reconstructions in Fig. 3 indicate that phytopathogenicity can be confined to a number of terminal clades throughout the tree and that these always reside within saprobic lineages. A maximum of seven transitions likely occurred in several lineages of the orders Pleosporales, Capnodiales and singular lineages in Myriangiales, Botryosphaeriales and Venturiaceae (also see in this volume; Crous et al. 2009a, Zhang et al. 2009). Several transitions to lichenisation have also occurred, although phylogenetic uncertainty may limit this to a minimum of two. Due to the use of lichenised Arthoniomycetes as outgroup a broader assessment is required to determine whether the Dothideomycetes evolved from a lichenised ancestor. Previous studies suggested that the saprobic habit is an ancestral trait but only with marginal support (Schoch et al. 2009a). Similar conclusions can be reached for the aquatic ecological characters – the majority of fresh water and marine clades reside within terrestrial clades as has been shown previously e.g. (Spatafora et al. 1998, Vijaykrishna et al. 2006). Transitions from a terrestrial life style to fresh water likely occurred at least three times and transitions to marine environments up to six times. Phylogenetic uncertainty for the placement of some marine clades can limit this to a minimum of four times (Fig. 2). Reversions from aquatic to terrestrial environments are rare, with one possible exception in the Lentitheciaceae where bambusicolous saprobes reside, nested within several fungi occurring in freshwater habitats (for additional details see Zhang et al. 2009; this volume). Phylogenetic resolution will have to improve to test this further.
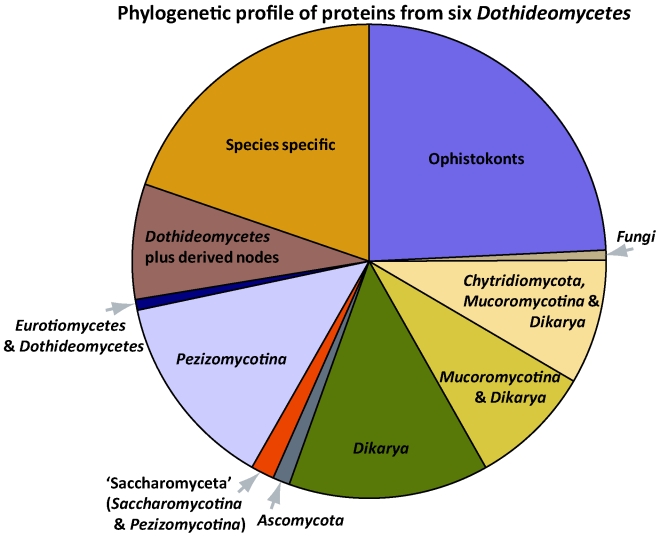
An analysis of recently released genomes was compared to consider whether genome composition reinforces phylogenetic support for Dothideomycetes (Fig. 4). Relative to a clustering analysis of proteins from 52 sequenced fungi and Drosophila melanogaster, about 5 515 protein coding genes from Dothideomycetes shared protein clusters with proteins from other dothideomycete fungi only. This comprises roughly 8–11 % of the protein coding genes in each of six sequenced Dothideomycetes. The species profile of each protein cluster was used to assign a phylogenetically informed designation. The profiles most frequently seen were those of the most conserved proteins, namely clusters designated as having a shared Ophistokont phylogenetic profile. Among the more derived nodes of the Dothideomycetes, protein clusters were observed that had a species composition that could reflect the result of selection pressure on more distantly related fungi that share the same niche.
Fig. 4.
Pie chart showing relative numbers of unique proteins per genome according to taxonomic classification.
A phylogenomic profile (Fig. 4) of the proteins from six Dothideomycetes from the two largest orders seen in Fig. 1 is presented (Mycosphaerella graminicola, Mycosphaerella fijiensis, Phaeosphaeria nodorum, Alternaria brassicicola, Pyrenophora tritici-repentis, Cochliobolus heterostrophus). The highest percentage of proteins (excluding species specific proteins) were conserved outside kingdom Fungi (Ophistokont node, 23 %), followed by proteins specific for the Dikarya (14 %) and the Pezizomycotina (13 %). This breakdown was also prevalent within other Pezizomycotina classes. Approximately 8 % of the proteins from the six Dothideomycetes were conserved across and within derived nodes in this class. Relative to this analysis 28 % of the proteins were specific to the Dothideomycetes (including species specific proteins). The other class containing loculoascomyetes, Eurotiomycetes, had 19.5 % proteins characterised as class specific. This means the percentage dothideomycete specific proteins were about 8.5 % more. Eurotiomycetes in the analysis were mostly human pathogens, with most having no known sexual state whereas the Dothideomycetes in the analysis were all plant pathogens and mostly with known sexual states. This breakdown of nutritional modes, although not comprehensive for these two classes, is somewhat representative. In Eurotiomycetes human pathogens are more diverse and plant pathogens uncommon, with the converse being true for Dothideomycetes. Both classes contain melanised species with similar morphologies and more comprehensive comparative studies need to expand sampling to incorporate species from the different nutritional modes for both classes.
Phylogenetic relationships
In the phylogram presented (Fig. 2) the two dothideomycete subclasses previously described based on presence or absence of pseudoparaphyses (Schoch et al. 2006) could be recovered with varying levels of bootstrap representation. Subclass Pleosporomycetidae previously included Pleosporales plus a single species, representing Mytilinidiaceae, namely Lophium mytilinum (Schoch et al. 2006). Taxon sampling for the Mytilinidiaceae was considerably expanded by Boehm et al. (2009b), with the addition of a number of new taxa, leading to the establishment of the Mytilinidiales. Likewise, extensive taxon sampling for the family Hysteriaceae led to a newly redefined Hysteriales also included in this subclass (Boehm et al. 2009a; this volume). It appears that persistent, hysteriaceous carbonaceous ascomata that dehisce via a longitudinal slit (e.g., hysterothecia) have evolved multiple times within Pleosporomycetidae (Mugambi & Huhndorf 2009,Mugambi & Huhndorf 2009). Pleosporomycetidae can be expanded to tentatively include Jahnulales (Fig. 2B) based on strong bootstrap support from RAxML analyses and morphology. Perithecioid ascomata and a hamathecium of wide cellular pseudoparaphyses are characteristic of Jahnulales (Inderbitzin et al. 2001, Pang et al. 2002; Shearer et al. 2009; this volume) and agree with diagnostic features for Pleosporomycetidae. We also recommend that the definition of the subclass be reassessed with more inclusive character sets. Also, Leptosphaerulina species characterised by the absence of pseudoparaphyses reside within the pseudoparaphysate Pleosporales (Fig. 2C; Silva-Hanlin & Hanlin 1999, Kodsueb et al. 2006), indicating that pseudoparaphyses could have been lost multiple times. It should be noted that the maturity of ascomata may play an important role in these assessments. Immature specimens may contain pseudoparaphyses that dehisce when mature and these characteristics need to be evaluated with more complete sampling of the numerous aparaphysate taxa still listed as incertae sedis. The second subclass, Dothideomycetidae, previously circumscribed based on the absence of pseudoparaphyses remains well supported (Fig. 2C).
The results of this study provided continued support for ten orders within class Dothideomycetes, namely Pleosporales, Hysteriales, Mytilinidiales, Patellariales, Botryosphaeriales, Jahnulales, Dothideales, Capnodiales, Myriangiales and Trypetheliales. The latter order was recently proposed (Aptroot et al. 2008) and represents the largest lichen forming clade in Dothideomycetes. Another recently proposed order, Botryosphaeriales includes only the single family, Botryosphaeriaceae. The analysis (Fig. 2B), however, shows strong support for a narrower interpretation of the Botryosphaeriaceae, typified by Botryosphaeria dothidea and related genera, excluding a separate clade of species residing in Guignardia (with Phyllosticta anamorphs). Bagnisiella examinens and Saccharata protea did not reside in either of the above clades, placed on early diverging branches. A more extensive taxon sampling is required to address the diversity in this order, which most likely will validate the separation of additional families. Another currently accepted order, Microthyriales, consisting of species occurring as saprobes or epiphytes on stems and leaves is represented in this study by only a single sample, Microthyrium microscopicum (Fig. 2C). Members of this order are poorly represented in culture and have unusual thyrothecial ascomata that have a scutate covering comprising a thin layer of radiating cells. This structure is generally lacking a basal layer and is quite unlike any morphologies in other orders. This positioning adjacent to the plant parasitic Venturiaceae and coprophilic Phaeotrichaceae, is unexpected but since the single representative of the Microthyriales is on a long branch this is a relationship that will require more intensive taxon sampling.
Additional families that could not be placed in an order are Tubeufiaceae and Gloniaceae (Fig. 2B). Species in Tubeufiaceae have superficial clustered ascomata and characteristic bitunicate asci with relatively long ascospores, often with helicosporous anamorphs (Kodsueb et al. 2008). Members of Tubeufiaceae, which frequently occur in freshwater habitats include anamorph genera, such as Helicoon and Helicodendron, and are ecologically classified as aeroaquatic species. A few teleomorph taxa such as Tubeufia asiana occur on submerged wood (Tsui et al. 2007), and Tubeufia paludosa occur on herbaceous substrates in wet habitats (Webster 1951). The Gloniaceae are saprobic, have dichotomously branched, laterally anastomosed pseudothecia that form radiating pseudo-stellate composites and dehisce by an inconspicuous, longitudinal, but evaginated slit. They reside sister to the saprobic Mytilinidiales but due to conspicuous morphological differences and moderate statistical support they are placed in Pleosporomycetidae incertae sedis (Boehm et al. 2009a, this volume).
Several other well supported clades representing families were evident in this study (Fig. 2). These include several families in Pleosporales, treated elsewhere (Zhang et al. 2009; this volume). Other clades have lower levels of support. For example Leptosphaeriaceae (Fig. 2A) have moderate bootstrap support and it is treated in the very broad sense here. There was also support for several newly described families treated in different papers within this volume. In Pleosporales these include Amniculicolaceae and Lentitheciaceae (Zhang et al. 2009; this volume). The Lindgomycetaceae (Shearer et al. 2009; this volume, Hirayama et al. 2010) encompassing a majority of species isolated from fresh water habitats. Two other novel families, Aigialaceae and Morosphaeriaceae include mainly marine species (Suetrong et al. 2009; this volume). In addition to these, the sampling of a wide diversity of fungi on bamboo yielded the description of Tetraplosphaeriaceae (Tanaka et al. 2009; this volume). Another novel family, Dissoconiaceae, is proposed by Crous et al. 2009 (this volume) for foliicolous commensalists on Eucalyptus leaves, some of which are putative hyper parasites and reside in Capnodiales.
Results of this study suggest that sampling within existing families also requires continued expansion as familial definitions in Dothideomycetes remains problematic. A paper focused on two families, with poor representation in molecular data sets, Melanommataceae and Lophiostomataceae addresses this in more detail (Mugambi & Huhndorf 2009,Mugambi & Huhndorf 2009; this volume). Numerous other clades in our tree remain without familial placement. This includes a diverse group in Capnodiales (Fig. 2C, clade C) a newly described group of hysteriaceous fungi in Pleosporales (Fig. 2A, clade G) and additional marine lineages (clades H, L, Fig. 2A). An interesting clade tentatively circumdescribed by Zhang et al. (2009; this volume) as Massariaceae contains bambusicolous fungi and appears related to the lichenised Arthopyreniaceae (Fig. 2A).
Finally, a clade including Corynespora anamorphs (clade K, Fig. 2A) is placed for the first time, but without clear relationship to any other currently defined families. The genus Corynespora includes anamorphic fungi with tretic, percurrent, and acropetal conidiogenesis. The melanised, pseudoseptate conidia have a pronounced hilum from which the conidial germ tube emerges and are borne apically from solitary, melanised conidiophores. Though nearly 100 species are described based on differences in morphology, considerable phenotypic plasticity within individual isolates complicates species recognition, and molecular analyses that may result in taxonomic clarification have not been done. Corynespora species fill a diversity of roles as saprobes, pathogens, and endophytes on and in woody and herbaceous plants, other fungi, nematodes, and human skin (Dixon et al. 2009). One of the species represented here, C. cassiicola is an important pathogen of rubber. The teleomorphic fungi Pleomassaria swidae (Pleomassariaceae; Tanaka et al. 2005) and Corynesporasca caryotae (Corynesporascaceae; Sivanesan 1996) have unnamed Corynespora species as anamorphs. In this study, species currently placed in Corynespora are not monophyletic and are positioned in at least two families: Massarinaceae and Clade K (Fig. 2A).
Anamorph taxa
The previously mentioned Dissoconiaceae relies on taxonomic descriptions based on anamorph characters. This is a theme that is expected to continue for mitosporic taxa in Dothideomycetes as molecular data accelerates their integration. The artificial nature of the “higher” taxa of anamorphs e.g., deuteromycetes (Kirk et al. 2001) is now well recognised, but the integration of anamorphs into the phylogenetic classification of teleomorphs remains a significant challenge in fungal systematics (Shenoy et al. 2007). The correlation of teleomorphs and anamorphs (Seifert et al. 2000) is not always predictive but it has been applied in some genera within Dothideomycetes, e.g. Botryosphaeria and Mycosphaerella (Crous et al. 2006, 2009b). However, numerous examples underscoring anamorph convergence can be found throughout the class e.g. Dictyosporium (Tsui et al. 2006, Kodsueb et al. 2008), Sporidesmium (Shenoy et al. 2006), Cladosporium (Crous et al. 2007b) and Phoma (Fig. 2A; Aveskamp et al. 2009, de Gruyter et al. 2009, Woudenberg et al. 2009) as well as Fusicoccum and Diplodia (Crous et al. 2006, Phillips et al. 2008). The use of large multigene phylogenies will be essential to bring taxonomic order to cryptic anamorph lineages.
Ecological diversity
Besides the unclassified diversity found in anamorphic genera, numerous ecological niches contain diverse lineages of fungi lacking systematically sampled molecular characters. Several examples of this knowledge gap can be found in papers in this volume. In this regard, the rock inhabiting fungi are amongst the least understood. These fungi exist ubiquitously as melanised, slow growing colonies and that usually do not produce generative structures. They subsist on bare rock surfaces and are consequently highly tolerant of the environmental stresses induced by lack of nutrients, water and extremes in radiation and temperature (Palmer et al. 1990, Sterflinger 1998, Ruibal 2004, Gorbushina et al. 2008). Members of this ecological guild are diverse and occur in two classes – Eurotiomycetes and Dothideomycetes. Ruibal et al. 2009 (this volume) present the results of an expanded sampling of rock-inhabiting fungi that include lineages residing within Dothideomycetes and sister class Arthoniomycetes. These rock inhabiting fungi can be placed in Capnodiales, Pleosporales, Dothideales and Myriangiales, as well as some unclassified lineages of Dothideomycetes. Interestingly, some associated lineages were without clear placement within either Arthoniomycetes or Dothideomycetes. The rock isolates included in Fig. 2C illustrate a subsection of genetic diversity seen in these extremophiles, in particular for the Capnodiales, with two rock isolates-rich lineages Teratosphaeriaceae and Clade C (Fig. 2C). A more detailed analysis (Ruibal et al. 2009; this volume) allows for the presentation of hypotheses related to evolution of pathogenicity and lichenisation because these modes of nutrition are often found in close proximity of rock inhabiting fungal lineages.
The lichenised fungi allied with the Dothideomycetes represent another poorly sampled group of fungi. Several lichenised species remain enigmatically placed after they were confirmed as members of Dothideomycetes based on DNA sequence data (Lumbsch et al. 2005, Del Prado et al. 2006). Although the number of species is comparatively small, their placement can play an important link in determining how transitions to and from lichenisation influenced dothideomycete evolution. Trypetheliaceae known for its anastomosing, branched pseudoparaphyses was until very recently still placed within Pyrenulales, an ascohymenial order in Eurotiomycetes, based on bitunicate asci and lense-shaped lumina in the ascospores (Del Prado et al. 2006). Attempts to resolve members of this family remain challenging as they tend to occur on long, rapidly evolving branches in our phylogenetic analyses, which often lead to artifacts. Nelsen et al. 2009 (this volume) demonstrate the occurrence of two additional lichen-forming lineages within Dothideomycetes representing the families Strigulaceae and Monoblastiaceae. The delineation of lichenised family Arthopyreniaceae should continue to be assessed given their placement with a clade containing bambusicolous fungi (Tanaka et al. 2009; this volume) and their non monophyly is also confirmed elsewhere (Nelsen et al. 2009; this volume). The relationship between the lichenised groups and bambusicolous genera Roussoella and Roussoellopsis (Didymosphaeriaceae; Ju et al. 1996, Lumbsch & Huhndorf 2007) is strongly supported, but their affinity is not fully understood due to their considerable morphological differences.
The fungi collected from marine and freshwater habitats contain yet more varied species that have not been assessed well within a molecular based framework. Their diversity is supported by the fact that whole orders (Jahnulales) and several families, already mentioned, almost exclusively consist of species collected from these environments. A recent assessment of marine fungi tallied a number of more than 500 species with more than a fifth of these suggested to reside in Dothideomycetes (Jones et al. 2009). The number for fungi from fresh water habitats is somewhat lower (about 170 taxa).
Despite similarities in their preferred medium for spore dispersal (water) an examination of phylogenetic diversity within Dothideomycetes indicates that these groups of fungi tend to reside in divergent parts of the tree (Figs 2, 3). However, some exceptions may occur: For example, members of Aigialaceae are weakly supported to share ancestry with members of freshwater clade Lindgomycetaceae (Raja et al. 2010). The Jahnulales represents another recently delineated aquatic lineage with an interesting mixture of fresh water and marine taxa. It was delineated based on molecular and morphological data (Inderbitzin et al. 2001, Pang et al. 2002) and now contains four genera and several species (Campbell et al. 2007). Previously, two anamorphic species in the Jahnulales, Xylomyces rhizophorae (described from mangrove wood of Rhizophora) and X. chlamydosporus have been reported from mangroves and thus saline habitats (Kohlmeyer & Volkmann-Kohlmeyer 1998). It has further been documented that X. chlamydosporus is the anamorph of Jahnula aquatica, a freshwater species (Sivichai, pers. comm.).
Marine Dothideomycetes generally exist in association with algae and plants in marine and brackish environments, usually with intertidal or secondary marine plants (e.g., mangroves). The majority of these fungi have been classified in families and genera that comprise mostly terrestrial species (e.g., Pleospora) and no definitive clades of marine Dothideomycetes have been identified. Here we find support for diverse aquatic lineages similar to the situation in Sordariomycetes. Papers by Suetrong et al. 2009 (this volume) and Shearer et al. 2009 (this volume) continue to address this disparity by using multigene phylogenies to describe several lineages within a class wide context. In contrast, many marine members of the Dothideomycetes await interrogation at the DNA sequence level, especially the genera Belizeana, Thalassoascus, Lautospora and Loratospora, all exclusively marine taxa.
The final environmentally defined group sampled in this volume is the bambusicolous fungi. More than 1 100 fungal species have been described or recorded worldwide from bamboo (Hyde et al. 2002). Furthermore, their ecological specialisation as pathogens, saprophytes, and endophytes has been relatively well documented (e.g. Hino 1961). However, relatively few studies based on DNA sequence comparisons have been undertaken for many bambusicolous fungi. Several unique lineages, e.g. the Katumotoa bambusicola-Ophiosphaerella sasicola clade in a freshwater lineage (Lentitheciaceae) and the Roussoella-Roussoellopsis clade close to lichen-forming families could be found (Fig. 2). Particularly, a new family Tetraplosphaeriaceae including five new genera characterised by a Tetraploa anamorph s. l. is introduced as a lineage of fungi with bamboo habitat (Tanaka et al. 2009; this volume). It is clear that much additional diversity within this group of fungi remains to be sampled using DNA sequence data
A number of other niches remain poorly discussed in this volume. Coprophilous fungi occur in three families Delitschiaceae, Phaeotrichaceae, and Sporormiaceae (Figs 2A, C). These families are not closely related and it is clear that the fimicolous life style has arisen more than once in the Dothideomycetes. Also, many species from these groups are not strictly dung-inhabiting, but can be found on other substrates like soil, wood, and plant-debris. Interestingly, some are human pathogens, plant endophytes and lichenicolous fungi. As is true throughout the Ascomycota, a change in substrate is apparently not a substantial evolutionary step in these taxa (Kruys & Wedin 2009).
Additional observations
Several orders e.g. Dothideales, Myriangiales and Microthyriales have not been treated using the extensive systematic sampling that is true for studies treated in this volume. However, individual smaller studies continue to provide interesting and surprising results. One such example is the first described meristematic and endoconidial species residing in Myriangiales (Fig. 2C) reported by Tsuneda et al. (2008). These Endosporium species were isolated from very different substrates such as: poplar twigs and a dead bird. They also have a close relationship to a single lineage of rock inhabiting fungi. The nutritional shifts represented by these closely related species correlate well with scenarios described by Ruibal et al. (2009; this volume) for rock inhabiting fungi. Another melanised meristematic fungus, Sarcinomyces crustaceus, isolated from pine trees appears in a similar position in a phylogeny presented in the aforementioned paper (Ruibal et al. 2009; this volume).
Another unusual species, Catinella olivacea is included in Fig. 2C, but without any clearly resolved position, diverging early to Dothideomycetidae. This species was initially placed in Leotiomycetes, due to their flattened apothecia, found on the underside of moist, well-decayed logs of hardwood. Asci are unitunicate but they appear to form after ascolocular development. As in the previous analysis, it was not possible to identify relationships between this species and any known order, although there are indications of a close relationship with the Dothideomycetidae (Greif et al. 2007).
The placement of the single asexual mycorrhizal lineage representing Cenococcum geophilum in the Dothideomycetes (LoBuglio et al. 1996), allied to members of the saprobic Gloniaceae is intriguing (Fig. 2B; Boehm et al. 2009a; this volume). No resolved placement for this species in Dothideomycetes has been possible in the past. The results of this study were also unexpected because no biological data suggest a connection to the family. Cenococcum is a fungus that is intensively used in environmental studies and this could suggest a very interesting biology for members of the ostensibly saprobic Gloniaceae. Results of this study advocate a more expansive sampling of Cenococcum in order to confirm this intriguing result.
CONCLUSIONS
One of the major obstacles in dothideomycete systematics remains the lack of a clear understanding of what species are members of the class based on morphology alone. Throughout most of the 20th Century, comparative morphological studies have been the only character on which to base phylogenetic relationships. The advent of large DNA-sequence data sets should allow for a substantially improved interpretation of morphological characters for this class of fungi. Studies in this volume and elsewhere have provided a clear understanding that many of the characters classically used in taxonomy and systematics of the group are homoplastic and not helpful for reconstructing phylogenetic relationships. Dothideomycete taxonomy also needs to keep pace with the rapid advances being made in phylogenetics, genomics and related fields. The important principle here is that our classification should communicate diversity accurately and allow dothideomycete biologists from disparate fields to have access to an agreed upon set of taxonomic names to aid communication. In addition, it should allow for a focus on under-sampled groups and clades (i.e. poorly sampled saprobes and others). A major task ahead will be to add asexual genera to present phylogenetic schemes, and integrate these into the existing familial and ordinal classification. As most of these asexual genera are in fact poly- and paraphyletic, their type species will need to be recollected to clarify their phylogenetic position. In addition to this, it appears that even some concepts of teleomorphic taxa will require extensive reconsideration. Finally, we should attempt to incorporate valuable biological information from past workers, such as the three mycologists to which this volume is dedicated, by reliably assessing culture and sequence identity. It is hoped that the papers in this volume will make a meaningful contribution towards these goals.
Acknowledgments
Authors from individual papers in this volume contributed to this work and specific acknowledgements to that regard can be found in individual papers. Work performed for this paper by the first author after 2008 was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. Part of this work was also funded by grants from NSF (DEB-0717476) to J. W. Spatafora (and C.L. Schoch until 2008) and (DEB-0732993) to J.W. Spatafora and B. Robbertse.
References
- Aptroot A, Lücking R, Sipman H, Umana L, Chaves J-L (2008). Pyrenocarpous lichens with bitunicate asci. A first assessment of the lichen biodiversity inventory in Costa Rica. Bibliotheca Lichenologica 97: 1–162. [Google Scholar]
- Arx von J, Müller E (1975). A re-evaluation of the bitunicate ascomycetes with keys to families and genera. Studies in Mycology 9: 1–159. [Google Scholar]
- Aveskamp MM, Verkley GJM, Gruyter J de, Murace MA, Perello A, et al. (2009). DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 101: 359–378. [DOI] [PubMed] [Google Scholar]
- Barr ME (1979). A classification of Loculoascomycetes. Mycologia 71: 935–957. [Google Scholar]
- Barr ME (1987). Prodromus to class Loculoascomycetes. M.E. Barr Bigelow, Amherst, Massachusetts.
- Berbee ML (1996). Loculoascomycete origins and evolution of filamentous ascomycete morphology based on 18S rRNA gene sequence data. Molecular Biology and Evolution 13: 462–470. [DOI] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW (1992). Two Ascomycete Classes Based on Fruiting-Body Characters and Ribosomal DNA Sequence. Molecular Biology and Evolution 9: 278–284. [DOI] [PubMed] [Google Scholar]
- Boehm EWA, Mugambi GK, Miller AN, Huhndorf SM, Marincowitz S, et al. (2009a). A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology 64: 49–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EWA, Schoch CL, Spatafora JW (2009b). On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479. [DOI] [PubMed] [Google Scholar]
- Campbell J, Ferrer A, Raja HA, Sivichai S, Shearer CA (2007). Phylogenetic relationships among taxa in the Jahnulales inferred from 18S and 28S nuclear ribosomal DNA sequences. Canadian Journal of Botany 85: 873–882. [Google Scholar]
- Chase MW, Fay MF (2009). Barcoding of Plants and Fungi. Science 325: 682–683. [DOI] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Groenewald JZ (2007a). Mycosphaerella is polyphyletic. Studies in Mycology 58: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Schubert K, Groenewald JZ (2007b). Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, et al. (2009). Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, et al. (2006). Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC, et al. (2009b). Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LJ, Schlub RL, Pernezny K, Datnoff LE (2009). Host Specialization and Phylogenetic Diversity of Corynespora cassiicola. Phytopathology 99: 1015–1027. [DOI] [PubMed] [Google Scholar]
- Eriksson OE (1981). The families of bitunicate ascomycetes. Opera Botanica 60: 1–220. [Google Scholar]
- Eriksson OE, Winka K (1997). Outline of Ascomycota. Myconet: www.umu.se/myconet/M_outline.html.
- Ertz D, Miadlikowska J, Lutzoni F, Dessein S, Raspe O, et al. (2009). Towards a new classification of the Arthoniales (Ascomycota) based on a three-gene phylogeny focussing on the genus Opegrapha. Mycological Research 113: 141–152. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G (2006). A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evolutionairy Biology 6: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, et al. (2006). Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 98: 1053–1064. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Kotlova ER, Sherstneva OA (2008). Cellular responses of microcolonial rock fungi to long-term desiccation and subsequent rehydration. Studies in Mycology 61: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruyter de J, Aveskamp MM, Woudenberg JHC, Verkley GJM, Groenewald JZ, Crous PW (2009). Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycological Research 113: 508–519. [DOI] [PubMed] [Google Scholar]
- Greif MD, Gibas CFC, Tsuneda A, Currah RS (2007). Ascoma development and phylogeny of an apothecioid dothideomycete, Catinella olivacea. American Journal of Botany 94: 1890–1899. [DOI] [PubMed] [Google Scholar]
- Gueidan C, Ruibal Villasenor C, Hoog GS de, Gorbushina AA, Untereiner WA, Lutzoni F (2008). A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Studies in Mycology 61: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T, Bioedit v 7.0.1. Isis Pharmaceuticals, 2004.
- Henssen A, Thor G (1994). Developmental morphology of the “Zwischengruppe” between Ascohymeniales and Ascoloculares. In: Ascomycete systematics. Problems and perspectives in the nineties (Hawksworth DL, ed.). Plenum Press, New York: 43–61.
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, et al. (2007). A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547. [DOI] [PubMed] [Google Scholar]
- Hino I (1961). Icones fungorum bambusicolorum japonicorum., Fuji Bamboo Garden, Gotenba.
- Hirayama K, Tanaka K, Raja HA, Miller AN, Shearer CA (2010). A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 102: In press. [DOI] [PubMed]
- Hyde KD, Zhou DQ, Dalisay T (2002). Bambusicolous fungi: A review. Fungal Diversity 9: 1–14. [Google Scholar]
- Inderbitzin P, Landvik S, Abdel-Wahab MA, Berbee ML (2001). Aliquandostipitaceae, a new family for two new tropical ascomycetes with unusually wide hyphae and dimorphic ascomata. American Journal of Botany 88: 52–61. [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. (2006). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Jones EBG, Sakayaroj J, Suetrong S, Somrithipol S, Pang KL (2009). Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Diversity 35: 1–187. [Google Scholar]
- Ju YM, Rogers JD, Huhndorf SM (1996). Valsaria and notes on Endoxylina, Pseudothyridaria, Pseudovalsaria, and Roussoella. Mycotaxon 58: 419–481. [Google Scholar]
- Katoh K, Asimenos G, Toh H (2009). Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology 537: 39–64. [DOI] [PubMed] [Google Scholar]
- Kauff F, Cox CJ, Lutzoni F (2007). WASABI: An Automated Sequence Processing System for Multigene Phylogenies. Systematic Biology 56: 523–531. [DOI] [PubMed] [Google Scholar]
- Kauff F, Lutzoni F (2002). Phylogeny of the Gyalectales and Ostropales (Ascomycota, Fungi): among and within order relationships based on nuclear ribosomal RNA small and large subunits. Molecular Phylogenetics and Evolution 25: 138–156. [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, David J, Stalpers JA (2001). Ainsworth and Bisby's Dictionary of the Fungi, 9th ed. CAB International, 1203pp. Wallingford, U.K.
- Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008). Ainsworth and Bisby's dictionary of the Fungi, 10th ed. CAB International, 2283pp. Wallingford, U.K.
- Kodsueb R, Dhanasekaran V, Aptroot A, Lumyong P, McKenzie EHC, et al. (2006). The family Pleosporaceae: intergeneric relationships and phylogenetic perspectives based on sequence analyses of partial 28S rDNA. Mycologia 98: 571–583. [DOI] [PubMed] [Google Scholar]
- Kodsueb R, Jeewon R, Vijaykrishna D, Mckenzie EHC, Lumyong P, et al. (2008). Systematic revision of Tubeufiaceae based on morphological and molecular data. Fungal Diversity 21: 105–145. [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B (1998). A new marine Xylomyces on Rhizophora from the Caribbean and Hawaii. Fungal Diversity 1: 159–164. [Google Scholar]
- Kruys Å, Wedin M (2009). Phylogenetic relationships and an assessment of traditionally used taxonomic characters in the Sporormiaceae (Pleosporales, Dothideomycetes, Ascomycota), utilizing multi-gene phylogenies. Systematics and Biodiversity 7: 465–478. [Google Scholar]
- Kuramae E, Robert V, Snel B, Weiß M, Boekhout T (2006). Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Research 6: 1213–1220. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Brown JM, Stanger-Hall K, Lemmon EM (2009). The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Systematic Biology 58: 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew EC, Aptroot A, Hyde KD (2000). Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Molecular Phylogenetics and Evolution 16: 392–402. [DOI] [PubMed] [Google Scholar]
- Lindemuth R, Wirtz N, Lumbsch HT (2001). Phylogenetic analysis of nuclear and mitochondrial rDNA sequences supports the view that loculoascomycetes (Ascomycota) are not monophyletic. Mycological Research 105: 1176–1181. [Google Scholar]
- Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T (2009). Rapid and Accurate Large-Scale Coestimation of Sequence Alignments and Phylogenetic Trees. Science 324: 1561–1564. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Hall BD (2004). Body plan evolution of ascomycetes, as inferred from an RNA polymerase II phylogeny. Proceedings of the National Academy of Sciences of the United States of America 101: 4507–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoBuglio KF, Berbee ML, Taylor JW (1996). Phylogenetic origins of the asexual mycorrhizal symbiont Cenococcum geophilum Fr. and other mycorrhizal fungi among the ascomycetes. Molecular Phylogenetics and Evolution 6: 287–294. [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Huhndorf S (2007). Whatever happened to the pyrenomycetes and loculoascomycetes? Mycological Research 111: 1064–1074. [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Lindemuth R (2001). Major lineages of Dothideomycetes (Ascomycota) inferred from SSU and LSU rDNA sequences. Mycological Research 105: 901–908. [Google Scholar]
- Lumbsch HT, Schmitt I, Lindemuth R, Miller A, Mangold A, et al. (2005). Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomyceta). Molecular Phylogenetics and Evolution 34: 512–524. [DOI] [PubMed] [Google Scholar]
- Luttrell ES (1951). Taxonomy of Pyrenomycetes. University of Missouri Studies 24: 1–120. [Google Scholar]
- Luttrell ES (1955). The ascostromatic Ascomycetes. Mycologia 47: 511–532. [Google Scholar]
- Lutzoni F, et al. (2004). Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany 91: 1446–1480. [DOI] [PubMed] [Google Scholar]
- McLaughlin DJ, Hibbett DS, Lutzoni F, Spatafora JW, Vilgalys R (2009). The search for the fungal tree of life. Trends in Microbiology 17: 488–497 [DOI] [PubMed] [Google Scholar]
- Miadlikowska J, Kauff F, Hofstetter V, Fraker E, Grube M, et al. (2006). New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA- and two protein-coding genes. Mycologia 98: 1088–1103. [PubMed] [Google Scholar]
- Miller JH (1949). A revision of the classification of the ascomycetes with special emphasis on the Pyrenomycetes. Mycologia 41: 99–127. [Google Scholar]
- Mugambi GK, Huhndorf SM (2009). Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. [Google Scholar]
- Mugambi GK, Huhndorf SM (2009). Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Studies in Mycology 64: 103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller E, Arx J von (1962). Die Gattungen der didymosporen Pyrenomyceten. Beitrage zur Kryptogamenflora der Schweiz 11: 1–992. [Google Scholar]
- Nannfeldt JA (1932). Studien über die Morphologie und Systematik der nicht-lichenisierten inoperculaten Discomyceten. In: Nova acta Regiae Societatis scientiarum upsalensis Almqvist & Wiksells, Uppsala.
- Nelsen MP, Lücking R, Grube M, Mbatchou JS, Muggia L, et al. (2009). Unravelling the phylogenetic relationships of lichenized fungi in Dothideomyceta. Studies in Mycology 64: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer FE, Staley JT, Ryan B (1990). Ecophysiology of microcolonial fungi and lichens on rocks in Northeastern Oregon. New Phytologist 116: 613–620. [Google Scholar]
- Pang KL, Abdel-Wahab MA, Sivichai S, El-Sharouney HM, Jones EBG (2002). Jahnulales (Dothideomycetes, Ascomycota): a new order of lignicolous freshwater ascomycetes. Mycological Research 106: 1031–1042. [Google Scholar]
- Prado R Del, Schmitt I, Kautz S, Palice Z, Lücking R, Lumbsch HT (2006). Molecular data place Trypetheliaceae in Dothideomycetes. Mycological Research 110: 511–520. [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, Johnston PR, Ramaley A, et al. (2008). Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21: 29–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja HA, Ferrer A, Miller AN, Shearer CA (2010). Freshwater Ascomycetes: Wicklowia aquatica, a new genus and species in the Pleosporales from Florida and Costa Rica. Mycoscience. In press.
- Robbertse B, Reeves JB, Schoch CL, Spatafora JW (2006). A phylogenomic analysis of the Ascomycota. Fungal Genetics and Biology 43: 715–725. [DOI] [PubMed] [Google Scholar]
- Ruibal C (2004). Isolation and characterization of melanized, slow-growing fungi from semiarid rock surfaces of central Spain and Mallorca. Ph.D. dissertation Universidad Autónoma de Madrid, Madrid, Spain.
- Ruibal C, Gueidan C, Selbmann L, Gorbushina AA, Crous PW, et al. (2009). Phylogeny of rock-inhabiting fungi related to Dothideomycetes. Studies in Mycology 64: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santesson (1952). Foliicolous lichens I. A revision of the taxonomy of the obligately foliicolous, lichenized fungi. Symbolae Botanicae Upsaliensis 12: 1–590. [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW (2006). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, et al. (2009a). The Ascomycota Tree of Life: A Phylum-wide Phylogeny Clarifies the Origin and Evolution of Fundamental Reproductive and Ecological Traits. Systematic Biology 58: 224–239. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Wang Z, Townsend JP, Spatafora JW (2009b). Geoglossomycetes cl. nov., Geoglossales ord. nov. and taxa above class rank in the Ascomycota Tree of Life. Persoonia 22: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Gams W, Crous PW, Samuels GJ (2000). Molecules, morphology and classification: towards monophyletic genera in Ascomycetes. Studies in Mycology 45: 1–4. [Google Scholar]
- Seifert KA (2009). Progress towards DNA barcoding of fungi. Molecular Ecology Resources 9: 83–89. [DOI] [PubMed] [Google Scholar]
- Seifert KA, Samson RA, Dewaard JR, Houbraken J, Levesque CA, et al. (2007). Prospects for fungus identification using C01 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences of the United States of America 104: 3901–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer CA, Raja HA, Miller AN, Nelson P, Tanaka K, et al. (2009). The molecular phylogeny of freshwater Dothideomycetes. Studies in Mycology 64: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy BD, Jeewon R, Hyde KD (2007). Impact of DNA sequence-data on the taxonomy of anamorphic fungi. Fungal Diversity 26: 1–54. [Google Scholar]
- Shenoy BD, Jeewon R, Wu WP, Bhat DJ, Hyde KD (2006). Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycological Research 110: 916–928. [DOI] [PubMed] [Google Scholar]
- Silva-Hanlin DMW, Hanlin RT (1999). Small subunit ribosomal RNA gene phylogeny of several loculoascomycetes and its taxonomic implications. Mycological Research 103: 153–160. [Google Scholar]
- Sivanesan A (1996). Corynesporasca caryotae gen. et sp. nov. with a Corynespora anamorph, and the family Corynesporascaceae. Mycologia 100: 783–788. [Google Scholar]
- Spatafora JW, Mitchell TG, Vilgalys R (1995). Analysis of genes coding for small-subunit rRNA sequences in studying phylogenetics of dematiaceous fungal pathogens. Journal of Clinical Microbiology 33: 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Sung G-H, Johnson D, Hesse C, O'Rourke B, et al. (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Volkmann-Kohlmeyer B, Kohlmeyer J (1998). Independent terrestrial origins of the Halosphaeriales (marine Ascomycota). American Journal of Botany 85: 1569–1580. [PubMed] [Google Scholar]
- Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Sterflinger K (1998). Temperature and NaCl- tolerance of rock-inhabiting meristematic fungi. Antonie Van Leeuwenhoek 74: 271–281. [DOI] [PubMed] [Google Scholar]
- Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-Kohlmeyer B, et al. (2009). Molecular systematics of the marine Dothideomycetes. Studies in Mycology 64: 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hirayama K, Yonezawa H, Hatakeyama S, Harada Y, et al. (2009). Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Studies in Mycology 64: 175–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Ooki Y, Hatakeyama S, Harada Y, Barr ME (2005). Pleosporales in Japan (5): Pleomassaria, Asteromassaria, and Splanchnonema. Mycoscience 46: 248–260. [Google Scholar]
- Tsui CKM, Berbee ML (2006). Phylogenetic relationships and convergence of helicosporous fungi inferred from ribosomal DNA sequences. Molecular Phylogenetics and Evolution 39: 587–597. [DOI] [PubMed] [Google Scholar]
- Tsui CKM, Berbee ML, Jeewon R, Hyde KD (2006). Molecular phylogeny of Dictyosporium and allied genera inferred from ribosomal DNA. Fungal Diversity 21: 157–166. [Google Scholar]
- Tsuneda A, Davey ML, Hambleton S, Currah RS (2008). Endosporium, a new endoconidial genus allied to the Myriangiales. Botany 86: 1020–1033. [Google Scholar]
- Vijaykrishna D, Jeewon R, Hyde KD (2006). Molecular taxonomy, origins and evolution of freshwater ascomycetes. Fungal Diversity 23: 351–390. [Google Scholar]
- Webster J (1951). Graminicolous Pyrenomycetes 1. The conidial stage of Tubeufia helicomyces. Transactions of the British Mycological Society 34: 304–308. [Google Scholar]
- Wernersson R, Pedersen AG (2003). RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Research 31: 3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ (2006). Missing data and the design of phylogenetic analyses. Journal of Biomedical Informatics 39: 34–42. [DOI] [PubMed] [Google Scholar]
- Winka K, Eriksson OE, Bang A (1998). Molecular evidence for recognizing the Chaetothyriales. Mycologia 90: 822–830. [Google Scholar]
- Woudenberg JHC, Aveskamp MM, Gruyter J de, Spiers AG, Crous PW (2009). Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. Thesis. Universty of Texas at Austin.
- Zhang Y, Fournier J, Pointing SB, Hyde KD (2008). Are Melanomma pulvis-pyrius and Trematosphaeria pertusa congeneric? Fungal Diversity 23: 351–390. [Google Scholar]
- Zhang Y, Schoch CL, Fournier J, Crous PW, Gruyter J de, et al. (2009). Multi-locus phylogeny of the Pleosporales: a taxonomic, ecological and evolutionary reevaluation. Studies in Mycology 64: 85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]