Abstract
The class Dothideomycetes (along with Eurotiomycetes) includes numerous rock-inhabiting fungi (RIF), a group of ascomycetes that tolerates surprisingly well harsh conditions prevailing on rock surfaces. Despite their convergent morphology and physiology, RIF are phylogenetically highly diverse in Dothideomycetes. However, the positions of main groups of RIF in this class remain unclear due to the lack of a strong phylogenetic framework. Moreover, connections between rock-dwelling habit and other lifestyles found in Dothideomycetes such as plant pathogens, saprobes and lichen-forming fungi are still unexplored. Based on multigene phylogenetic analyses, we report that RIF belong to Capnodiales (particularly to the family Teratosphaeriaceae s.l.), Dothideales, Pleosporales, and Myriangiales, as well as some uncharacterised groups with affinities to Dothideomycetes. Moreover, one lineage consisting exclusively of RIF proved to be closely related to Arthoniomycetes, the sister class of Dothideomycetes. The broad phylogenetic amplitude of RIF in Dothideomycetes suggests that total species richness in this class remains underestimated. Composition of some RIF-rich lineages suggests that rock surfaces are reservoirs for plant-associated fungi or saprobes, although other data also agree with rocks as a primary substrate for ancient fungal lineages. According to the current sampling, long distance dispersal seems to be common for RIF. Dothideomycetes lineages comprising lichens also include RIF, suggesting a possible link between rock-dwelling habit and lichenisation.
Keywords: Arthoniomycetes, Capnodiales, Dothideomycetes, evolution, extremotolerance, multigene phylogeny, rock-inhabiting fungi
INTRODUCTION
The Dothideomycetes constitute the largest class of ascomycetes with approximately 19 000 species, which are currently classified in 11 orders and 90 families (Kirk et al. 2008). This class is ecologically diverse, with many pathogens or saprobes on plants, some coprophilous species, and a few lichen-forming fungi (Schoch et al. 2009b; this volume). Early studies have shown that a large part of the non-lichenised, slow-growing melanised fungi isolated from rock surfaces (here referred to as rock-inhabiting fungi) also belong to this class (Sterflinger et al. 1997, 1999). Subsequent sampling efforts revealed a higher diversity of species than expected for these rock-inhabiting fungi (Ruibal 2004, Ruibal et al. 2005, 2008, Selbmann et al. 2005, 2008).
Rock-inhabiting fungi (RIF) are peculiar organisms that apparently lack sexual reproductive structures and form compact, melanised colonies on bare rock surfaces (Fig. 1). Although very common, RIF have often been overlooked due to their small size, their slow growth and the lack of diagnostic features. First discovered in hot and cold deserts (Krumbein & Jens 1981, Friedmann 1982, Staley et al. 1982), RIF are now known to be ubiquitous on hard surfaces, in extreme as well as in temperate climates (Urzì et al. 1995, Sterflinger & Prillinger 2001, Gorbushina 2007, Gorbushina & Broughton 2009). RIF are well adapted to nutrient-poor and dry habitats where they are particularly successful colonisers due to restricted competition with other microbes (Gorbushina 2007) and their extremotolerance.
Fig 1.
Rock-inhabiting fungi related to Dothideomycetes. A–C: sampling localities (photos C. Ruibal and L. Selbmann). A. Metamorphic black slate from Atazar, Central Mountain System, Spain. B. Limestone from Cala Sant Vicenç, Serra de Tramuntana, Mallorca, Spain. C. Sandstone from Alatna Valley, McMurdo Dry Valleys, Antarctica. D–G: Coniosporium apollinis, a rock-inhabiting species from the Mediterranean region (CBS 100213, photos C. Gueidan). D. Colony on MEA. E. Melanised torulose hyphae. F. Hypha disarticulating into bi- to multi-cellular clumps; G. Meristematic growth. H–J: Antarctic rock-inhabiting fungi (photos L. Selbmann). H. RIF growing on a crystal of sandstone. I. Melanised hypha of Friedmanniomyces endolithicus. J. Meristematic growth of Cryomyces antarcticus. K–L: Cystocoleus ebeneus, a lichenised species assigned to Capnodiales (photos L. Muggia). K. Microfilamentous thallus. L. Melanised hyphae of the mycobiont forming a furrow around the filamentous algae. Scale bars: D = 2 mm, E–G and I–J = 10 μm, H = 0.5 mm, K–L = 20 μm.
Extremotolerance comprises some specific universally present adaptations that enable these fungi to tolerate surprisingly wide ranges of temperatures, irradiation and osmotic stresses (Palmer et al. 1990, Sterflinger 1998, Gorbushina et al. 2003, Ruibal 2004, Onofri et al. 2007, Gorbushina et al. 2008). Melanisation protects cells against UV radiations (Dadachova & Casadevall 2008), whereas the typical isodiametrical (meristematic) growth form ensures an optimal volume: surface ratio and, therefore, allows them to survive extreme temperatures and desiccation (Wollenzien et al. 1995). These oligotrophic organisms are able to rely only on sparse, airborne nutrients available on rock surfaces. Their growth on these substrates is limited, and, for some of them, the production of internal asexual spores further allows to save energy. All adaptations contribute to the amazing survival capabilities of RIF in hostile habitats. The environmental tolerance of these fungi, and, in some cases, their capacity to penetrate minerals, make them an attractive subject for studies in microbial ecophysiology and applied research, such as biodeterioration of monuments and exobiology (Gorbushina et al. 1993, Diakumaku et al. 1995, Wollenzien et al. 1997, Gorbushina et al. 2002, Gorbushina 2003, Onofri et al. 2008).
Sterflinger et al. (1997) provided the first molecular evidence of RIF phylogenetic affiliations, and they are known to belong to two groups of ascomycetes, namely Dothideomycetes and Eurotiomycetes (de Hoog et al. 1999, Sterflinger et al. 1999, Ruibal 2004, Ruibal et al. 2005, 2008, Sert et al. 2007a). In Eurotiomycetes, multigene phylogenetic analyses have shown that RIF cluster in early diverging lineages of Chaetothyriales, whereas two species seem to be more closely related to the lichenised order Verrucariales, the sister group of Chaetothyriales (Gueidan et al. 2008). Gueidan et al. (2008) also demonstrated that the most recent common ancestor of both lichenised Verrucariales and pathogen-rich Chaetothyriales was probably a rock-inhabiting fungus. It was hypothesised that adaptations to life in extreme conditions might have been a prerequisite for the evolution of human pathogenicity (de Hoog 1993, Haase et al. 1999, Gueidan et al. 2008) and lichenisation in this class (Gueidan et al. 2008). In contrast, despite the high diversity of RIF within Dothideomycetes, only very few human pathogens are known from this class of Ascomycota (de Hoog et al. 2000). Alternatively, associations with plants and in particular plant pathogenicity are very common (Schoch et al. 2006, Arzanlou et al. 2007, Crous et al. 2007a, b, c, 2009; this volume). Additionally, lichenised species also appeared to be nested within Dothideomycetes (Lutzoni et al. 2004, James et al. 2006, Del Prado et al. 2006, Muggia et al. 2008, Nelsen et al. 2009). Presently no strong phylogenetic hypothesis is available to assess the placement of RIF within Dothideomycetes. Moreover, no studies have investigated phylogenetic relationships among RIF, lichenforming fungi and plant-associated fungi within Dothideomycetes. Our main goal was to infer phylogenetic relationships of RIF within Dothideomyceta, a lineage including Dothideomycetes and Arthoniomycetes, to explore more specifically their diversity, origins and evolution.
MATERIAL AND METHODS
Taxon and gene sampling
Representative taxa of most of the main orders and families of Dothideomyceta (Dothideomycetes and Arthoniomycetes) were sampled. Two separate sets of data matrices were assembled. The first set (three-gene analysis; Table 1 - see online Supplementary Information) is composed of 182 taxa (including 102 rock-inhabiting strains) for which DNA sequences of three ribosomal genes have been obtained: the large and small subunits of the nuclear ribosomal RNA gene (nucLSU and nucSSU, respectively) and the small subunit of the mitochondrial ribosomal RNA gene (mtSSU). Because this first set of data matrices included only ribosomal genes, low phylogenetic confidence was expected for deep relationships within Dothideomyceta. To overcome this problem, a second set of data matrices was assembled (five-gene analysis; Table 1 in Supplementary Information) consisting of DNA sequences of five loci from 113 taxa (including 40 rock-inhabiting strains): the largest and second largest subunits of the RNA polymerase II (RPB1 and RPB2, respectively), nucLSU, nucSSU and mtSSU. The outgroup for the three-gene analysis included Hyphozyma lignicola, Symbiotaphrina buchneri and S. kochii, whereas only the latter two species were selected as outgroup for the five-gene analysis. These species were chosen because they constituted a sister group to Dothideomyceta in a previous study (Schoch et al. 2009a).
Table 1.
Taxon and gene sampling for the three- and five- gene analyses. Geographical origins are also mentioned for RIF. A dash indicates missing sequences. Newly produced sequences are shown in bold. A column also indicates if taxa were included in the three-gene (3) or in both three- and five-gene analyses (3 & 5).
|
Taxon |
Collection # |
Additional information |
Order |
LSU |
SSU |
mtSSU |
RPB2 |
RPB1 |
Analysis |
|
| Hyphozyma lignicola | CBS 325.93 | Outgroup | AF353595 | AJ496239 | — | 3 | ||||
| Symbiotaphrina buchneri | CBS 6902 | Outgroup, AFTOL 1836 | FJ176887 | FJ176831 | — | FJ238370 | FJ238442 | 3 & 5 | ||
|
Symbiotaphrina kochii |
CBS 250.77 |
Outgroup, AFTOL 1902
|
AY227719
|
FJ176833
|
—
|
GU397369
|
FJ238443
|
3 & 5
|
||
|
Arthoniomycetes |
LSU |
SSU |
mtSSU |
RPB2 |
RPB1 |
Analysis |
||||
| Arthonia caesia | — | AFTOL 775 | Arthoniales | FJ469668 | — | FJ469671 | FJ469670 | FJ772241 | 3 & 5 | |
| Dendrographa leucophaea | — | AFTOL 308 | Arthoniales | AY548810 | AY548803 | AY548811 | EU704017 | — | 3 & 5 | |
| Dendrographa minor | — | AFTOL 355 | Arthoniales | AF279382 | AF279381 | GU561843 | AY641034 | GU561849 | 3 & 5 | |
| Lecanactis abietina | — | AFTOL 305 | Arthoniales | AY548812 | AY548805 | AY548813 | AH013900 | GU561850 | 3 & 5 | |
| Opegrapha dolomitica | — | AFTOL 993 | Arthoniales | — | DQ883706 | — | DQ883714 | DQ883717 | 3 & 5 | |
| Roccella fuciformis | — | AFTOL 126 | Arthoniales | AY584654 | AY584678 | EU704082 | DQ782866 | — | 3 & 5 | |
| Roccellographa cretacea | — | AFTOL 93 | Arthoniales | DQ883696 | DQ883705 | FJ772240 | DQ883713 | DQ883716 | 3 & 5 | |
| Schismatomma decolorans | — | AFTOL 307 | Arthoniales | AY548815 | AY548809 | AY548816 | DQ883715 | — | 3 & 5 | |
|
Simonyella variegata |
—
|
AFTOL 80
|
Arthoniales |
—
|
AY584669
|
AY584631
|
DQ782861
|
DQ782819
|
3 & 5
|
|
|
Dothideomycetes |
LSU |
SSU |
mtSSU |
RPB2 |
RPB1 |
Analysis |
||||
| Botryosphaeria dothidea | CBS 115476 | AFTOL 946 | Botryosphaeriales | DQ678051 | DQ677998 | FJ190612 | DQ677944 | EU186063 | 3 & 5 | |
| Guignardia bidwellii | CBS 237.48 | AFTOL 1618 | Botryosphaeriales | DQ678085 | DQ678034 | — | DQ677983 | — | 3 & 5 | |
| Macrophomina phaseolina | CBS 227.33 | AFTOL 1783 | Botryosphaeriales | DQ678088 | DQ678037 | FJ190645 | DQ677986 | — | 3 & 5 | |
| Neofusicoccum ribis | CBS 115475 | AFTOL 1232 | Botryosphaeriales | DQ678053 | DQ678000 | — | DQ677947 | — | 3 & 5 | |
| Capnodium coffeae | CBS 147.52 | AFTOL 939 | Capnodiales, Capnodiaceae | DQ247800 | DQ247808 | FJ190609 | DQ247788 | DQ471162 | 3 & 5 | |
| Capnodium salicinum | CBS 131.34 | AFTOL 937 | Capnodiales, Capnodiaceae | DQ678050 | DQ677997 | — | 3 | |||
| Microxyphium citri | CBS 451.66 | Capnodiales, Capnodiaceae | GU301848 | GU296177 | — | GU371727 | GU357750 | 3 & 5 | ||
| Scorias spongiosa | CBS 325.33 | AFTOL 1594 | Capnodiales, Capnodiaceae | DQ678075 | DQ678024 | FJ190643 | DQ677973 | — | 3 & 5 | |
| Cladosporium cladosporioides | CBS 170.54 | AFTOL 1289 | Capnodiales, Davidiellaceae | DQ678057 | DQ678004 | FJ190628 | DQ677952 | EU186064 | 3 & 5 | |
| Cladosporium sp. | CBS 180.53 | AFTOL 1035 | Capnodiales, Davidiellaceae | AY016367 | AY016351 | AY350576 | DQ677945 | — | 3 & 5 | |
| Davidiella tassiana | CBS 399.80 | AFTOL 1591 | Capnodiales, Davidiellaceae | DQ678074 | DQ678022 | — | DQ677971 | — | 3 & 5 | |
| Cercospora beticola | CBS 116456 | AFTOL 1788 | Capnodiales, Mycosphaerellaceae | DQ678091 | DQ678039 | FJ190647 | 3 | |||
| Mycosphaerella fijiensis | OSC 100622 | AFTOL 2021 | Capnodiales, Mycosphaerellaceae | DQ678098 | DQ767652 | FJ190656 | DQ677993 | — | 3 & 5 | |
| Mycosphaerella graminicola | CBS 292.38 | AFTOL 1615 | Capnodiales, Mycosphaerellaceae | DQ678084 | DQ678033 | DQ677982 | DQ677982 | — | 3 & 5 | |
| Mycosphaerella punctiformis | CBS 113265 | AFTOL 942 | Capnodiales, Mycosphaerellaceae | DQ470968 | DQ471017 | FJ190611 | DQ470920 | DQ471165 | 3 & 5 | |
| Capnobotryella renispora | CBS 214.90 | Capnodiales, Teratosphaeriaceae | EU019248 | Y18698 | — | — | — | 3 & 5 | ||
| Catenulostroma abietis | CBS 459.93 | AFTOL 2210 | Capnodiales, Teratosphaeriaceae | DQ678092 | DQ678040 | FJ190648 | — | GU357797 | 3 & 5 | |
| CBS 110890; | ||||||||||
| Catenulostroma microsporum | CPC 1832 | Capnodiales, Teratosphaeriaceae | EU019255 | GU214520 | — | 3 | ||||
| Hortaea werneckii | CBS 107.67 | mtSSU from CBS 708.76 | Capnodiales, Teratosphaeriaceae | EU019270 | Y18693 | GU561844 | 3 | |||
| Teratosphaeria associata | CBS 112224 | ex Teratosphaeria fibrillosa | Capnodiales, Teratosphaeriaceae | GU301874 | GU296200 | — | — | GU357744 | 3 & 5 | |
| Teratosphaeria destructans | CBS 111370 | Capnodiales, Teratosphaeriaceae | GU214702 | GU214702 | — | 3 | ||||
| Teratosphaeria juvenalis | CBS 110906 | Capnodiales, Teratosphaeriaceae | AY720715 | FJ493217 | — | 3 | ||||
| Capnodiales sp. 1 | CBS 101364 | ex Anisomeridium consobrinum | Capnodiales, incertae sedis | GU323215 | GU561840 | — | GU561853 | — | 3 & 5 | |
| Devriesia strelitziae | CBS 122379 | Capnodiales, incertae sedis | GU296146 | GU301810 | GU561845 | GU371738 | — | 3 & 5 | ||
| Mycosphaerella eurypotami | JK 5586J | Capnodiales, incertae sedis | GU301852 | GU479761 | — | GU371722 | GU561851 | 3 & 5 | ||
| Tripospermum myrti | CBS 437.68 | Capnodiales, incertae sedis | GU323216 | — | GU561846 | GU561854 | GU561852 | 3 & 5 | ||
| Columnosphaeria fagi 1 | CBS 171.93 | AFTOL 1582 | Dothideales | AY016359 | AY016342 | — | DQ677966 | — | 3 & 5 | |
| Columnosphaeria fagi 2 | CBS 584.75 | AFTOL 912 | Dothideales | DQ470956 | DQ471004 | FJ713608 | DQ470906 | DQ471148 | 3 & 5 | |
| Delphinella strobiligena | CBS 735.71 | AFTOL 1257 | Dothideales | DQ470977 | DQ471029 | — | DQ677951 | DQ471175 | 3 & 5 | |
| Dothidea insculpta | CBS 189.58 | AFTOL 921 | Dothideales | DQ247802 | DQ247810 | FJ190602 | AF107800 | DQ471154 | 3 & 5 | |
| Dothiora cannabinae | CBS 737.71 | AFTOL 1359 | Dothideales | DQ470984 | DQ479933 | FJ190636 | DQ470936 | DQ471182 | 3 & 5 | |
| Stylodothis puccinioides | CBS 193.58 | AFTOL 902 | Dothideales | AY004342 | AY016353 | — | — | FJ238427 | 3 & 5 | |
| Sydowia polyspora | CBS 116.29 | AFTOL 1300 | Dothideales | DQ678058 | DQ678005 | FJ190631 | DQ677953 | — | 3 & 5 | |
| Gloniopsis praelonga | CBS 112415 | Hysteriales | FJ161173 | FJ161134 | — | — | — | 3 & 5 | ||
| Rhytidhysterium rufulum | CBS 306.38 | Hysteriales | FJ469672 | AF164375 | — | — | FJ238444 | 3 & 5 | ||
| Elsinoë centrolobi | CBS 222.50 | AFTOL 1854 | Myriangiales | DQ678094 | DQ678041 | FJ190651 | — | — | 3 & 5 | |
| Elsinoë phaseoli | CBS 165.31 | AFTOL 1855 | Myriangiales | DQ678095 | DQ678042 | FJ190652 | — | — | 3 & 5 | |
| Myriangium duriaei | CBS 260.36 | AFTOL 1304 | Myriangiales | DQ678059 | AY016347 | AY571389 | DQ677954 | — | 3 & 5 | |
| Phaeosclera dematoides | CBS 157.81 | Myriangiales | GU301858 | GU296184 | — | — | GU357764 | 3 & 5 | ||
| Lophium mytilinum | CBS 269.34 | AFTOL 1609 | Mytilinidiales | DQ678081 | DQ678030 | GU456342 | DQ677979 | — | 3 & 5 | |
| Mytilinidion resinicola | CBS 304.34 | Mytilinidiales | FJ161185 | FJ161145 | — | FJ161101 | — | 3 & 5 | ||
| Hysteropatella clavispora | CBS 247.34 | AFTOL 1305 | Patellariales | AY541493 | DQ678006 | AY571388 | DQ677955 | — | 3 & 5 | |
| Hysteropatella elliptica | CBS 935.97 | AFTOL 1790 | Patellariales | DQ767657 | EF495114 | FJ190649 | DQ767647 | — | 3 & 5 | |
| Patellaria atrata | CBS 958.97 | Patellariales | GU301855 | GU296181 | — | DQ767647 | GU357749 | 3 & 5 | ||
| Arthopyrenia salicis | CBS 368.94 | mtSSU from GenBank | Pleosporales | AY538339 | AY538333 | AY538345 | — | FJ941893 | 3 & 5 | |
| Bimuria novae—zelandiae | CBS 107.79 | AFTOL 931 | Pleosporales | AY016356 | AY016338 | FJ190605 | DQ470917 | DQ471159 | 3 & 5 | |
| Dendryphiella arenaria | CBS 181.58 | AFTOL 995 | Pleosporales | DQ470971 | DQ471022 | FJ190617 | DQ470924 | DQ842036 | 3 & 5 | |
| Leptosphaeria maculans | DAOM 229267 | AFTOL 277 | Pleosporales | DQ470946 | DQ470993 | — | DQ470894 | DQ471136 | 3 & 5 | |
| Pleospora herbarum | CBS 541.72 | AFTOL 940 | Pleosporales | DQ247804 | DQ247812 | FJ190610 | DQ247794 | DQ471163 | 3 & 5 | |
| Preussia terricola | DAOM 230091 | AFTOL 282 | Pleosporales | AY544686 | AY544726 | AY544754 | DQ470895 | DQ471137 | 3 & 5 | |
| Sirodesmium olivaceum | CBS 395.59 | Pleosporales | GU250894 | GU250915 | GU250904 | GU250947 | GU250958 | 3 & 5 | ||
| Westerdykella cylindrica | CBS 454.72 | AFTOL 1037 | Pleosporales | AY004343 | AY016355 | AF346430 | DQ470925 | DQ471168 | 3 & 5 | |
| Pleosporales sp. 1 | CBS 101277 | ex Thelenella luridella | Pleosporales | — | GU456309 | — | GU456361 | — | 3 & 5 | |
| Pleosporales sp. 2 | AFTOL 101 | ex Anisomeridium polypori | Pleosporales | — | DQ782877 | — | DQ782864 | DQ782822 | 3 & 5 | |
| Astrothelium cinnamomeum | AFTOL 110 | ex Trypethelium sp. | Trypetheliaceae | AY584652 | AY584676 | AY584632 | AY584690 | DQ782824 | 3 & 5 | |
| Laurera megasperma | AFTOL 2094 | Trypetheliaceae | FJ267702 | GU561841 | GU561847 | GU561855 | — | 3 & 5 | ||
| Trypethelium nitidiusculum | AFTOL 2099 | Trypetheliaceae | FJ267701 | GU561842 | GU561848 | GU561856 | — | 3 & 5 | ||
| Helicomyces roseus | CBS 283.51 | AFTOL 1613 | Tubeufiaceae | DQ678083 | DQ678032 | — | DQ677981 | — | 3 & 5 | |
| Tubeufia cerea | CBS 254.75 | AFTOL 1316 | Tubeufiaceae | DQ470982 | DQ471034 | FJ190634 | DQ470934 | DQ471180 | 3 & 5 | |
| Tubeufia paludosa | CBS 245.49 | AFTOL 1580 | Tubeufiaceae | DQ767654 | DQ767649 | — | DQ767643 | — | 3 & 5 | |
| Cystocoleus ebeneus | L348 | RPB2 from L344; RPB1 from L343 | Dothideomycetes, incertae sedis | EU048580 | EU048573 | EU048586 | GU214293 | GU214204 | 3 & 5 | |
| Farlowiella carmichaelina | CBS 206.36 | AFTOL 1787 | Dothideomycetes, incertae sedis | AY541492 | AY541482 | — | DQ677989 | — | 3 & 5 | |
| Kirschsteiniothelia aethiops 1 | CBS 109.53 | AFTOL 925 | Dothideomycetes, incertae sedis | AY016361 | AY016344 | FJ190604 | DQ470914 | DQ471157 | 3 & 5 | |
| Kirschsteiniothelia aethiops 2 | DAOM 231155 | AFTOL 273 | Dothideomycetes, incertae sedis | DQ678046 | DQ677996 | FJ190590 | DQ677940 | — | 3 & 5 | |
| Phaeotrichum benjaminii | CBS 541.72 | AFTOL 1184 | Dothideomycetes, incertae sedis | AY004340 | AY016348 | — | DQ677946 | — | 3 & 5 | |
| Racodium rupestre | L424 | RPB1 from L341 | Dothideomycetes, incertae sedis | EU048582 | EU048577 | EU048589 | — | GU214205 | 3 & 5 | |
| Sarcinomyces crustaceus | CBS 156.89 | Dothideomycetes, incertae sedis | GU250893 | — | GU250905 | GU250948 | GU250959 | 3 & 5 | ||
|
Tyrannosorus pinicola |
CBS 124.88 |
AFTOL 1235
|
Dothideomycetes, incertae sedis |
DQ470974
|
DQ471025
|
FJ190620
|
DQ470928
|
DQ471171
|
3 & 5
|
|
|
Rock—inhabiting fungi |
LSU |
SSU |
mtSSU |
RPB2 |
RPB1 |
Analysis |
Locality |
|||
| Coniosporium apollinis | CBS 352.97 | ex-type strain | Dothideomycetes, incertae sedis | GU250895 | GU250916 | GU250906 | GU250949 | — | 3 & 5 | Greece |
| Coniosporium apollinis | CBS 100213 | Dothideomycetes, incertae sedis | GU250896 | GU250917 | GU250907 | GU250950 | GU250960 | 3 & 5 | Greece | |
| Coniosporium apollinis | CBS 100214 | Dothideomycetes, incertae sedis | GU250897 | GU250918 | GU250908 | GU250951 | — | 3 & 5 | Greece | |
| Coniosporium apollinis | CBS 100218 | Dothideomycetes, incertae sedis | GU250898 | GU250919 | GU250909 | GU250952 | GU250961 | 3 & 5 | Greece | |
| Coniosporium apollinis | CBS 109860 | Dothideomycetes, incertae sedis | GU250899 | GU250920 | GU250910 | GU250953 | GU250962 | 3 & 5 | Spain | |
| Coniosporium apollinis | CBS 109865 | Dothideomycetes, incertae sedis | GU250900 | GU250921 | GU250911 | GU250954 | GU250963 | 3 & 5 | Greece | |
| Coniosporium apollinis | CBS 109867 | Dothideomycetes, incertae sedis | GU250901 | — | GU250912 | GU250955 | GU250964 | 3 & 5 | Greece | |
| Coniosporium uncinatum | CBS 100212 | Dothideomycetes, incertae sedis | GU250902 | GU250922 | GU250913 | GU250956 | — | 3 & 5 | Italy | |
| Coniosporium uncinatum | CBS 100219 | ex-type strain | Dothideomycetes, incertae sedis | GU250903 | GU250923 | GU250914 | GU250957 | GU250965 | 3 & 5 | France, Paris |
| rock isolate TRN 5 | CBS 118762 | Ruibal et al. (2008) | Capnodiales, Teratosphaeriaceae | GU323956 | GU323988 | GU324017 | — | GU324051 | 3 & 5 | Central Spain |
| rock isolate TRN 11 | CBS 118281 | Ruibal et al. (2008) | Dothideales | GU323957 | — | GU324018 | — | GU324052 | 3 & 5 | Central Spain |
| rock isolate TRN 42 | CBS 117958 | Ruibal et al. (2008) | Capnodiales, Davidiellaceae | GU323958 | — | GU324019 | — | GU324053 | 3 & 5 | Central Spain |
| rock isolate TRN 43 | CBS 117950 | Ruibal et al. (2008) | Capnodiales, Davidiellaceae | GU323959 | GU323989 | GU324020 | 3 | Central Spain | ||
| rock isolate TRN 44 | CBS 118324 | Ruibal et al. (2008) | Capnodiales, Davidiellaceae | GU323960 | GU323990 | GU324021 | 3 | Central Spain | ||
| rock isolate TRN 49 | — | Ruibal et al. (2008) | Pleosporales | — | AY843233 | — | 3 | Central Spain | ||
| rock isolate TRN 62 | CBS 118305 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323961 | GU323991 | GU324022 | — | GU324054 | 3 & 5 | Mallorca |
| rock isolate TRN 66 | CBS 118306 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323962 | GU323992 | GU324023 | — | GU324055 | 3 & 5 | Mallorca |
| rock isolate TRN 77 | CBS 118287 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323963 | GU323993 | GU324024 | GU324066 | GU324057 | 3 & 5 | Mallorca |
| rock isolate TRN 79 | CBS 117930 | Ruibal et al. (2005) | Capnodiales, Teratosphaeriaceae | GU323964 | GU323994 | GU324025 | 3 | Mallorca | ||
| rock isolate TRN 80 | CBS 118286 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323965 | GU323995 | GU324026 | — | GU324056 | 3 & 5 | Mallorca |
| rock isolate TRN 87 | CBS 118290 | Ruibal et al. (2005) | Capnodiales, Capnodiaceae | GU323966 | GU323996 | GU324027 | — | GU324058 | 3 & 5 | Mallorca |
| rock isolate TRN 111 | CBS 118294 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323967 | GU323997 | GU324028 | — | GU324059 | 3 & 5 | Mallorca |
| rock isolate TRN 119 | CBS 118250 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323968 | — | GU324029 | 3 | Mallorca | ||
| rock isolate TRN 122 | CBS 117931 | Ruibal et al. (2005) | Capnodiales, Teratosphaeriaceae | GU323969 | GU323998 | GU324030 | 3 | Mallorca | ||
| rock isolate TRN 123 | CBS 117932 | Ruibal et al. (2005) | Capnodiales, Teratosphaeriaceae | GU323970 | GU323999 | GU324031 | GU324067 | GU324060 | 3 & 5 | Mallorca |
| rock isolate TRN 124 | CBS 118283 | Ruibal et al. (2005) | Capnodiales, Teratosphaeriaceae | GU323971 | GU324000 | GU324032 | — | GU324061 | 3 & 5 | Mallorca |
| rock isolate TRN 129 | CBS 117933 | Ruibal et al. (2005) | Capnodiales, Teratosphaeriaceae | GU323972 | GU324001 | GU324033 | 3 | Mallorca | ||
| rock isolate TRN 137 | CBS 118300 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323973 | GU324002 | GU324034 | — | GU324062 | 3 & 5 | Mallorca |
| rock isolate TRN 138 | CBS 118301 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323974 | GU324003 | GU324035 | GU324068 | GU324063 | 3 & 5 | Mallorca |
| rock isolate TRN 142 | CBS 118302 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323975 | GU324004 | GU324036 | GU324069 | — | 3 & 5 | Mallorca |
| rock isolate TRN 152 | CBS 118346 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323976 | GU324005 | GU324037 | 3 | Mallorca | ||
| rock isolate TRN 153 | CBS 118330 | Ruibal et al. (2005) | Capnodiales, incertae sedis | GU323977 | GU324006 | GU324038 | GU324070 | — | 3 & 5 | Mallorca |
| rock isolate TRN 211 | CBS 117937 | Ruibal et al. (2008) | Capnodiales, Teratosphaeriaceae | GU323978 | GU324007 | GU324039 | 3 | Central Spain | ||
| rock isolate TRN 213 | — | Ruibal et al. (2008) | related to Arthoniales | — | GU324008 | GU324040 | 3 | Central Spain | ||
| rock isolate TRN 221 | — | Ruibal et al. (2008) | Pleosporales | — | AY843241 | — | 3 | Central Spain | ||
| rock isolate TRN 235 | CBS 118605 | Ruibal et al. (2008) | Myriangiales | GU323979 | — | GU324041 | GU324071 | — | 3 & 5 | Central Spain |
| rock isolate TRN 245 | CBS 117940 | Ruibal et al. (2008) | Capnodiales, Teratosphaeriaceae | GU323980 | GU324009 | GU324042 | 3 | Central Spain | ||
| rock isolate TRN 267 | CBS 118769 | Ruibal et al. (2008) | Dothideomycetes, incertae sedis | — | GU324010 | GU324043 | GU324072 | — | 3 & 5 | Central Spain |
| rock isolate TRN 268 | CBS 119305 | Ruibal et al. (2008) | Dothideales | GU323981 | — | GU324044 | — | — | 3 & 5 | Central Spain |
| rock isolate TRN 279 | CBS 117943 | Ruibal et al. (2008) | Capnodiales, Teratosphaeriaceae | GU323983 | GU324012 | GU324046 | 3 | Central Spain | ||
| rock isolate TRN 434 | — | Ruibal et al. (2008) | Pleosporales | — | AY843260 | — | 3 | Central Spain | ||
| rock isolate TRN 437 | CBS 118327 | Ruibal et al. (2008) | Dothideomycetes, incertae sedis | GU323984 | GU324013 | GU324047 | 3 | Central Spain | ||
| rock isolate TRN 452 | — | Ruibal et al. (2008) | related to Arthoniales | GU323985 | GU324014 | GU324048 | 3 | Central Spain | ||
| rock isolate TRN 456 | — | Ruibal et al. (2008) | related to Arthoniales | GU323986 | GU324015 | GU324049 | — | GU324065 | 3 & 5 | Central Spain |
| rock isolate TRN 499 | — | Ruibal et al. (2008) | Pleosporales | — | AY843278 | — | 3 | Central Spain | ||
| rock isolate TRN 529 | — | Ruibal et al. (2008) | related to Arthoniales | GU323987 | GU324016 | GU324050 | — | — | 3 & 5 | Central Spain |
| rock isolate A6 | — | Gorbushina (unpublished) | Dothideomycetes, incertae sedis | GU250924 | GU250932 | — | GU250939 | — | 3 & 5 | Turkey |
| rock isolate A35 | CBS 123158 | Gorbushina (unpublished) | Coniosporium uncinatum | GU250925 | GU250933 | — | — | GU250943 | 3 & 5 | Crimea |
| rock isolate A73 | — | Gorbushina (unpublished) | Capnodiales, incertae sedis | GU250926 | GU250934 | — | GU250940 | GU250944 | 3 & 5 | Greece |
| rock isolate AN1 | — | Gorbushina (unpublished) | Capnodiales, Davidiellaceae | GU250927 | GU250935 | — | GU250941 | — | 3 & 5 | Israel, Negev |
| rock isolate AN13 | CBS 125207 | Gorbushina (unpublished) | Dothideomycetes, incertae sedis | GU250928 | GU250936 | — | GU250942 | GU250945 | 3 & 5 | Israel, Negev |
| rock isolate S2 | — | Gorbushina (unpublished) | Capnodiales, incertae sedis | GU250931 | — | — | — | GU250946 | 3 & 5 | Slovenia |
| rock isolate DVA4 | — | Staley et al. (1982) | Dothideomycetes, incertae sedis | GU250929 | GU250937 | — | 3 | U.S.A., Arizona | ||
| rock isolate DVA7 | — | Staley et al. (1982) | Dothideomycetes, incertae sedis | GU250930 | GU250938 | — | 3 | U.S.A., Arizona | ||
| rock isolate CCFEE 451 | — | Selbmann et al. (2005, 2008) | Capnodiales, incertae sedis | GU250360 | GU250314 | GU250403 | 3 | Antarctica | ||
| rock isolate CCFEE 453 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | GU250361 | GU250315 | GU250404 | 3 | Antarctica | ||
| rock isolate CCFEE 456 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | — | GU250316 | GU250405 | 3 | Antarctica | ||
| rock isolate CCFEE 502 | — | Selbmann et al. (2005, 2008) | Capnodiales, Teratosphaeriaceae | GU250363 | GU250318 | GU250406 | 3 | Antarctica | ||
| rock isolate CCFEE 514 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | — | GU250319 | GU250407 | 3 | Antarctica | ||
| rock isolate CCFEE 515 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | — | GU250320 | GU250408 | 3 | Antarctica | ||
| rock isolate CCFEE 524 | — | Selbmann et al. (2005, 2008) | Friedmanniomyces endolithicus | GU250364 | DQ066715 | GU250409 | — | — | 3 & 5 | Antarctica |
| rock isolate CCFEE 534 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | — | DQ066713 | GU250410 | 3 | Antarctica | ||
| rock isolate CCFEE 536 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | GU250365 | GU250321 | GU250411 | — | — | 3 & 5 | Antarctica |
| rock isolate CCFEE 670 | — | Selbmann et al. (2005, 2008) | Friedmanniomyces endolithicus | GU250366 | GU250322 | GU250412 | 3 | Antarctica | ||
| rock isolate CCFEE 690 | — | Selbmann et al. (2005, 2008) | Cryomyces antarcticus | — | GU250323 | GU250413 | 3 | Antarctica | ||
| rock isolate CCFEE 5018 | — | Selbmann et al. (2005, 2008) | Capnodiales, Davidiellaceae | — | GU250324 | GU250414 | 3 | Antarctica | ||
| rock isolate CCFEE 5176 | — | Selbmann et al. (2005, 2008) | related to Arthoniales | — | GU250325 | — | 3 | Antarctica | ||
| rock isolate CCFEE 5180 | — | Selbmann et al. (2005, 2008) | Friedmanniomyces endolithicus | GU250367 | GU250326 | GU250415 | 3 | Antarctica | ||
| rock isolate CCFEE 5184 | — | Selbmann et al. (2005, 2008) | Friedmanniomyces simplex | GU250368 | DQ066716 | GU250416 | 3 | Antarctica | ||
| rock isolate CCFEE 5187 | CBS 116302 | Selbmann et al. (2005, 2008) | Cryomyces minteri | GU250369 | DQ066714 | GU250417 | — | — | 3 & 5 | Antarctica |
| rock isolate CCFEE 5205 | — | Selbmann et al. (2005, 2008) | Capnodiales, incertae sedis | GU250370 | GU250327 | GU250418 | 3 | Antarctica | ||
| rock isolate CCFEE 5211 | — | Selbmann et al. (2005, 2008) | Capnodiales, Davidiellaceae | GU250371 | GU250328 | GU250419 | — | — | 3 & 5 | Antarctica |
| rock isolate CCFEE 5264 | — | Selbmann et al. (2008) | Recurvomyces mirabilis | GU250372 | GU250329 | — | 3 | Antarctica | ||
| rock isolate CCFEE 5284 | — | Selbmann (unpublished) | related to Arthoniales | GU250373 | GU250330 | — | 3 | Antarctica | ||
| rock isolate CCFEE 5299 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | GU250374 | — | — | 3 | Antarctic Peninsula | ||
| rock isolate CCFEE 5303 | — | Selbmann (unpublished) | related to Arthoniales | — | GU250331 | — | 3 | Antarctica | ||
| rock isolate CCFEE 5319 | — | Selbmann et al. (2008) | Elasticomyces elasticus | GU250375 | GU250332 | — | 3 | Antarctica on lichens | ||
| rock isolate CCFEE 5320 | CBS 122540 | Selbmann et al. (2008) | Elasticomyces elasticus | GU250376 | GU250333 | GU250420 | — | — | 3 & 5 | Antarctica on lichens |
| rock isolate CCFEE 5322 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250377 | GU250334 | — | 3 | Antarctica on lichens | ||
| rock isolate CCFEE 5388 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | GU250380 | GU250337 | GU250422 | 3 | Alps | ||
| rock isolate CCFEE 5389 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250381 | GU250338 | GU250423 | 3 | Alps | ||
| rock isolate CCFEE 5398 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | GU250382 | GU250339 | — | 3 | Alps | ||
| rock isolate CCFEE 5401 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250383 | GU250340 | GU250424 | 3 | Alps | ||
| rock isolate CCFEE 5410 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250384 | GU250341 | GU250425 | 3 | Andes | ||
| rock isolate CCFEE 5413 | — | Selbmann (unpublished) | Dothideomycetes, incertae sedis | GU250385 | GU250342 | GU250426 | 3 | Alps | ||
| rock isolate CCFEE 5414 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | GU250386 | GU250343 | — | 3 | Alps | ||
| rock isolate CCFEE 5416 | — | Selbmann (unpublished) | Dothideomycetes, incertae sedis | GU250387 | GU250344 | GU250427 | 3 | Alps | ||
| rock isolate CCFEE 5456 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | GU250388 | GU250345 | GU250428 | 3 | Alps | ||
| rock isolate CCFEE 5457 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250389 | GU250346 | GU250429 | 3 | Alps | ||
| rock isolate CCFEE 5458 | — | Selbmann (unpublished) | Capnodiales, Davidiellaceae | — | GU250347 | GU250430 | 3 | Alps | ||
| rock isolate CCFEE 5459 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250390 | GU250348 | GU250431 | 3 | Alps | ||
| rock isolate CCFEE 5460 | — | Selbmann (unpublished) | Dothideomycetes, incertae sedis | GU250391 | GU250349 | GU250432 | 3 | Alps | ||
| rock isolate CCFEE 5466 | — | Selbmann (unpublished) | Dothideomycetes, incertae sedis | GU250392 | GU250350 | GU250433 | 3 | Alps | ||
| rock isolate CCFEE 5467 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250393 | GU250351 | — | 3 | Alps | ||
| rock isolate CCFEE 5476 | — | Selbmann (unpublished) | close to Cryomyces | GU250394 | GU250352 | GU250434 | 3 | Alps | ||
| rock isolate CCFEE 5489 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250395 | — | GU250435 | 3 | Antarctica | ||
| rock isolate CCFEE 5490 | — | Selbmann (unpublished) | Elasticomyces elasticus | — | GU250353 | — | 3 | Antarctica | ||
| rock isolate CCFEE 5499 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250398 | GU250355 | GU250436 | 3 | Alps | ||
| rock isolate CCFEE 5501 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250399 | GU250356 | GU250437 | 3 | Aconcagua, Andes | ||
| rock isolate CCFEE 5502 | — | Selbmann (unpublished) | Capnodiales, incertae sedis | GU250400 | GU250357 | GU250438 | 3 | Aconcagua, Andes | ||
| rock isolate CCFEE 5508 | — | Selbmann (unpublished) | Capnodiales, Teratosphaeriaceae | GU250401 | GU250358 | — | 3 | Aconcagua, Andes | ||
| rock isolate D007 09 | — | Selbmann (unpublished) | related to Arthoniales | GU250402 | GU250359 | — | 3 | Antarctica |
DNA isolation and sequencing
Different laboratories contributed data using various protocols, but most DNA sequence information was produced as follows: genomic DNA was isolated from cultures grown on MEA. Fungal biomass was transferred to a tube with 500 μL of TES buffer and ground with a micro-pestle for 1–2 min, with or without silica-mix (2/3 silica-gel, 1/3 Celite® 545). A volume of 140 μL of 5 M NaCl was then added, followed by 65 μL of 10 % (w/v) CTAB (cetyltrimethylammoniumbromid). After an incubation of 30 min at 65 °C, 700 μL of (24:1) chloroform/isoamylalcohol was added, the tubes were mixed carefully by hand, stored on icy water for 30 min, and centrifuged for 10 min at 4 °C (10 000 x g). The supernatant was recovered and the genomic DNA precipitated using isopropanol. After washing the pellets with 70 % ethanol, they were dried in a vacuum centrifuge and re-suspended in 60 μL of TE buffer (protocol modified from Möller et al. 1992).
Six regions covering five genes were amplified: nucLSU, nucSSU, mtSSU, RPB1 region A–D, RPB2 region 5–7, and RPB2 region 7–11 (see table 2 for primers used). Genomic DNA (1 μL of a 1/10 or 1/100 dilution) was added to a PCR mix comprising 2.5 μL of PCR buffer (buffer IV with 15 mM MgCl2, Abgene, Epsom, U.K.), 2.5 μL of dNTPs (2 mM), 2.5 μL of BSA (10 mg/mL), 2.0 μL of primers (10 μM), 0.15 μL Taq polymerase (5 U/μL, Denville, Metuchen NJ, U.S.A.), and water for a total volume of 25 μL. Amplification cycles for nucLSU, nucSSU and RPB1 (same conditions applied for RPB2) are described in Gueidan et al. (2007), and in Zoller et al. (1999) for mtSSU. The PCR products were purified using Microcon PCR cleaning kits (Millipore, Billerica MA, U.S.A.). Sequencing was carried out using Big Dye Terminator Cycle sequencing Kits (ABI PRISM version 3.1, Perkin-Elmer, Applied Biosystems) on ABI 3730xl DNA Analyzers (Applied Biosystems, Foster City CA, U.S.A.) from the Duke Center for Evolutionary Genomics (Durham NC, U.S.A.) and the Hubrecht Institute (Utrecht, Netherlands).
Table 2.
List of primers for the five genes used in this study (RPB2 was amplified in two regions).
| Gene regions | PCR primers | Additional primers used for sequencing |
|---|---|---|
| nucLSU | LR0Ra, LR7b | LR3, LR3R, LR5, LR5R, LR6, LR6Rb |
| nucSSU | nssu131c, NS24d | nssu1088, nssu1088R, nssu897R, nssu634c, SR11Re, NS23, NS22d, SR7R, SR7, SR10Rf |
| mtSSU | mtSSU1, mtSSU3Rg | mtSSU2, mtSSU2Rg |
| RPB1 region A—D | RPB1-AFh, RPB1-6R1asci | — |
| RPB2 region 5-7 | RPB2-5F, RPB2-7cRj | — |
| RPB2 region 7-11 | RPB2-7cF, RPB2-11aRj | — |
Rehner & Samuels (1994), bVilgalys & Hester (1990), cKauff & Lutzoni (2002), dGargas & Taylor (1992), eSpatafora et al. (1995), fVilgalys (unpubl.; www.biology.duke.edu/fungi/mycolab/primers.htm), gZoller et al. (1999), hHall (unpubl.; http://faculty.washington.edu/benhall/), iHofstetter et al. (2007), jLiu et al. (1999).
Alignments and phylogenetic analyses
Sequences were assembled and edited using Sequencher (Gene Codes Corporation, Ann Arbor MI, U.S.A.). Manual alignments were performed using MacClade v. 4.08 (Maddison & Maddison 2003). Ambiguous regions (sensu Lutzoni et al. 2000) and introns were delimited manually and excluded from the alignments. Congruence was tested using a 70 % reciprocal bootstrap criterion (Mason-Gamer & Kellogg 1996, Reeb et al. 2004). For the three-gene dataset, the test was performed using Compat (Kauff & Lutzoni 2002) on all possible gene pairs (mtSSU vs. nucSSU, mtSSU vs. nucLSU, and nucLSU vs. nucSSU) and based on bootstrap consensus trees. Bootstrap trees were obtained using Neighbor-Joining bootstrap analyses with Maximum Likelihood distances in PAUP v. 4.0b10 (Swofford 2003). Models of molecular evolution were estimated using the Akaike Information Criterion implemented in Modeltest v. 3.7 (Posada & Crandall 1998). For the five-gene dataset, congruence was also tested using a 70 % reciprocal bootstrap criterion, but the comparison was done manually based on trees obtained with 500 bootstrap replicates using RAxML VI-HPC (Stamatakis et al. 2005, 2008) on the Cipres Web Portal (www.phylo.org/sub_sections/portal/). Taxa or sequences responsible for incongruence were removed from the dataset, and the markers were combined. Final phylogenetic analyses of the three-gene and five-gene datasets were performed using RAxML on the Cipres Web Portal. The ML search followed a GTRMIX model of molecular evolution applied to the following nine partitions: RPB1 first, second and third codon positions, RPB2 first, second and third codon positions, nucLSU, nucSSU and mtSSU. Support values were obtained with bootstrap analyses of 1 000 pseudoreplicates using RAxML.
RESULTS
DNA sequence alignments
Not all markers were recovered or available for all taxa. For the three-gene dataset, 20 nucLSU, 11 nucSSU and 54 mtSSU sequences were missing. Among the 182 taxa, 119 had sequences for three genes, 61 for two genes, and 12 for one gene (Table 1 in Supplementary Information). After exclusion of ambiguous regions and introns, the combined dataset included 3 274 characters (1 106 for nucLSU, 1 616 for nucSSU and 552 for mtSSU). Among these, 2 063 were constant while 931 were parsimony-informative. For the five-gene dataset, missing data comprised 5 nucLSU, 8 nucSSU, 30 mtSSU, 48 RPB1 and 30 RPB2 sequences. Among the 113 taxa, 32 had sequences for five genes, 46 for four genes, 30 for 3 genes, and 5 for 2 genes (Table 1 in Supplementary Information). After exclusion of ambiguous regions and introns, the combined dataset included 6 045 characters (1 133 for nucLSU, 1 607 for nucSSU, 593 for mtSSU, 1 011 for RPB1 and 1 701 for RPB2). Among these, 2 912 were constant while 2 693 were parsimony-informative.
Phylogenetic inference
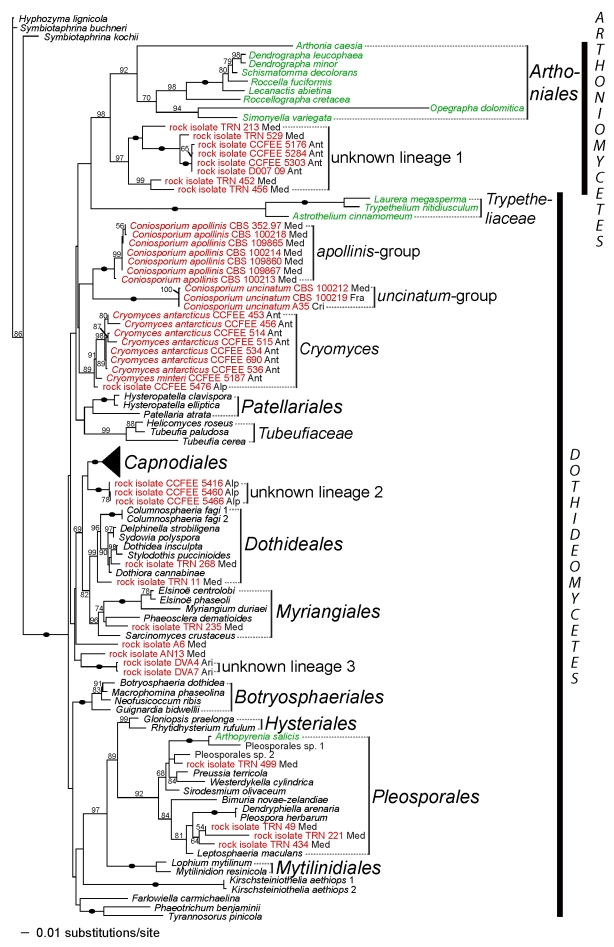
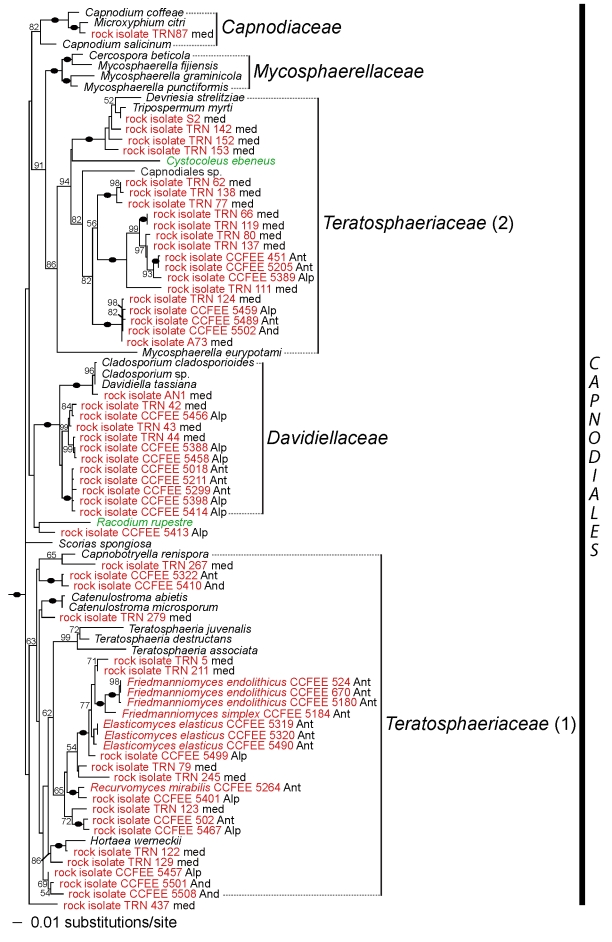
For the three-gene analysis (Figs 2, 3), results show that, within the two classes Dothideomycetes and Arthoniomycetes, rock-inhabiting fungi belong to 13 groups, either well-known orders or families, or lineages that have not previously been characterised. Among the rock-inhabiting fungi clustering with well-known groups of Dothideomycetes, two strains are found in the order Dothideales, four in the order Pleosporales, one in Myriangiales, 12 forming a monophyletic group sister to the remaining members of Davidiellaceae, and one in the family Capnodiaceae. The family Teratosphaeriaceae is not monophyletic in this analysis (also see Crous et al. 2009; this volume). In a first group including the generic type Teratosphaeria fibrillosa (Teratosphaeriaceae 1, Fig. 3), many rock-inhabiting strains are present, including taxa from the three genera Friedmanniomyces, Elasticomyces and Recurvomyces. The second group (Teratosphaeriaceae 2, Fig. 3), including the three leaf-colonising species Devriesia strelitziae, Mycosphaerella eurypotami and Tripospermum myrti, an unknown species of Capnodiales, the lichen species Cystocoleus ebeneus as well as 20 undescribed rock inhabiting strains, is supported as sister to the family Mycosphaerellaceae (91 % bootstrap). The two rock-inhabiting species Coniosporium uncinatum and C. apollinis are well supported (100 % bootstrap), but their sister relationship is not. Neither these two species of Coniosporium nor the Antarctic genus Cryomyces can be assigned to any known family or order sampled here. Amongst the unknown lineages, one does not seem to be part of Dothideomycetes (lineage 1, Fig. 2), and appears as sister to Arthoniomycetes (98 % bootstrap). Due to the lack of support for many deep internodes, it is not possible to determine if lineages 2 and 3 can be accommodated by the expansion of known groups of Dothideomycetes, or if the recognition of new taxonomical entities are needed. Finally, the rock isolates A6, AN13, TRN 437 and CCFEE 5413 do not significantly cluster with any other taxa.
Fig. 2.
Phylogenetic placement of 102 rock-inhabiting strains within Dothideomyceta (Dothideomycetes and Arthoniomycetes). The tree is based on a Maximum Likelihood analysis of the combined nucLSU, nucSSU and mtSSU (three-gene analysis). A black oval on a branch indicates a bootstrap support value of 100 %. Other bootstrap values ≥ 50 % are shown below or above branches. RIF are highlighted in red and lichens in green. Geographical origins are also labeled for RIF (Alp = Alps, And = Andes, Ant = Antarctica, Ari = Arizona desert, Cri = Crimea, Fra = France, Med = Mediterranean region, including Greece, Israel, Italy, Slovenia, Spain and Turkey). Phylogenetic relationships within Capnodiales are detailed in Fig. 3.
Fig. 3.
Phylogenetic placement of RIF within the order Capnodiales. The tree is based on a Maximum Likelihood analysis of the combined nucLSU, nucSSU and mtSSU (three-gene analysis). A black oval on a branch indicates a bootstrap support value of 100 %. Other bootstrap values ≥ 50 % are shown below or above branches. RIF are highlighted in red and lichens in green. Geographical origins are also labeled for RIF (Alp = Alps, And = Andes, Ant = Antarctica, Ari = Arizona desert, Cri = Crimea, Fra = France, Med = Mediterranean region, including Greece, Israel, Italy, Slovenia, Spain and Turkey).
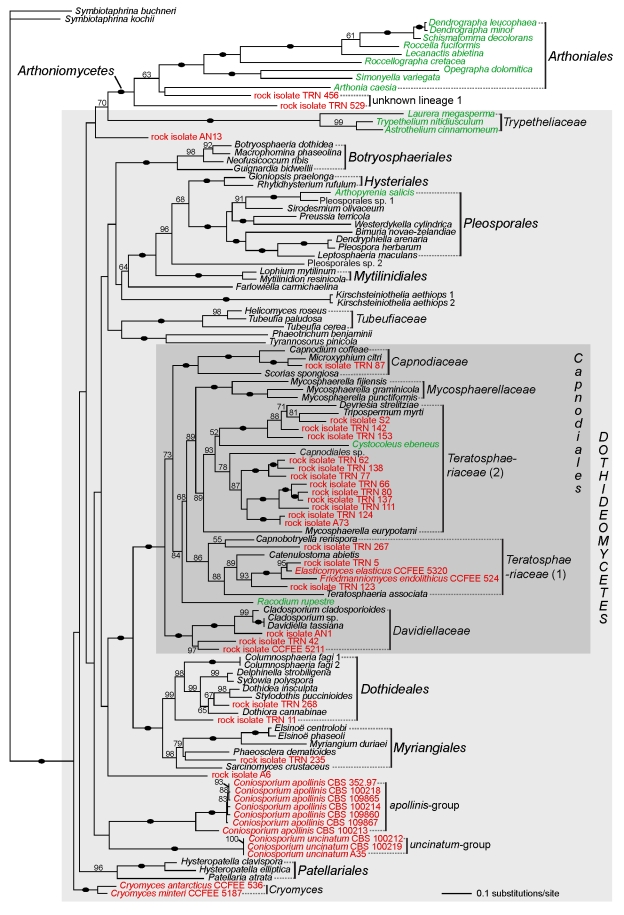
With the five-gene analysis (Fig. 4), the inferred deep branching pattern within Dothideomyceta is still poorly supported, but additional well-supported nodes are recovered (e.g., Capnodiaceae as sister to the lineage including Mycosphaerellaceae and Teratosphaeriaceae, and the monophyly of Teratosphaeriaceae 1). As in the three-gene analysis, the sister relationship between lineage 1 and Arthoniomycetes obtains high support (100 % bootstrap), even though the two rock-inhabiting strains included do not seem to form a monophyletic group. The placement of the lichen family Trypetheliaceae as sister to Arthoniomycetes (70 % bootstrap) might be an artifact, as this relationship was not recovered in any other studies (Del Prado et al. 2006, Spatafora et al. 2006, Nelsen et al. 2009). Within Dothideomycetes, the orders Dothideales and Myriangiales form a sister group (100 % bootstrap), and are sister to the well-supported Capnodiales (100 % bootstrap), which includes most of the rock-inhabiting strains. Within Capnodiales, the second group of Teratosphaeriaceae (Teratosphaeriaceae 2; Fig. 4) is still supported as sister to Mycosphaerellaceae (89 % bootstrap). Other lineages comprising exclusively RIF (Cryomyces, Coniosporium uncinatum, and C. apollinis) do not significantly cluster with any known group of Dothideomycetes.
Fig. 4.
Phylogenetic relationships of rock-inhabiting lineages with known groups of Dothideomyceta based on a Maximum Likelihood analysis of the combined nucLSU, nucSSU, mtSSU, RPB1 and RPB2 (five-gene analysis). A black dot on a branch indicates a bootstrap support value of 100 %. Other bootstrap values ≥ 50 % are shown below or above the branches. RIF are highlighted in red and lichens in green.
DISCUSSION
Species diversity in Dothideomycetes
The Dothideomycetes are very diverse in term of species, some of which are well known for their pathogenicity on crops (e.g., Mycosphaerella fijiensis, the agent of the leaf spot disease of banana, or Leptosphaeria maculans, the agent of the blackleg disease of cabbage). Whilst many species are associated with plants (either as pathogens or as epiphytes), saprobic, coprophilous, lichen-forming and rock-inhabiting fungi are also present in this class. The importance of RIF in term of species richness is still under-investigated. A thorough sampling of dothideomycetous RIF from few localities in Mallorca and Central Spain formed the basis of the analyses described here (Ruibal 2004, Ruibal et al. 2005, 2008). RIF from Antarctica, the Alps and the Andes (Selbmann et al. 2005, 2008), as well as the Arizona and Negev deserts (Staley et al. 1982, A.A. Gorbushina, unpubl. data) extended the geographical range of the sampled taxa. Finally, isolates from monuments in the Mediterranean area supplemented the sampling (Gorbushina et al. 1996, Sterflinger et al. 1997, Volkmann & Gorbushina 2006). In comparison to known RIF habitats (Gorbushina 2007), our sampling was very restricted and does not permit a realistic overview of fungal diversity on rock surfaces. Nevertheless, an impressive number of rock-inhabiting species is already evident. Our data show that rock-inhabiting fungi are not only present in well-known orders, such as Capnodiales or Pleosporales, but also in novel lineages (e.g., lineage 1, Fig. 2). Moreover, very few species with overlapping distribution were recovered from neighbouring geographical localities in Mallorca and Central Spain (Ruibal et al. 2005, 2008). Therefore, we can hypothesise that species richness within Dothideomycetes remains woefully underestimated, and that many more species will need to be described within this class in the future, especially for fungi colonising rocky substrates.
Classification of rock fungi related to Dothideomycetes
Although very diverse within Dothideomycetes, RIF have not been included in recent phylogenetic studies of this class (Lumbsch et al. 2001, Schoch et al. 2006). Only very few of these rock-inhabiting species have been taxonomically described (Sterflinger et al. 1997, Bills et al. 2005, Sert et al. 2007b), and the molecular marker available for most of these species (ITS) does not allow their inclusion in large-scale phylogenetic analyses. The few attempts to produce phylogenies involving RIF have shown that they belong to two diverse classes of Ascomycota, namely Eurotiomycetes (particularly the order Chaetothyriales) and Dothideomycetes (preponderantly the orders Capnodiales, Dothideales and Pleosporales) (Sterflinger et al. 1999, Ruibal 2004, Ruibal et al. 2005, 2008).
Our results confirm the placement of RIF in the same orders of Dothideomycetes, although some lineages are shown to belong to additional groups. Based on our results, many RIF should be classified within Dothideales, Pleosporales and Capnodiales, the latter order holding the largest number in rock-colonising species. The genera Elasticomyces and Recurvomyces, as well as the Antarctic genus Friedmanniomyces, were previously attributed to Capnodiales based on nucSSU data (Selbmann et al. 2008). Our multigene analyses confirm this placement, and show that these three genera belong to Teratosphaeriaceae s. str., the family currently showing the highest diversity in RIF (Fig. 3). We also showed that one RIF (TRN 235) previously thought to be related to the Dothideales (Ruibal et al. 2008) actually belongs to Myriangiales, along with Sarcinomyces crustaceus, a species similarly melanised and meristematic, but isolated from plant material (Sigler et al. 1981).
Several well-supported groups of RIF could not be attributed to any known families and orders according to our data. As a consequence, Cryomyces should still be considered as Dothideomycetes incertae sedis, as no close relationship was recovered for this enigmatic Antarctic genus (Selbmann et al. 2005). The positions of RIF-rich genera Coniosporium and Sarcinomyces are also problematic. Previous studies placed them either in Dothideales or Chaetothyriales based on ITS or nucSSU data (de Leo et al. 1999, Sterflinger et al. 1999, Sert et al. 2007a). Yet, the limited taxon and gene sampling on which these analyses were based was probably insufficient to demonstrate clear phylogenetic relationships. Our results show that Coniosporium apollinis (including the type strain CBS 352.97), C. uncinatum (including the type strain CBS 100219) and Sarcinomyces crustaceus belong to Dothideomyceta (Fig. 4). However, a previous multigene analysis showed that two other species, Coniosporium perforans and Sarcinomyces petricola, belong to Chaetothyriales (Gueidan et al. 2008). These anamorphic genera are therefore not monophyletic, and additional research is required to clarify their status.
Among lineages lacking known reference taxa, two groups seem to belong to Dothideomycetes (unknown group 2, a lineage comprising RIF from the Alps, and unknown group 3, a lineage including strains isolated in Arizona; Fig. 2). Another unknown group (lineage 1) clusters outside Dothideomycetes, sister to the Arthoniales (Figs 2, 4). A previous study had noted the problematic placement of this latter group (Ruibal et al. 2008). Many lineages including RIF still need to be named. In the past, several melanised meristematic species and genera have been described such as Lichenothelia (Hawksworth 1981; see also Henssen 1987), which could potentially correspond to some of these RIF lineages. However, little is known about these formerly named taxa, and no molecular data or cultures are available for many of them. Naming RIF will therefore require an extensive study of both rock-inhabiting species and formerly described melanised meristematic species, whether they grow on rock or not.
Rock surfaces: “terroirs” for ancient lineages or reservoirs for plant-associated fungi?
Despite the prevailing extreme conditions, rock surfaces host a large variety of specialised fungi. Fungal colonisation of subaerial rocks can be explained by two non-exclusive hypotheses. Firstly, atmosphere-exposed rock substrates could constitute “terroirs” for ancient fungal lineages. Rock surfaces were among the first terrestrial substrates available for living organisms on earth (Gorbushina & Broughton 2009). It is therefore likely that, early on, some species became adapted to colonise rock surfaces. RIF are persistent to different types of physical stress, but are poor competitors and surrender to more combative organisms (Gorbushina et al. 2008). Increasing competition with other rock-inhabiting organisms living under more permissive conditions may have restricted some of these ancient, morphologically reduced, slow-growing, fungal relicts to extreme habitats. The presence of lineages comprising exclusively RIF that diverged early in the evolution of Dothideomyceta (e.g., Cryomyces and lineage 1, Fig. 2) supports this hypothesis of rock surfaces as substrates for ancient fungal lineages.
Secondly, rock surfaces could form reservoirs for plant-associated or saprobic fungi. Through spore or propagule dispersal, some species of various unrelated groups of plant pathogens, epiphytes or saprobes can reach rock substrates. Their ability to survive in these environments will depend on some key features, namely oligotrophy, melanisation and pleiomorphism (or diversity of growth forms, amongst which meristematic growth). Under extreme conditions prevailing on rock surfaces, fungi possessing these key features can survive due to their slow, meristematic, clumpy growth and thick-walled, heavily melanised cells. These key features seem to have evolved several times in Dothideomycetes, allowing different lineages to colonise rock substrates. In Dothideales, phyllosphere fungi such as Aureobasidium pullulans and relatives, which have a filamentous or yeast-like growth under moist conditions, but convert to a meristematic form when colonising inert substrates, have also been isolated from rock surfaces (Ruibal et al. 2008). The family Teratosphaeriaceae s. l. is another example of a group in which some leaf-colonising species can also grow meristematically and form dark, thick-walled cells. According to our results, this family (as traditionally delimited; i.e., including Teratosphaeriaceae 1 and 2) is also extremely diverse in RIF (Fig. 3). Rocks supporting growth of subaerial biofilms (Gorbushina & Broughton, 2009) may be viewed as a reservoir for all types of melanised meristematic fungi, from where other habitats can be re-colonised. Survival of new comers is probably additionally facilitated by the existing microbial community on rocks (Gorbushina & Broughton 2009) in a fashion known for immigrant bacteria on leaf surfaces (Monier & Lindow 2005).
Alternatively, rock-colonising lichens may supply buffered environments and refugia for RIF or organisms otherwise occupying other niches (Selbmann et al. 2010). Recent studies have shown that lichens harbour an amazing diversity of ascomycetous endophyte-like (endolichenic) fungi (Arnold et al. 2009), and phylogenetic relatedness was found between some endolichenic fungi isolated from saxicolous lichens and RIF (Harutyunyan et al. 2008). If in most cases, species from rock surfaces can still go back to their primary habitats, in some cases, these fungi keep specialising and get trapped in these extreme habitats. This may be the case for groups with no close relationships with plant-associated fungi, such as the genus Friedmanniomyces (Fig. 3).
Geographical distribution of rock-inhabiting fungi
The large majority of rock-inhabiting strains isolated thus far originated from rocks in the Mediterranean region or Antarctica (Sterflinger et al. 1999, Ruibal 2004, Ruibal et al. 2005, 2008 Selbmann et al. 2005, 2008). In Antarctica, RIF tend to grow within rocks, together with the cryptoendolithic lichen communities, finding shelter from extreme conditions prevailing on rock surfaces. In the Mediterranean area, RIF tend to grow on the rock surface or in cracks, causing damages to the substrate (e.g., biopitting of marble). Despite differences in temperature, they share similar morphological and physiological adaptations, such as melanisation, meristematic growth and oligotrophism.
Similarly to previous studies (Selbmann et al. 2005, Ruibal et al. 2008), our results show that Antarctic RIF often share an evolutionary history with RIF from semi-arid areas. In our study, RIF sampled in geographically disjoint localities (Antarctica versus Mediterranean region) cluster together in Davidiellaceae, the two groups of Teratosphaeriaceae, and unknown lineage 1 (Figs 2, 3). In some cases, Antarctic and Mediterranean strains are even phylogenetically very closely related, showing a recent common evolutionary history (e.g., in Teratosphaeriaceae 2, the Mediterranean rock isolates TRN 124 and A73 with the Antarctic strain CCFEE 5489). Likewise, some strains of Recurvomyces mirabilis and Elasticomyces elasticus have been recorded in the Antarctic as well as in high peaks of the Alps and Andes (Selbmann et al. 2008). Therefore, it seems that an efficient mechanism of dispersal, most probably wind-mediated (Gorbushina et al. 2007, Gorbushina & Broughton 2009), have led to a colonisation spanning different continents.
Rock-dwelling habit and evolution of lichenisation
Most of the diversity in lichen-forming fungi is found in Lecanoromycetes, a large and diverse class of ascomycetes including approximately 14 000 species (Miadlikowska et al. 2006, Kirk et al. 2008). Yet, the classes Lichinomycetes (with the single order Lichinales), Eurotiomycetes (with the orders Pyrenulales and Verrucariales), Arthoniomycetes (with the single order Arthoniales), and Dothideomycetes also include lichens. Although Lichinales, Pyrenulales, Verrucariales and Arthoniales are monophyletic lineages containing mostly lichenised species, lichens in Dothideomycetes seem to encompass a broader phylogenetic spectrum: the Trypetheliaceae, a family of mostly tropical bark-colonising lichens, forms a monophyletic group within Dothideomycetes (Del Prado et al. 2006, Nelsen et al. 2009, Schoch et al. 2009a). Arthopyrenia salicis, a corticolous, temperate lichen species nests within the order Pleosporales (Del Prado et al. 2006, Nelsen et al. 2009). Two melanised micro-filamentous lichens, Cystocoleus ebeneus and Racodium rupestre, were assigned to the order Capnodiales (Muggia et al. 2008, Nelsen et al. 2009). Finally, the two lichen families Strigulaceae (mostly leaf-colonising tropical species) and Monoblastiaceae (temperate and tropical species) are now shown to belong to Dothideomycetes (Nelsen et al. 2009; this volume).
Whether these lichen lineages, that are unrelated to Lecanoromycetes, originated from independent gains of lichenisation is not clear (Lutzoni et al. 2001, James et al. 2006, Gueidan et al. 2008, Arnold et al. 2009, Schoch et al. 2009a, b). Within Eurotiomycetes, phylogenetic data suggest that the lineage including Pyrenulales and Verrucariales possibly results from an independent gain of lichenisation (Gueidan et al. 2008, Schoch et al. 2009a). Phylogenetic data suggest that lichens in Verrucariales may have evolved from rock-inhabiting fungi (Gueidan et al. 2008), a result in agreement with experimental data demonstrating that some RIF and one melanised lichen-colonising fungus could form associations with lichen-associated algae (Gorbushina et al. 2005, Brunauer et al. 2007). This rock-inhabiting ancestor may have evolved associations with epilithic microalgae in order to get a more constant supply in nutrients. If the evolution of fungal-algal associations occurred in Eurotiomycetes, it most likely also occurred in different fungal groups. It is therefore interesting to see if in Dothideomycetes, where rock fungi are so diverse, similar transitions in lifestyles can be suggested.
Although many lichenised species in Dothideomycetes are either corticolous or only secondarily or occasionally saxicolous, Cystocoleus ebeneus and Racodium rupestre are true rock inhabitants. Amongst lichens in Dothideomycetes, these two species are the most likely to have evolved from a rock-inhabiting ancestor. They share substrate preference and some morphological features, such as their melanised hyphae, with RIF. Strikingly, in our result, Cystocoleus ebeneus is nested within a lineage comprising almost exclusively RIF (Teratosphaeriaceae 2, Fig. 3). Racodium rupestre is also related to a RIF, but this relationship is not supported (Fig. 3). This result agrees with a rock-inhabiting ancestor for these two lichenised species, but further data will however be necessary to test this hypothesis. Also of interest is the close phylogenetic relationship between the lichen order Arthoniales and the lineage 1 of RIF (Figs 2, 4). Although mostly corticolous or foliicolous, Arthoniales also comprises saxicolous species (Ertz et al. 2009). Further data is needed to explore the relationships between saxicolous species of Arthoniales and RIF. In conclusion, these preliminary results suggest that there might be a link between rock-dwelling habit and lichenisation. However, additional taxon and gene sampling are needed to confirm the phylogenetic placements of some of the lichenised taxa and to clarify their relationships to RIF. Only then the hypothesis of RIF as ancestors of lichenised lineages can be adequately tested.
SUPPLEMENTARY INFORMATION
Acknowledgments
Work performed by C.R. at Duke University was supported by a NSF AToL grant (AFTOL, DEB-0228725) to F.L. Work performed by C.L.S. after 2008 was supported in part by the Intramural Research Program of the National Institutes of Health (National Library of Medicine), and until 2008 by a grant from NSF (DEB-0717476). Work performed by A.A.G. was funded by grants from the National Swiss Foundation (31003A-122513) and the Deutsche Forschungsgemeinschaft (DFG Go 897/3). Work performed by L.S. at the CBS was funded by a Synthesys grant. The authors would like to acknowledge the Italian National Program for Antarctic Research (PNRA) for supporting the collection of samples, the Italian National Antarctic Museum “Felice Ippolito” for supporting the Culture Collection of Fungi From Extreme Environments (CCFEE) and the Alpine guides A. Serafini and M. Heltai for collecting rock samples in the Alps and Aconcagua, respectively. Thanks to William Broughton for his editorial help and to the technical staff of the CBS for their assistance with the cultures.
References
- Arnold AE, Miadlikowska J, Higgins KL, Sarvate SD, Gugger P, Way A, Hofstetter V, Kauff F, Lutzoni F (2009). A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Systematic Biology 58: 283–297. [DOI] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin H-D, Crous PW (2007). Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills GF, Collado J, Ruibal C, Peláez F, Platas G (2005). Hormonema carpetanum, sp. nov., a new lineage of dothideaceous black yeasts from Spain. Studies in Mycology 50: 149–157. [Google Scholar]
- Brunauer G, Blaha J, Hager A, Türk R, Stocker-Wörgötter E, Grube M (2007). An isolated lichenicolous fungus forms lichenoid structures when co-cultured with various coccoid algae. Symbiosis 44: 127–136. [Google Scholar]
- Crous PW, Braun U, Groenewald JZ (2007a). Mycosphaerella is polyphyletic. Studies in Mycology 58: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Schubert K, Groenewald JZ (2007b). Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, Hoog GS de, Groenewald JZ (2009). Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schubert K, Braun U, Hoog GS de, Hocking AD, Shin H-D, Groenewald JZ (2007c). Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Studies in Mycology 58: 185–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadachova E, Casadevall A (2008). Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Current Opinion in Microbiology 11: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prado R, Schmitt I, Kautz S, Palice Z, Lücking R, Lumbsch HT (2006). Molecular data place Trypetheliaceae in Dothideomycetes. Mycological Research 110: 511–520. [DOI] [PubMed] [Google Scholar]
- Diakumaku E, Gorbushina AA, Krumbein WE, Panina L, Soukharjeski S (1995). Black fungi in marble and limestones – an aesthetical, chemical and physical problem for the conservation of monuments. The Science of the Total Environment 167: 295–304. [Google Scholar]
- Ertz D, Miadlikowska J, Lutzoni F, Dessein S, Raspé O, Vigneron N, Hofstetter V, Diederich P (2009). Towards a new classification of the Arthoniales (Ascomycota) based on a three-gene phylogeny focusing on the genus Opegrapha. Mycological Research 113: 141–152. [DOI] [PubMed] [Google Scholar]
- Friedmann EI (1982). Endolithic microorganisms in the Antarctic cold desert. Science 215: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Gargas A, Taylor JW (1992). Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia 84: 589–592. [Google Scholar]
- Gorbushina AA (2003). Methodologies and techniques for detecting extraterrestrial (microbial) life. Microcolonial fungi: survival potential of terrestrial vegetative structures. Astrobiology 3: 543–554. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA (2007). Life on the rocks. Environmental Microbiology 9: 1613–1631. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Beck A, Schulte A (2005). Microcolonial rock inhabiting fungi and lichen photobionts: evidence for mutualistic interactions. Mycological Research 109: 1288–1296. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Broughton WJ (2009). Microbiology of the atmosphere-rock interface: how biological interactions and physical stresses modulate a sophisticated microbial ecosystem. Annual Review of Microbiology 63: 431–450. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Kort R, Schulte A, Lazarus D, Schnetger B, Brumsack HJ, Broughton WJ, Favet J (2007). Life in Darwin's dust: inter-continental transport and survival of microbes in the nineteenth century. Environmental Microbiology 9: 2911–2922. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Kotlova ER, Sherstneva OA (2008). Cellular responses of microcolonial rock fungi to long-term desiccation and subsequent rehydration. Studies in Mycology 61: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbushina AA, Krumbein WE, Hamman CH, Panina L, Soukharjevski S, Wollensien U (1993). Role of black fungi in color-change and biodeterioration of antique marbles. Geomicrobiology Journal 11: 205–221. [Google Scholar]
- Gorbushina AA, Krumbein WE, Volkmann M (2002). Rock surfaces as life indicators: new ways to demonstrate life and traces of former life. Astrobiology 2: 203–213. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Panina LK, Vlasov DY, Krumbein WE (1996). Fungi deteriorating Chersonessus marbles. Mycologia i Fitopatologija 30: 23–28. [Google Scholar]
- Gorbushina AA, Whitehead K, Dornieden T, Niesse A, Schulte A, Hedges JI (2003). Black fungal colonies as units of survival: hyphal mycosporines synthesized by rock-dwelling microcolonial fungi. Canadian Journal of Botany 81: 131–138. [Google Scholar]
- Gueidan C, Roux C, Lutzoni F (2007). Using a multigene analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycological Research 111: 1147–1170. [DOI] [PubMed] [Google Scholar]
- Gueidan C, Ruibal C, Hoog GS de, Gorbushina A, Untereiner WA, Lutzoni F (2008). A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Studies in Mycology 61: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase G, Sonntag L, Melzer-Krick B, Hoog GS de (1999). Phylogenetic inference by SSU-gene analysis of members of the Herpotrichiellaceae with special reference to human pathogenic species. Studies in Mycology 43: 80-97. [Google Scholar]
- Harutyunyan S, Muggia L, Grube M (2008). Black fungi in lichens from seasonally arid habitats. Studies in Mycology 61: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL (1981). Lichenothelia, a new genus for the Microthelia aterrima group. Lichenologist 13: 141–153. [Google Scholar]
- Henssen A (1987). Lichenothelia, a genus of microfungi on rocks. In: Progress and problems in lichenology in the eighties (Peveling E, ed.), Bibliotheca Lichenologica 25: 257–293. [Google Scholar]
- Hofstetter V, Miadlikowska J, Kauff F, Lutzoni F (2007). Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: a case study of the Lecanoromycetes (Ascomycota). Molecular Phylogenetics and Evolution 44: 412–426. [DOI] [PubMed] [Google Scholar]
- Hoog GS de (1993). Evolution of black yeasts: possible adaptation to the human host. Antonie van Leeuwenhoek 63: 105–109. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ (2000). Atlas of clinical fungi. 2nd edition. CBS, Utrecht.
- Hoog GS de, Zalar P, Urzì C, Leo F de, Yurlova NA, Sterflinger K (1999). Relationships of dothideaceous black yeasts and meristematic fungi based on 5.8S and ITS2 rDNA sequence comparison. Studies in Mycology 43: 31–37. [Google Scholar]
- James TY, Kauff F, Schoch C, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, et al. (2006). Reconstructing the early evolution of the fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Kauff F, Lutzoni F (2002). Phylogeny of the Gyalectales and Ostropales (Ascomycota, Fungi): among and within order relationships based on nuclear ribosomal RNA small and large subunits. Molecular Phylogenetics and Evolution 25: 138–156. [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008). The dictionary of the Fungi. 10th edition. CAB International, Wallingford, U.K.
- Krumbein WE, Jens K (1981). Biogenic rock varnishes of the Negev desert (Israel), an ecological study of iron and manganese transformation by cyanobacteria and fungi. Oecologia 50: 25–38. [DOI] [PubMed] [Google Scholar]
- Leo F de, Urzì C, Hoog GS de (1999). Two Coniosporium species from rock surfaces. Studies in Mycology 43: 70–79. [Google Scholar]
- Liu YJ, Whelen S, Hall BD (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Lindemuth R (2001). Major lineages of Dothideomycetes (Ascomycota) inferred from SSU and LSU rDNA sequences. Mycological Research 105: 901–908. [Google Scholar]
- Lutzoni F, Kauff F, Cox C, McLaughlin D, Celio G, et al. (2004). Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany 91: 1446-1480. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Pagel M, Reeb V (2001). Major fungal lineages are derived from lichen symbiotic ancestors. Nature 41: 937–940. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Wagner P, Reeb V, Zoller S (2000). Integrating ambiguously aligned regions of DNA sequences in phylogenetic analyses without violating positional homology. Systematic Biology 49: 628–651. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (2003). MacClade: analysis of phylogeny and character evolution. V. 4.06. Sinauer, Sunderland, Massachusetts.
- Mason-Gamer R, Kellogg E (1996). Testing for phylogenetic conflict among molecular datasets in the tribe Triticeae (Graminae). Systematic Biology 45: 524–545. [Google Scholar]
- Miadlikowska J, Kauff F, Hofstetter V, Fraker E, Grube M, et al. (2006). New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA- and two protein-coding genes. Mycologia 98: 1088–1103. [PubMed] [Google Scholar]
- Möller EM, Bahnweg G, Sandermann H, Geiger HH (1992). A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Research 20: 6115–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE (2005). Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microbial Ecology 49: 343–352. [DOI] [PubMed] [Google Scholar]
- Muggia L, Hafellner J, Wirtz N, Hawksworth DL, Grube M (2008). The sterile microfilamentous lichenized fungi Cystocoleus ebeneus and Racodium rupestre are relatives of plant pathogens and clinically important dothidealean fungi. Mycological Research 112: 50–56. [DOI] [PubMed] [Google Scholar]
- Nelsen MP, Lücking R, Grube M, Mbatchou JS, Muggia L, Rivas Plata E, Lumbsch HT (2009). Unravelling the phylogenetic relationships of lichenised fungi in Dothideomyceta. Studies in Mycology 64: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofri S, Barreca D, Selbmann L, Isola D, Rabbow E, Horneck G, Vera JPP de, Hatton J, Zucconi L (2008). Resistance of Antarctic black fungi and cryptoendolithic communities to simulated space and Mars conditions. Studies in Mycology 61: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofri S, Selbmann L, Hoog GS de, Grube M, Barreca D, Ruisi S, Zucconi L (2007). Evolution and adaptation of fungi at the boundaries of life. Advances in Space Research 40: 1657–1664. [Google Scholar]
- Palmer FE, Staley JT, Ryan B (1990). Ecophysiology of microcolonial fungi and lichens on rocks in Northeastern Oregon. New Phytologist 116: 613–620. [Google Scholar]
- Posada D, Crandall KA (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Reeb V, Roux C, Lutzoni F (2004). Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32: 1036–1060. [DOI] [PubMed] [Google Scholar]
- Rehner SA, Samuels GJ (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634. [Google Scholar]
- Ruibal C (2004). Isolation and characterization of melanized, slow-growing fungi from semiarid rock surfaces of central Spain and Mallorca. Ph.D. dissertation. Universidad Autónoma de Madrid/Merck, Sharp & Dohme de España, Madrid, Spain.
- Ruibal C, Platas G, Bills GF (2005). Isolation and characterization of melanized fungi from limestone formations in Mallorca. Mycological Progress 4: 23–38. [Google Scholar]
- Ruibal C, Platas G, Bills GF (2008). High diversity and morphological convergence among melanised fungi from rock formations in the Central Mountain System of Spain. Persoonia 21: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZ, Barrès B, Boehm EWA, et al. (2009b). A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW (2006). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1043–1054. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, et al. (2009a). The Ascomycota Tree of Life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic Biology 58: 224–239. [DOI] [PubMed] [Google Scholar]
- Selbmann L, Hoog GS de, Mazzaglia A, Friedmann EI, Onofri S (2005). Fungi at the edge of life: cryptoendolithic black fungi from Antarctic desert. Studies in Mycology 51: 1–32. [Google Scholar]
- Selbmann L, Hoog GS de, Zucconi L, Isola D, Ruisi S, Gerrits van den Ende AHG, Ruibal C, Leo F de, Urzì C, Onofri S (2008). Drought meets acid: three new genera in a dothidealean clade of extremotolerant fungi. Studies in Mycology 61: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbmann L, Zucconi L, Ruisi S, Grube M, Cardinale M, Onofri S (2010). Culturable bacteria associated with Antarctic lichens: affiliation and psychrotolerance. Polar Biology 33: 71–83. [Google Scholar]
- Sert HB, Sümbül H, Sterflinger K (2007a). Microcolonial fungi from antique marbles in Perge/Side/Termessos (Antalya/Turkey). Antonie van Leeuwenhoek 91: 217–227. [DOI] [PubMed] [Google Scholar]
- Sert HB, Sümbül H, Sterflinger K (2007b). Sarcinomyces sideticae, a new black yeast from historical marble monuments in Side (Antalya, Turkey). Botanical Journal of the Linnean Society 154: 373–380. [Google Scholar]
- Sigler L, Tsuneda A, Carmichael JW (1981). Phaeotheca and Phaeosclera, two new genera of dematiaceous hyphomycetes and redescription of Sarcinomyces Lindner. Mycotaxon 12: 449–467. [Google Scholar]
- Spatafora JW, Mitchell TG, Vilgalys R (1995). Analysis of genes coding for small-subunit rRNA sequences in studying phylogenetics of dematiaceous fungal pathogens. Journal of Clinical Microbiology 33: 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, O'Rourke B, Serdani M, et al. (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Staley JT, Palmer F, Adams JB (1982). Microcolonial fungi: common inhabitants on desert rocks? Science 215: 1093–1095. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J (2008). A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology 75: 758–771. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H (2005). RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463. [DOI] [PubMed] [Google Scholar]
- Sterflinger K (1998). Temperature and NaCl-tolerance of rock-inhabiting meristematic fungi. Antonie van Leeuwenhoek 74: 271–281. [DOI] [PubMed] [Google Scholar]
- Sterflinger K, Baere R de, Hoog GS de, Watcher R de, Krumbein WE, Haase G (1997). Coniosporium perforans and C. apollinis, two new rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie van Leeuwenhoek 72: 349–363. [DOI] [PubMed] [Google Scholar]
- Sterflinger K, Hoog GS de, Haase G (1999). Phylogeny and ecology of meristematic ascomycetes. Studies in Mycology 43: 5–22. [Google Scholar]
- Sterflinger K, Prillinger H (2001). Molecular taxonomy and biodiversity of rock fungal communities in an urban environment (Vienna, Austria). Antonie van Leeuwenhoek 80: 275–286. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2003). PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
- Urzì C, Wollenzien U, Criseo G, Krumbein WE (1995). Biodiversity of the rock inhabiting microbiota with special reference to black fungi and black yeasts. In: Microbial diversity and ecosystem function (Allsopp D, Colwell RR, Hawksworth DL, eds). CAB International, Wallingford, U.K.: 289–302.
- Vilgalys R, Hester M (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann M, Gorbushina AA (2006). A broadly applicable method for extraction and characterisation of mycosporines and mycosporine-like amino acids of terrestrial, marine and freshwater origin. FEMS Microbiology Letters 255: 286–295. [DOI] [PubMed] [Google Scholar]
- Wollenzien U, Hoog GS de, Krumbein WE, Uijthof JMJ (1997). Sarcinomyces petricola, a new microcolonial fungus from marble in the Mediterranean basin. Antonie van Leeuwenhoek 71: 281–288. [DOI] [PubMed] [Google Scholar]
- Wollenzien U, Hoog GS de, Krumbein WE, Urzì C (1995). On the isolation of microcolonial fungi occurring on and in marble and other calcareous rocks. The Science of the Total Environment 167: 287–294. [Google Scholar]
- Zoller S, Scheidegger C, Sperisen C (1999). PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. The Lichenologist 31: 511–516. [Google Scholar]