Abstract
PTEN is a tumour suppressor with phosphatase activity in vitro against both lipids and proteins and other potential non-enzymatic mechanisms of action. Although the importance of PTEN’s lipid phosphatase activity in regulating the PI3K signalling pathway is recognised, the significance of PTEN’s other mechanisms of action is currently unclear. Here, we describe the systematic identification of a PTEN mutant, PTEN Y138L, with activity against lipid, but not soluble substrates. Using this mutant we provide evidence for the interfacial activation of PTEN against lipid substrates. We also show that when re-expressed at physiological levels in PTEN null U87MG glioblastoma cells the protein phosphatase activity of PTEN is not required to regulate cellular PtdInsP3 levels or the downstream protein kinase Akt/PKB. Finally, in 3D Matrigel cultures of U87MG cells similarly re-expressing PTEN mutants, both the protein and lipid phosphatase activities were required to inhibit invasion, but either activity alone significantly inhibited proliferation, albeit only weakly for the protein phosphatase activity. Our data provides a novel tool to address the significance of PTEN’s separable lipid and protein phosphatase activities and suggest that both activities act to suppress proliferation and act together to suppress invasion.
Keywords: PTEN, PI 3-kinase, phosphoinositide, cancer, TPIP, Akt
Introduction
PTEN is one of the most frequently lost tumour suppressors in human cancers, that in culture and in vivo, appears to inhibit cell proliferation, survival and growth, and in some cell types, motility (Keniry & Parsons, 2008; Salmena et al., 2008). PTEN is a lipid phosphatase that inhibits phosphoinositide 3-kinase (PI3K)-dependent signalling, by metabolising the PI3K product, PtdInsP3. Although PTEN’s PtdInsP3 phosphatase activity appears to mediate many of its cellular effects, PTEN has additional potential mechanisms of action, including robust protein phosphatase activity and proposed non-enzymatic mechanisms (Mahimainathan & Choudhury, 2004; Myers et al., 1997; Okumura et al., 2005; Raftopoulou et al., 2004; Shen et al., 2007). Several studies have reported lipid phosphatase-independent effects of PTEN affecting cell migration, and also contributing to other functions potentially related to tumour suppression (Freeman et al., 2003; Gildea et al., 2004; Leslie et al., 2007; Park et al., 2002; Raftopoulou et al., 2004; Shen et al., 2007; Trotman et al., 2007). A wealth of correlative data connects the regulation of PI3K signalling by PTEN with its physiological functions and tumour suppressor activity (Sansal & Sellers, 2004). However, given the apparent number and diversity of both PTEN’s normal functions and of the effects of PTEN loss on tumour development in different tissues (Chow & Baker, 2006; Suzuki et al., 2008), it seems quite possible that other mechanisms of action may account for some of PTEN’s functions and tumour suppressor activities in some tissues.
Experiments to reveal the mechanisms and significance of PTEN’s separable activities have been greatly assisted by the use of functionally selective mutants. In particular, many studies have used PTEN G129E, a mutation that was identified in two Cowden disease families (Liaw et al., 1997) and shows remarkable selectivity, displaying greatly reduced lipid phosphatase activity whilst retaining full protein phosphatase activity in vitro (Furnari et al., 1998; Myers et al., 1998). Here we describe the systematic generation of a PTEN mutant with the converse specificity, ie, retaining lipid phosphatase whilst lacking significant protein phosphatase activity. We then describe experiments using both mutants to reveal the contribution of these two separable activities to PTEN’s activity in several cell based assays of proliferation, invasion and migration.
Results
The identification of a PTEN mutant with lipid but not protein phosphatase activity
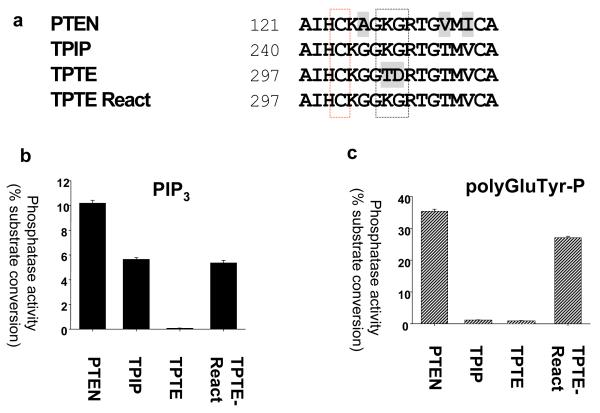
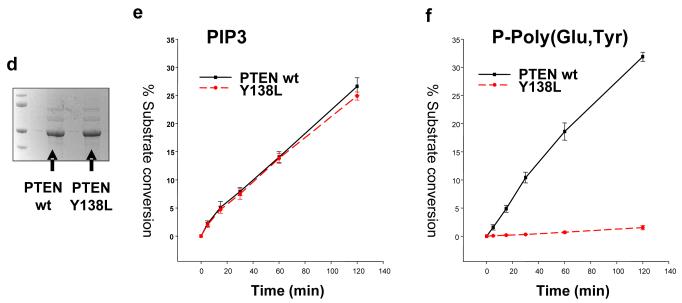
The proteins encoded in the human genome that are most closely related to PTEN are TPIP and TPTE. These are expressed almost exclusively in the testis (Tapparel et al., 2003; Walker et al., 2001), although their physiological functions are unclear. Our previous studies of the lipid phosphatase activity of TPIP and TPTE revealed that TPIP has similar PtdInsP3 phosphatase activity to PTEN, and that while TPTE lacks detectable activity in the assays used, it could be reactivated through mutation, producing an active phosphatase termed ‘TPTE-reactivated’ (Leslie et al., 2007; Walker et al., 2001). However, when the protein phosphatase activity of these proteins was investigated, we obtained two surprising results (Figure 1). Firstly, in contrast to PTEN, TPIP was found to lack detectable protein phosphatase activity against the phosphorylated tyrosine/glutamic acid polymer polyGluTyr-P. Secondly, despite its extremely close sequence similarity with TPIP, TPTE-reactivated displayed robust activity against both the lipid PtdInsP3 and this peptide substrate polyGluTyr-P. This identification of a PTEN-related phosphatase lacking protein phosphatase activity, suggests that protein phosphatase activity may have been selected for during evolution. The protein phosphatase activity displayed by TPTE-reactivated has interesting implications for TPTE function, but given its close relationship to TPIP, suggests a systematic approach for the mutation of PTEN to produce a mutant enzyme with only lipid phosphatase activity. Therefore, at each of the 11 residues at which TPIP differs from TPTE and PTEN through the phosphatase domain and N-terminal region of the C2 domain, we mutated the PTEN sequence to the corresponding amino acid in TPIP (See Figure S1). In an initial screen, these 11 mutants were expressed in bacteria as GST-PTEN fusion proteins, purified and assayed against both PtdInsP3 and polyGluTyr-P in vitro. This screen identified one mutant, PTEN Y138L which displayed similar PtdInsP3 phosphatase activity to wild-type PTEN, but lacked detectable activity against polyGluTyr-P (Figure S2). Further kinetic analysis of PTEN Y138L revealed that the lipid phosphatase activity against 3-33P PtdInsP3 is essentially indistinguishable from wild-type PTEN, whereas activity against polyGluTyr-P is extremely weak, being approximately 3% of wild-type activity, and frequently undetectable (Figures 1, 2 and S2).
Figure 1.
Both TPIP and PTEN Y138L have lipid but not protein phosphatase activity (a) An alignment of the active site P-loop of PTEN, TPIP and TPTE is shown, with the PTEN catalytic cysteine residue boxed in red. The black boxing shows the threonine and aspartic acid residues at which TPTE differs from the active phosphatases PTEN and TPIP in this region, and the mutation of these residues performed in reactivated TPTE (called TPTE-R) (Leslie et al., 2007). (b and c) PTEN and the cytosolic regions of TPIP and TPTE (wild-type and reactivated) were expressed as GST-fusion proteins and purified. The activity of the indicated proteins was assayed using either 33P radiolabelled PtdIns(3,4,5)P3 (b) or phosphorylated polyGluTyr (c) as substrate. Data is shown as mean activity and standard deviation from triplicate assays. (d) Wild-type and Y138L PTEN proteins were expressed in bacteria as GST-fusion proteins, affinity purified and the tag removed by proteolysis before analysis of these preparations by electrophoresis and staining with Coomassie. (e and f) These wild-type and Y138L PTEN proteins were assayed against 33P radiolabelled PtdIns(3,4,5)P3 (e) or phosphorylated polyGluTyr (f) for the indicated times and activity shown as the mean activity and standard deviation from triplicate assays.
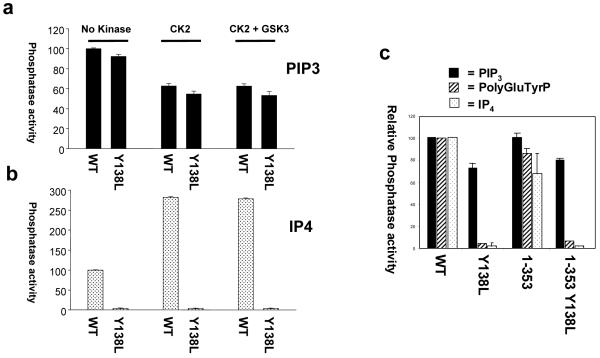
Figure 2.
PTEN Y138L lacks activity against polyGluTyrP and Ins(1,3,4,5)P4 and this substrate specificity is not affected by the C-terminal phosphorylated tail. (a and b) Purified tagless PTEN wild-type and Y138L proteins were phosphorylated with purified recombinant CK2 and GSK3 as indicated, before assaying against PtdInsP3 or Ins(1,3,4,5)P4. Activity data are represented as the mean activity relative to unphosphorylated wild-type + SD from triplicate assays. (c) Mutant GST-PTEN proteins (wt and Y138L) lacking the unstructured C-terminal tail, termed PTEN 1-353, were expressed, purified and assayed against radiolabelled PtdInsP3, polyGluTyrP and Ins(1,3,4,5)P4. Activity data are represented as the mean activity relative to wild-type + the range/2 from duplicate assays.
PTEN Y138L has activity against lipid but not soluble substrates
The activity of PTEN Y138L was tested against the soluble inositol phosphate substrate, Ins(1,3,4,5)P4 (the headgroup of PtdInsP3) and although the wild-type enzyme was strongly active, PTEN Y138L lacked detectable activity (Figure 2b and c). This result suggested that the effect of the Y138L mutation was not to alter the precise conformation of the active site to accommodate the headgroup of PtdInsP3 yet exclude phosphotyrosine, but might alter the conformation of the phosphatase on a larger scale, favouring lipid substrates presented at a membrane interface over soluble substrates. It seemed possible that the unstructured C-terminal tail of PTEN might be involved in this selective activity, as especially when phosphorylated, this tail is believed to bind into the basic surfaces of the PTEN phosphatase and C2 domains, interfering with membrane binding (Das et al., 2003; Gil et al., 2007; Ning et al., 2006; Vazquez et al., 2001). We therefore tested the effects of phosphorylation of PTEN with CK2, and also with GSK3, on the activity of wild-type PTEN and PTEN Y138L against both PtdInsP3 and Ins(1,3,4,5)P4. These experiments showed, as might be predicted from previous experiments using mutant proteins (Ning et al., 2006), that phosphorylation of bacterially expressed PTEN with CK2 caused a modest reduction in activity against PtdInsP3 and a larger increase in activity against the soluble headgroup Ins(1,3,4,5)P4. However, phosphorylated PTEN Y138L showed a similar decrease in PtdInsP3 phosphatase activity, but still lacked detectable activity against Ins(1,3,4,5)P4 (Figure 2b). We therefore tested the activity of wild type and Y138L mutant PTEN proteins lacking the C-terminal tail due to a stop codon at position 354. This showed that PTEN Y138L 354stop still lacked activity against PolyGluTyr-P and Ins(1,3,4,5)P4, whilst retaining lipid phosphatase activity (Figure 2c). The substrate specificity of PTEN Y138L therefore may suggest that its active site can only be available for substrate binding when the enzyme interacts with a lipid surface.
Defining the requirements for the lipid and protein phosphatase activities of PTEN in cell based assays
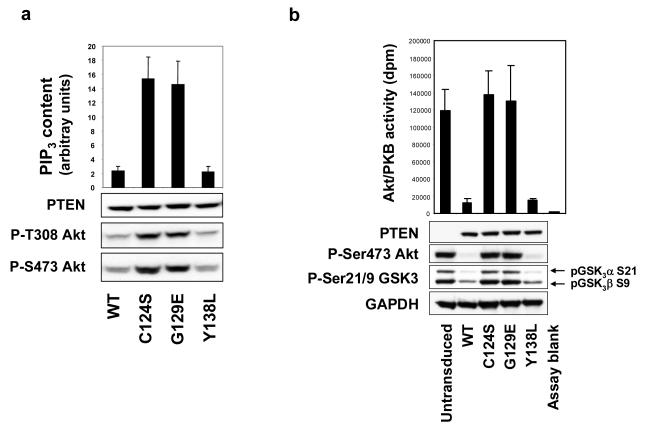
To analyse the effects of PTEN’s separable lipid and protein phosphatase activities on cellular signalling and behaviour, we first used the well characterised PTEN null glioblastoma cell line, U87MG. We expressed PTEN in these cells using a lentiviral delivery system able to engineer expression of PTEN in almost all cells in target populations, even when used at titres required to express PTEN at levels broadly equivalant to those in several cells expressing endogenous PTEN. We re-expressed wild-type PTEN and the three mutant proteins, PTEN C124S (phosphatase dead), PTEN G129E (protein phosphatase only) and PTEN Y138L (as described, lipid phosphatase only) in U87MG cells to analyse their effects on cellular PtdInsP3 levels. These data showed a strict correlation with the lipid phosphatase activity of the expressed proteins, as wild-type and Y138L enzymes both reduced cellular PtdInsP3 by approximately 80%, whereas C124S and G129E had no significant effect (Figure 3a). We also similarly analysed the effects of these PTEN proteins on the phosphorylation of the PtdInsP3-dependent protein kinase Akt/PKB. In these experiments, expression of PTEN C124S or PTEN G129E had no detectable effect on Akt phosphorylation, whereas expression of either wild-type PTEN or PTEN Y138L at approximately physiological levels greatly reduced the phosphorylation of Akt (Figures 3b and S4). Performing these experiments many times over a range of expression levels, the wild-type and Y138L proteins were indistinguishable in their ability to suppress Akt phosphorylation (Figures 3 and S4 (Maccario et al., 2007)) .As part of this project, we also addressed the effects of PTEN activity on the phosphorylation of some of its proposed substrates, FAK, PDGFR and SHC. However, we found that these proteins appeared to have the same level of tyrosine phosphorylation in U87MG cells regardless of PTEN expression (Figure S5).
Figure 3.
PTEN Y138L reduces cellular PtdInsP3 levels and Akt activity and phosphorylation with similar efficacy to wild-type PTEN. (a and b) PTEN null U87MG cells were transduced with lentiviruses encoding GFP, PTEN or mutant PTEN (C124S, G129E or Y138L) as indicated, and lysed after 24 hours. Cellular PtdInsP3 content was measured using a TR-FRET sensor complex by comparison with a standard curve of known PtdInsP(3,4,5)3 concentrations as previously described (Gray et al., 2003). Data is presented as the mean PIP3 content + SD in arbitrary units from quadruplicate samples. PTEN expression and Akt phosphorylation were investigated by western blotting of total cell lysates using total and phospho-specific antibodies and Akt enzyme activity in immunoprecipitates was assayed in vitro against a peptide substrate. Data is represented as the mean dpm radiolabelled phosphate incorporated into the peptide substrate + SD from triplicate samples. Experiments to measure PtdInsP3 content and Akt phosphorylation were performed on two and eight occasions respectively with similar results.
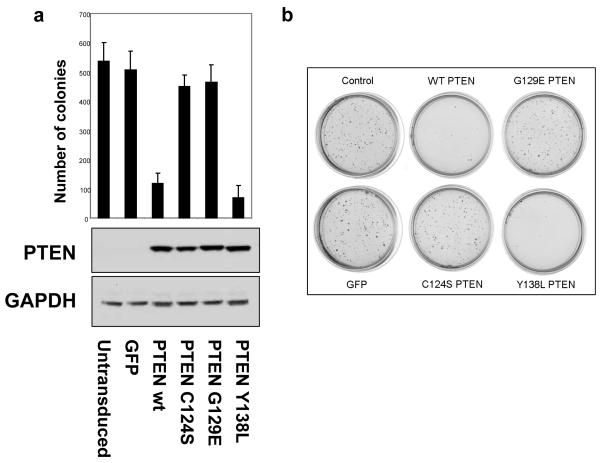
PTEN has been previously shown in cells cultured in soft agar, to suppress proliferation and colony formation (Cheney et al., 1998; Davies et al., 1998; Furnari et al., 1997) and that this ability appeared to require lipid phosphatase activity. Sham transduced U87MG cells and those expressing GFP form colonies suspended in agarose, whereas expression of wild-type PTEN caused an almost complete suppression of colony formation. Phosphatase dead PTEN C124S and PTEN G129E, each had no inhibitory effect on colony formation, whereas PTEN Y138L inhibited colony formation in a very similar fashion to the wild-type enzyme (Figure 4). Over four experiments, the mean number of colonies formed by cells expressing PTEN Y138L was slightly smaller than the number formed by cells expressing wild-type PTEN, but this difference was not significant. These data strongly correlate the ability of PTEN to suppress colony formation in soft agar with its lipid phosphatase activity.
Figure 4.
The effects of PTEN re-expression on U87MG cell growth in soft agar. (a and b) U87MG cells were transduced with recombinant lentiviruses encoding wild type PTEN and PTEN point mutants, tryprinised 24 later, and then grown in 0.35 % agarose for 21 days. Cells were fixed with 2% paraformaldehyde and the number of colonies counted. In parallel, samples of transduced cells were seeded onto standard culture dishes and lysed after 24 hours to determine the expression of PTEN by western blotting. (a) The numerical data shown are derived from 4 independent experiments, each performed in triplicate (+ SEM). Data were analysed by ANOVA and a Tukey’s pairwise comparison, showing that the data for PTEN wt and Y138L differ significantly from that for GFP, PTEN C124S and PTEN G129E, but there were no significant differences within these two groups. (b) An image of a representative data set is shown.
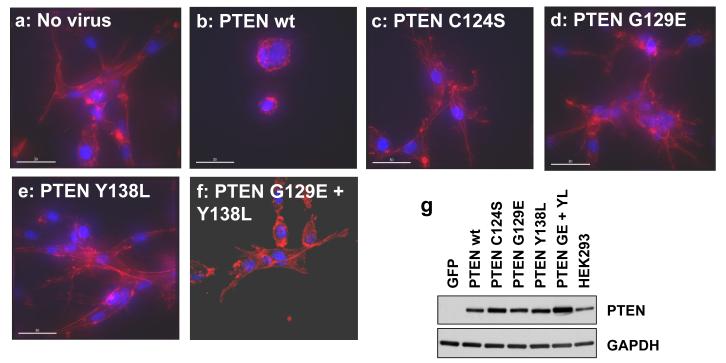
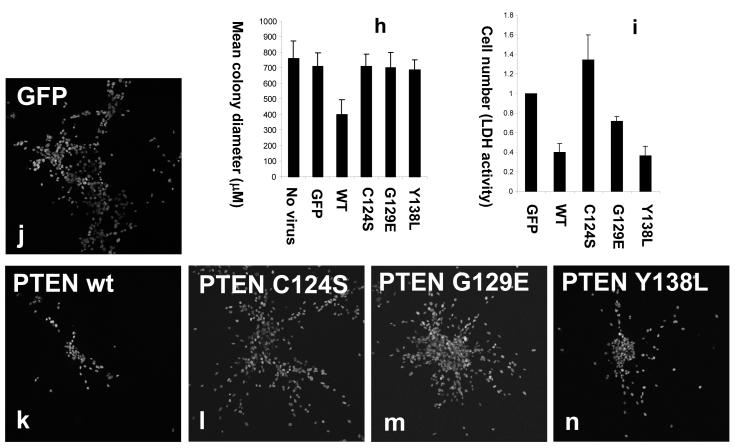
In vivo, the invasion of glial tumours frequently proceeds along white matter tracts and blood vessels, following the endothelial cell basement membrane (de Bouard et al., 2002; Goldbrunner et al., 1998). We therefore established an assay for the proliferation and invasion of cultured U87MG cells suspended in the reconstituted basement membrane extract, Matrigel, with cell morphology being visualized by staining for DNA and filamentous actin. In this assay, after 3 days in culture, sham transduced cells and those expressing GFP or phosphatase dead PTEN C124S proliferated to form colonies that spread through the surrounding matrix (Figure 5) in contrast to cells proliferating in agarose where they grow as nearly spherical clusters. The expression of wild-type PTEN at near physiological levels both suppressed the invasion of these cells, causing them to grow initially as rather tightly packed balls of cells, with few invading the surrounding matrix frequently as strings of cells, in manner different from other cells in these experiments (Figure 5). Neither expression of PTEN G129E nor of PTEN Y138L was sufficient to block the invasion of these cells into the surrounding matrigel, even when these two mutants were expressed additively together in the same cells (Figure 5 a-g). Consistent with the efficacy only of wild-type PTEN in the regulation of this invasive cell morphology, when colony size was measured after 7 days in 3D culture, addressing the spread of cells into the surrounding matrix, again, only wild-type PTEN significantly inhibited colony spread (Figure 5h). Microscopy also revealed other consistent differences in the morphology of these colonies, in particular, cells expressing PTEN G129E generally formed a more dense central mass of cells, with fewer cells invading deep into the surrounding matrix (Figure 5 j-n). We also investigated the effects of PTEN expression on net cell proliferation in these cultures, using an LDH enzyme assay based method (Figure 5i). The reproducible differences in colony morphology made unbiased counting of stained nuclei challenging although similar results were obtained (data shown in Figure S6) and Matrigel also appeared to interfere with several other proliferation assays methods. This work showed that both wild-type PTEN and PTEN Y138L were able to strongly suppress cellular proliferation, but with PTEN G129E mediating a weaker but still significant effect (Figures 5i and S6). Over expression of either PTEN wt or PTEN Y138L was able to cause an almost complete block in colony formation, apparently through cell death, but this effect was not evident at physiological expression levels (data not shown).
Figure 5.
The effects of PTEN re-expression on U87MG cell proliferation and invasion in 3D culture in matrigel. U87MG cells were transduced for 24 hours with recombinant lentiviruses encoding wild type PTEN and PTEN point mutants. Cells were trypsinised, then either seeded at low density in matrigel (a-f and h-n) or plated for 24 hours further 2D culture on plastic to verify expression of PTEN by western blotting alongside a sample of HEK293 cells expressing endogenous PTEN (g). (a-f) Cells were grown for 3 days, fixed with 2 % paraformaldehyde and stained with rhodamine phalloidin and DAPI to visualize filamentous actin and DNA respectively. Samples were examined and photographed by widefield deconvolution microscopy (a-e) or confocal microscopy (f). Similar data were obtained by both methods. White scale bars represent 30μM. (j-n). Cells were grown for 5 days, fixed with 2 % paraformaldehyde, stained with DAPI and examined by confocal microscopy. Numerical data shown in (h) represent the mean colony size of sets of 5 day colonies measured as the longest observable axis of colonies photographed at low magnification by phase contrast microscopy. An assessment of cell number shown in (i) was measured by an LDH enzyme assay of matrix/cell extracts after 10 days growth and is shown as mean + SEM.
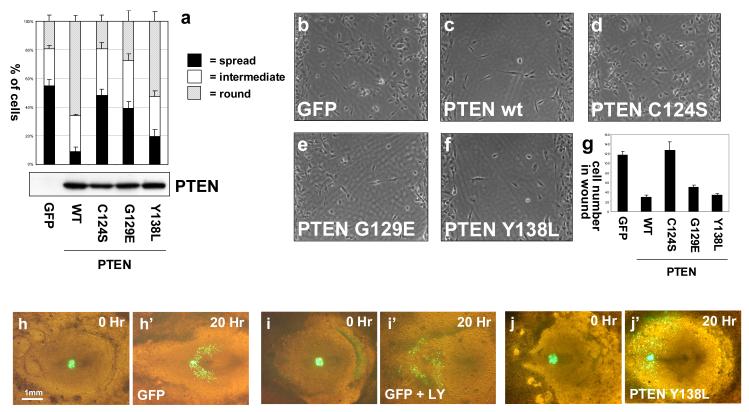
We also investigated the described effects of PTEN re-expression in U87MG cells on cell spreading and migration (Tamura et al., 1998). For cell spreading assays, cells transduced with lentiviruses encoding PTEN were trypsinised thoroughly before allowing attachment and spreading on culture substrates and as described, wild-type PTEN caused a delay in the ability of cells to spread. In this assay, the ability to inhibit spreading correlated with PTEN’s lipid phosphatase activity, as PTEN Y138L was almost as efficient as the wild-type enzyme, but PTEN C124S and PTEN G129E had little or no effect (Figure 6). Cell migration assays from a confluent monolayer into a 500μm defined cell free gap showed that PTEN retards the rate of migration into this gap. As has been previously described, PTEN G129E was also able to inhibit glioblastoma cell migration (Raftopoulou et al., 2004; Tamura et al., 1998), but interesting, this ability was shared by PTEN Y138L, suggesting that either lipid or protein phosphatase activity are sufficient to mediate a similar effect on cell migration in this assay (Figure 6). Interestingly in these experiments, migration of cells expressing wild-type PTEN was clearly observed, and this migration rate over the period of the experiment appeared little different with cells expressing either PTEN G129E or PTEN Y138L, suggesting that, at least using this assay format, the inhibitory effects of PTEN’s activities against lipid and soluble substrates may not be additive.
Figure 6.
The regulation of cell spreading, and cell migration by PTEN. (a) Trypsinised U87MG cells expressing GFP or different PTEN proteins were allowed to adhere onto collagen IV coated culture slides for 30 minutes before fixation. Data are presented as the mean fractions of each cell population blind scored as spread, intermediate or round, + SEM from 5 randomly selected fields from a representative experiment. This experiment has been repeated twice on collagen and twice on plastic with similar results. Equal expression of the PTEN mutants was verified by Western blotting and the same set of transduced cells was used in the following experiments addressing adgherent cell migration. (b-f) Trypsinised U87MG cells expressing GFP or different PTEN proteins were seeded at confluence and allowed to adhere to matrigel-treated plastic before the culture inserts were removed to leave a defined 500μm cell-free gap. Cells were then allowed to migrate over a 24 hour period, before cells were fixed and photographed. (g) Quantitation represents the mean number of cells in a central area of the gap from 6 randomly selected fields +/− SEM. This experiment was performed three times with similar results. (h-j) PTEN Y138L interferes with directed cell migration in the developing chick embryo, but does not block cell motility. Cell migration assays were performed using cells expressing GFP or GFP-PTEN Y138L as previously described (Leslie et al., 2007; Yang et al., 2002).
We went on to analyse the effects of PTEN Y138L on cell migration in vivo, using a previously established assay for mesoderm cell migration away from the primitive streak in the early chick embryo (Yang et al., 2002). In our previous studies, we found that the protein phosphatase activity of PTEN appeared to contribute to a block on the epithelial to mesenchymal transition (EMT) that these cells undergo before they exit the primitive streak, whereas the lipid phosphatase activity of PTEN had no effect on this EMT, but interfered with the directed nature of the subsequent migration of these cells through the embryo (Leslie et al., 2007). In this assay, in agreement with these previous conclusions, expression of GFP-PTEN Y138L did not affect primitive streak cell EMT, but interfered with the directed cell migration of cells through the embryo (Figure 6 i-k). Finally, attempting to identify effects of PTEN on well characterized signaling pathways, we used a panel of phospho-specific antibodies to investigate adherent U87MG cell cultures expressing wild-type and mutant PTEN proteins at approximately 2-3 fold above the level seen in HEK293 cells (Figure S8). These experiments showed robust effects of either wild-type or Y138L PTEN expression, suppressing Akt phosphorylation and also increasing IRS1 levels, as has been previously described in these cells (Lackey et al., 2007). However, other effects, and any apparently mediated by PTEN G129E implicating protein phosphatase activity, were not observed.
Discussion
Given the significance of PTEN as a tumour suppressor and as a regulator of many physiological processes, it is important that we determine its mechanisms of action in these specific functions. This is particularly true in tumour suppression, as intense efforts are going into the development of PI3K inhibitors for the treatment of cancer, with several currently in clinical trials. These agents are likely to be given to patients with tumours in which PTEN function is believed to be compromised, although as yet, in many tumour types, it is still possible that PI3K independent pathways may be of great importance. Here we have developed a PTEN mutant, PTEN Y138L that retains wild-type PtdInsP3 lipid phosphatase activity, but in contrast to the wild-type enzyme, PTEN Y138L lacks activity against a model peptide substrate, polyGluTyrP and the soluble PtdInsP3 headgroup, Ins(1,3,4,5)P4. We have then used this mutant to reveal several significant aspects to PTEN function.
The activity of the PTEN-related phosphatase, TPIP, towards lipids but not the tested peptide substrate indicates that the protein phosphatase activity of PTEN is not a necessary but inconsequential by-product of a PTP phosphatase domain with a large basic active site capable of accommodating PtdInsP3. Instead, this suggests a functional requirement for PTEN’s protein phosphatase activity. Similarly, the selective activity of PTEN Y138L against the lipid PtdInsP3, but not the identical soluble headgroup, Ins(1,3,4,5)P4, strongly suggests to us that the active site of PTEN Y138L is only available for substrate binding when the enzyme interacts with a lipid surface. It is possible that this conformational regulation does not apply to wild-type PTEN, but we feel this is unlikely, and that a more probable case is that PTEN can exist in at least 3 conformations: a ‘lipid active’ conformation in contact with membrane surfaces, a ‘soluble active’ conformation in solution, that has evolved to utilize protein substrates, and an inactive conformation (Leslie et al., 2008). This hypothetical model would require that PTEN Y138L and TPIP do not stably take up the ‘soluble active’ conformation and also fits well with data regarding the interfacial activation of PTEN on acidic membrane surfaces rich in PtdIns(4,5)P2 (Campbell et al., 2003; Das et al., 2003; Iijima et al., 2004; McConnachie et al., 2003; Redfern et al., 2008; Walker et al., 2004). It also seems possible that only one active conformation exists that is greatly stabilized by membrane surface interaction. However, in this latter case it seems to us harder to explain why if PTEN Y138L was less stable in a single active conformation in solution, it would not also have lower lipid phosphatase activity on a membrane surface. It may also be relevant to these ideas that our expression experiments lead us to believe that the PTEN Y138L protein could be generally slightly less stable than wild-type PTEN, as we consistently required more plasmid or viral expression vector (usually 2-3 fold) to achieve the same level of protein expression and PTEN Y138L was slightly less stable to thermal denaturation than the wild-type enzyme (Figure S7).
Secondly, the similarity between wild-type PTEN and PTEN Y138L in terms of their PtdInsP3 phosphatase activity in vitro and of their effects on both cellular PtdInsP3 levels and Akt/PKB phosphorylation strongly suggest that the protein phosphatase activity of PTEN is not required in order for PTEN to efficiently metabolize cellular PtdInsP3. Since autodephosphorylation has been proposed as a mechanism regulating PTEN conformation and activity that may be important in some settings (Raftopoulou et al., 2004), this seems potentially significant.
Finally, we have used the two functionally selective mutants, PTEN G129E and Y138L, to address the relative importance of the lipid and protein phosphatase activities of PTEN in the regulation of different cellular processes, when expressed at physiological levels in PTEN null U87MG cells. This combination of these two mutants has allowed us to identify not only processes that can be affected by PTEN’s protein phosphatase activity, but to identify processes that appear to require both activities acting together. This work has identified examples of process that appear to be inhibited solely or largely via the lipid phosphatase activity (proliferation in soft agar and cell spreading), processes that can be significantly inhibited by either activity (proliferation and adherent cell migration) and those that require both activities in concert for efficient inhibition (invasion in Matrigel). Also, together with previously published experiments (Leslie et al., 2007), although not in U87MG cells, we show that the epithelial to mesenchymal transition of cells in the chick embryo primitive streak is inhibited solely via the protein phosphatase activity of PTEN.
We find that the ability of PTEN to inhibit colony formation in a classic soft agar assay correlates closely with its lipid phosphatase activity. However, using a novel matrigel invasion/proliferation assay we show that both the lipid and protein phosphatase activities contribute significantly to the inhibition of both invasive morphology and proliferation in this setting. Although it has been well established that the protein phosphatase activity of PTEN can mediate a suppression of glial cell migration (Park et al., 2002; Raftopoulou et al., 2004; Tamura et al., 1998), effects on proliferation have not been previously described. This is perhaps because these effects are not evident in assays such as the classical soft agar assays and because they appear much weaker than those mediated via the lipid phosphatase activity. Indeed the contrasting results obtained from cells in soft agar/serum and those in Matrigel agree well with previous data implicating integrin signalling in the stimulation of signalling pathways inhibited by the protein phosphatase activity of PTEN (Dey et al., 2008; Tamura et al., 1998). Several proteins have been proposed as potential substrates for PTEN, including FAK, SHC, PDGF-R, β-catenin, 5-HT2c-R, and SMAD3 (Gu et al., 1999; Hjelmeland et al., 2005; Ji et al., 2006; Mahimainathan & Choudhury, 2004; Tamura et al., 1998; Vogelmann et al., 2005) and several of these would seem plausible mediators of effects on glial cell migration, invasion and proliferation. However, to date none of these have been validated with great confidence, and our experiments failed to find support for these proteins as substrates. It will require significant effort to investigate these possibilities conclusively and it seems most likely to us that significant substrates for PTEN’s protein phosphatase activity remain unidentified. The use of functionally selective mutants has identified biological processes that appear to be controlled independently by PTEN’s protein phosphatase activity (adherent cell migration) or require both activities (invasion in matrigel). These tools should also assist in the difficult challenges of investigating these mechanisms and identifying the substrates responsible.
Although a clear picture of the significance of PTEN as a regulator of cellular inositol phosphates is yet to emerge, (Deleu et al., 2006; Orchiston et al., 2004), the lack of activity of either PTEN G129E or PTEN Y138L against Ins(1,3,4,5)P4 would argue against these molecules as significant substrates, at least in mediating the effects we observe here on proliferation and migration. In the crystal structure of PTEN, tyrosine 138 is located in a pocket of the phosphatase domain, well away from the active site, giving few clues to explain the substrate selectivity of the Y138L mutant (Lee et al., 1999). To our knowledge, only one missense mutation has been described targeting tyrosine 138, (Y138C in a small cell lung cancer case) (http://www.sanger.ac.uk/genetics/CGP/cosmic).
Our work provides a novel tool that in the future should allow the determination of the significance of PTEN’s lipid and protein phosphatase activities in many assay systems. The data also show that when re-expressed in PTEN null glioblastoma cells at a physiological expression level and these cells are grown in Matrigel, both the lipid and protein phosphatase activities of PTEN independently contribute an inhibition of cellular proliferation and that both activities are required in concert to achieve efficient inhibition of invasion. Considering this, it seems possible that the importance of PTEN’s protein phosphatase activity may have been underestimated.
Materials and Methods
Cell culture
U87MG cells were originally purchased from the ECACC, and cultured in MEM and 10% FBS (Sigma). 293T cells were kindly provided by Michelle West and Colin Watts (Division of Cell Biology and Immunology, University of Dundee) and grown in DMEM, 10% FBS. PTEN proteins were expressed in U87MG cells using a lentiviral expression system developed in Mary Collins’ laboratory (Ikeda et al., 2003). Non-replicative lentiviruses were produced by triple transfection of 1.5 million 293T cells in a 60mm dish with 1.5μg pHR-SIN based shuttle vectors, 1μg pHR-CMV 8.2 deltaR packaging vector and 1μg pCMV VSV-G envelope vector using Trans-IT transfection reagent (Mirus). Transfected cells were maintained in category 2 containment following UK home office and local guidelines. Media was changed after 16hours and after a further 48 hours, the 4mls of viral supernatant was harvested, centrifuged for 5 minutes at 100 × g to pellet any cells and frozen at −80° in aliquots. Batches of viral supernatants were cross-compared by transduction of U87MG cells supplemented with 8μg/ml polybrene and western blotting for their efficiency in driving PTEN expression. Initial experiments transducing U87MG cells with simultaneously prepared virus preparations encoding GFP or PTEN indicated that even at low levels of virus at which resultant PTEN expression was approximately that of several cultured cell lines (around 1% culture volume), expression of GFP appeared relatively homogeneous and evident in at least 95% of the target cell populations.
Cell based assays
For 3D matrigel cultures, 50 μl of matrigel was spread on a 16 mm diameter coverslip in the well of a 12 well plate and allowed to form a gel. 2000 U87MG cells were seeded onto the matrigel in 50 μl of growth medium and allowed to attach for 2 hours. The medium was then removed and cells overlaid with a further 50 μl of matrigel. Once the second layer of matrigel had gelled, the culture was overlaid with 1 ml of growth medium and grown for 3, 7 or 10 days. The medium was removed, cultures washed twice with PBS, and cells fixed in 2 % paraformaldehyde in PBS for 30 minutes at room temperature. Cells were then washed twice with PBS and permeabilised with TBS-T (25mM Tris-HCl, 150mM NaCl, 0.1% Tween-20, pH 7.5) for 10 minutes. Cells were stained with rhodamine phalloidin (10 nM) and DAPI (1 μg/ml) for 10 minutes, washed twice with TBS-T, once with TBS and mounted using Vectashield. Cells were examined using either a SP2 Leica confocal microscope or an Applied Precision DV3 widefield deconvolution microscope equipped with a 20X 0.7 NA oil immersion lense. 2D projections were created and nuclei counted using Volocity imaging software (Improvision, Perkin Elmer). Data were analysed by ANOVA and Tukey’s pairwise comparison was used to identify groups that were significantly different.
Cell proliferation was also measured after 10 days growth in matrigel using an LDH activity assay kit (Roche). 3D matrigel cultures were washed twice in PBS, scraped into Eppendorf tubes and lysed in 200 μl of PBS containing 1% Triton X-100 and complete protease inhibitors on ice for 30 minutes. Samples were then centrifuged for 5 minutes at 5000 × g at 4°C to pellet the matrigel and the supernatant recovered to a fresh tube. 5 μl of each sample was added to 50 μl of assay reagent in the wells of 96 well plate, incubated for 1 hour at room temperature in the dark, and the absorbance measured at 490 nm using a spectrophotometer.
Additional methods are described in the Supplementary section.
Supplementary Material
Acknowledgments
We would like to thank Sam Swift and Paul Appleton from the Dundee Light Microscopy Facility for their help with image acquisition and analysis. We would like to thank Hilary McLauchlan, James Hastie and their staff in the DSTT (University of Dundee) for provision of purified antibodies and recombinant protein kinases and Steven Hubbard for advice regarding data analysis. NL is an RCUK Academic Fellow. Work in the Inositol Lipid Signalling laboratory is funded by the Medical Research Council, the Association for International Cancer Research and the pharmaceutical companies of the DSTT consortium (Astra Zeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck KGaA and Pfizer).
Footnotes
Conflict of Interest Statement The research funding of the Inositol Lipid Signalling laboratory includes a research grant from a pharmaceutical consortium (Astra Zeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck Serono and Pfizer). These companies have not been directly involved in, or influenced, this work.
References
- Campbell RB, Liu F, Ross AH. J Biol Chem. 2003;278:33617–20. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- Cheney IW, Johnson DE, Vaillancourt MT, Avanzini J, Morimoto A, Demers GW, Wills KN, Shabram PW, Bolen JB, Tavtigian SV, Bookstein R. Cancer Res. 1998;58:2331–4. [PubMed] [Google Scholar]
- Chow LM, Baker SJ. Cancer Lett. 2006;241:184–96. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Das S, Dixon JE, Cho W. Proc Natl Acad Sci U S A. 2003;100:7491–6. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MA, Lu Y, Sano T, Fang X, Tang P, LaPushin R, Koul D, Bookstein R, Stokoe D, Yung WK, Mills GB, Steck PA. Cancer Res. 1998;58:5285–90. [PubMed] [Google Scholar]
- de Bouard S, Christov C, Guillamo JS, Kassar-Duchossoy L, Palfi S, Leguerinel C, Masset M, Cohen-Hagenauer O, Peschanski M, Lefrancois T. J Neurosurg. 2002;97:169–76. doi: 10.3171/jns.2002.97.1.0169. [DOI] [PubMed] [Google Scholar]
- Deleu S, Choi K, Pesesse X, Cho J, Sulis ML, Parsons R, Shears SB. Cell Signal. 2006;18:488–98. doi: 10.1016/j.cellsig.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Dey N, Crosswell HE, De P, Parsons R, Peng Q, Su JD, Durden DL. Cancer Res. 2008;68:1862–71. doi: 10.1158/0008-5472.CAN-07-1182. [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, Liu X, Wu H. Cancer Cell. 2003;3:117–30. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Huang HJ, Cavenee WK. Cancer Res. 1998;58:5002–8. [PubMed] [Google Scholar]
- Furnari FB, Lin H, Huang HS, Cavenee WK. Proc Natl Acad Sci U S A. 1997;94:12479–84. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Andres-Pons A, Pulido R. Cell Death Differ. 2007;14:395–9. doi: 10.1038/sj.cdd.4402073. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Herlevsen M, Harding MA, Gulding KM, Moskaluk CA, Frierson HF, Theodorescu D. Oncogene. 2004;23:6788–97. doi: 10.1038/sj.onc.1207599. [DOI] [PubMed] [Google Scholar]
- Goldbrunner RH, Bernstein JJ, Tonn JC. Microsc Res Tech. 1998;43:250–7. doi: 10.1002/(SICI)1097-0029(19981101)43:3<250::AID-JEMT7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Gray A, Olsson H, Batty IH, Priganica L, Downes C Peter. Anal Biochem. 2003;313:234–45. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- Gu J, Tamura M, Pankov R, Danen EH, Takino T, Matsumoto K, Yamada KM. J Cell Biol. 1999;146:389–404. doi: 10.1083/jcb.146.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland AB, Hjelmeland MD, Shi Q, Hart JL, Bigner DD, Wang XF, Kontos CD, Rich JN. Cancer Res. 2005;65:11276–81. doi: 10.1158/0008-5472.CAN-05-3016. [DOI] [PubMed] [Google Scholar]
- Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. J Biol Chem. 2004;279:16606–13. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Takeuchi Y, Martin F, Cosset FL, Mitrophanous K, Collins M. Nat Biotechnol. 2003;21:569–72. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- Ji SP, Zhang Y, Van Cleemput J, Jiang W, Liao M, Li L, Wan Q, Backstrom JR, Zhang X. Nat Med. 2006;12:324–9. doi: 10.1038/nm1349. [DOI] [PubMed] [Google Scholar]
- Keniry M, Parsons R. Oncogene. 2008;27:5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- Lackey J, Barnett J, Davidson L, Batty IH, Leslie NR, Downes CP. Oncogene. 2007;26:7132–42. doi: 10.1038/sj.onc.1210520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Cell. 1999;99:323–34. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Oncogene. 2008;27:5464–76. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Yang X, Downes CP, Weijer CJ. Curr Biol. 2007;17:115–125. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Nature Genetics. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Maccario H, Perera NM, Davidson L, Downes CP, Leslie NR. Biochem J. 2007;405:439–44. doi: 10.1042/BJ20061837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahimainathan L, Choudhury GG. J Biol Chem. 2004;279:15258–68. doi: 10.1074/jbc.M314328200. [DOI] [PubMed] [Google Scholar]
- McConnachie G, Pass I, Walker SM, Downes CP. Biochem J. 2003;371:947–55. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. Proc Natl Acad Sci U S A. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, M.H. W, Parsons R, Tonks NK. Proc Natl Acad Sci U S A. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K, Miller LC, Laidlaw HA, Burgess LA, Perera NM, Downes CP, Leslie NR, Ashford ML. Embo J. 2006;25:2377–87. doi: 10.1038/sj.emboj.7601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Zhao M, Depinho RA, Furnari FB, Cavenee WK. Proc Natl Acad Sci U S A. 2005;102:2703–6. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchiston EA, Bennett D, Leslie NR, Clarke RG, Winward L, Downes CP, Safrany ST. J Biol Chem. 2004;279:1116–22. doi: 10.1074/jbc.M310933200. [DOI] [PubMed] [Google Scholar]
- Park MJ, Kim MS, Park IC, Kang HS, Yoo H, Park SH, Rhee CH, Hong SI, Lee SH. Cancer Res. 2002;62:6318–22. [PubMed] [Google Scholar]
- Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Science. 2004;303:1179–81. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- Redfern RE, Redfern D, Furgason ML, Munson M, Ross AH, Gericke A. Biochemistry. 2008;47:2162–71. doi: 10.1021/bi702114w. [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Sansal I, Sellers WR. J Clin Oncol. 2004;22:2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Nakano T, Mak TW, Sasaki T. Cancer Sci. 2008;99:209–13. doi: 10.1111/j.1349-7006.2007.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Science. 1998;280:1614–7. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- Tapparel C, Reymond A, Girardet C, Guillou L, Lyle R, Lamon C, Hutter P, Antonarakis SE. Gene. 2003;323:189–99. doi: 10.1016/j.gene.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. Cell. 2007;128:141–56. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR. J Biol Chem. 2001;276:48627–30. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- Vogelmann R, Nguyen-Tat MD, Giehl K, Adler G, Wedlich D, Menke A. J Cell Sci. 2005;118:4901–12. doi: 10.1242/jcs.02594. [DOI] [PubMed] [Google Scholar]
- Walker SM, Downes CP, Leslie NR. Biochem J. 2001;360:277–83. doi: 10.1042/0264-6021:3600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP. Biochem J. 2004;379:301–7. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. Dev Cell. 2002;3:425–37. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.