Abstract
Background
Somatic mutation in the X-linked phosphatidylinositol glycan class A gene (PIG-A) causes glycosyl phosphatidylinositol anchor deficiency in human patients with paroxysmal nocturnal hemoglobinuria.
Design and Methods
We produced an animal model of paroxysmal nocturnal hemoglobinuria by conditional Pig-a gene inactivation (Pig-a−/−) in hematopoietic cells; mice carrying two lox sites flanking exon 6 of the Pig-a gene were bred with mice carrying the transgene Cre-recombinase under the human c-fes promoter. We characterized the phenotypic and functional properties of glycosyl phosphatidylinositol-deficient and glycosyl phosphatidylinositol-normal hematopoietic cells from these Pig-a−/− mice using gene expression microarray, flow cytometry, bone marrow transplantation, spectratyping, and immunoblotting.
Results
In comparison to glycosyl phosphatidylinositol-normal bone marrow cells, glycosyl phosphatidylinositol-deficient bone marrow cells from the same Pig-a−/− animals showed up-regulation of the expression of immune function genes and contained a significantly higher proportion of CD8 T cells. Both characteristics were maintained when glycosyl phosphatidylinositol-deficient cells were transplanted into lethally-irradiated recipients. Glycosyl phosphatidylinositol-deficient T cells were inactive, showed pronounced Vβ5.1/5.2 skewing, had fewer γ-interferon-producing cells after lectin stimulation, and contained fewer CD4+CD25+FoxP3+ regulatory T cells. However, the levels of T-cell receptor signaling proteins from glycosyl phosphatidylinositol-deficient cells were normal relative to glycosyl phosphatidylinositol-normal cells from wild type animals, and cells were capable of inducing target cell apoptosis in vitro.
Conclusions
Deletion of the Pig-a gene in hematopoietic cells does not cause frank marrow failure but leads to the appearance of clonally-restricted, inactive yet functionally competent CD8 T cells.
Keywords: Pig-a deletion, paroxysmal nocturnal hemoglobinuria, glycosyl phosphatidylinositol, T-cell mediated immunity
Introduction
Acquired somatic mutation in the X-linked phosphatidylinositol glycan class A gene (PIG-A) is required for the development of paroxysmal nocturnal hemoglobinuria (PNH):1–4 a single inactivating mutation in a hematopoietic stem cell can lead to a progeny of blood cells that lack all surface proteins utilizing a glycosyl phosphatidylinositol (GPI)-anchor motif to attach to the cell membrane.5–9 PNH is strongly associated with bone marrow failure, as underlined by deficient hematopoiesis in sensitive progenitor assays of PNH marrow,10,11 and a large proportion of patients with immune-mediated aplastic anemia have expanded PNH clones.12 Although PNH clonal expansion is associated with histocompatibility antigens13 and is predictive of hematologic response to immunosuppressive treatment in some series,14 it is still unclear how PIG-A mutations are related to the development of bone marrow failure and why clonal expansion occurs under this specific circumstance.
In order to understand the pathophysiology of PNH, efforts have been directed toward constructing animal models carrying germline deletions of the PIG-A gene. Early attempts at Pig-a knock-outs were stymied because high-contribution chimeric embryos died early in utero, while low-contribution chimeric mice survived with very small proportions of GPI-deficient (GPI−) cells.15–18 In order to avoid early embryonic lethality, conditional Pig-a gene deletion models have been generated using the Cre-lox system to target Pig-a inactivation in specific cell types.19–22 These mice were not anemic, showed a stable proportion of PNH cells, and did not develop clinical evidence of marrow aplasia or signs of PNH.19,22 Of interest, the proportion of GPI− CD4+ T cells decreased slowly over time while the proportion of GPI− CD8+ T cells increased in the peripheral blood,20 lymph nodes and spleen of these animals.19
We have produced a mouse model for conditional Pig-a gene deletion in hematopoietic cells by cross-breeding mice carrying two lox sites flanking exon 6 of Pig-a gene (Pig-aflox) with mice carrying the transgene Cre-recombinase under the human c-fes promoter (Fes-Cre).22,23 As expected, the resultant Pig-a-deficient (Pig-a−/−) mice have hematopoietic cells lacking expression of GPI-linked proteins on the cell surface. These mice had normal blood and marrow cell counts, but showed an abnormally prominent over-representation of GPI− cells in the T-cell population, especially in CD8 T cells, in hematopoietic tissues such as blood, marrow, and spleen. We, therefore, focused, our attention on the characterization of GPI− cells in this mouse model, revealing their cellular, molecular and functional properties.
Design and Methods
Animals and genotyping
The Pig-aflox mice were provided by Dr T Kinoshita (Osaka University, Japan) and the Fes-Cre transgenic mice were from Dr P Pandolfi (Memorial Sloan-Kettering Cancer Center, NY, USA). All animal study protocols were approved by the National Heart, Lung, and Blood Institute’s Animal Care and Use Committee. Genotyping analysis for Pig-a and Cre was performed by polymerase chain reaction (PCR) using specific sets of primers, as previously described.19,24
Cell analysis, sorting and transplantation
Peripheral blood was obtained by orbital sinus bleeding. Bone marrow cells were extracted from femora and tibiae. Antibodies for murine CD3 (145-2C11), CD4 (RM4-5), CD8 (5.3-6.7), CD11a (2D7), CD11b (M1/70), CD24 (M1/69), CD44 (IM7), CD45R (RA3-6B2), CD48 (HM48-1), Gr1/Ly6-G (RB6-8C5), Ter 119 (TER-119), interferon-γ (XMG1), and the T-cell receptor (TCR) Vβ screening panel were obtained from BD Biosciences (San Diego, CA, USA). Anti-FoxP3 (FJK-16s) was obtained from e-Biosciences (San Diego, CA, USA). Bone marrow cells from female Pig-a−/− donors were incubated with 10 nM aerolysin (Protox Biotech, Victoria, BC, Canada) at 37°C for 30 min.25 The residual GPI− bone marrow cells were then transplanted into lethally-irradiated B6 recipients (TR-GPI−). Bone marrow cells from Pig-a+/+ donors were used as controls (TR-GPI+).
RNA extraction and microarray analysis
Total RNA was extracted from sorted GPI+, GPI−, TR-GPI+, TR-GPI−, and wild-type(WT)-GPI+ bone marrow cells using RNeasy mini kits (Qiagen, Valencia, CA, USA). The cRNA labeling and hybridizations were performed according to Affymetrix’s protocols. The primary CEL files were deposited in the public database Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with accession number GSE14561.
CDR3 size distribution analysis (CDR3 skewing)
Sorted GPI−CD8+ and GPI+CD8+ cells from splenocytes of male Pig-a−/0 and female Pig-a+/− mice were extracted using RNAqueous-Micro kits (Ambion, Austin, TX, USA). cDNA amplification was performed by PCR with a hex-labeled constant region and Vβ 5.1 and Vβ 5.2 specific primers.26 CDR3 size distribution analysis was performed as previously described.27,28
Immunoblotting and in vitro lymphocyte cytotoxicity assay
Sorted cells were lysed and analyzed by immunoblotting in order to detect the expression levels of TCR-associated ζ-chain, Lck, and ZAP-70. Sorted GPI+ and GPI− T cells from bone marrow and splenocytes of female Pig-a−/− mice were used as effector cells in an in vitro cytotoxicity assay.
Data analysis
Microarray analysis was performed using Affymetrix mouse 430 2.0 chips. Principal component analysis, one-way analysis of variance, and post-hoc t-tests were applied to evaluate each probe set. Probesets/genes were selected with thresholds of 2-fold change, false discovery rate less than 10% and percentage of present calls using the MAS5 algorithm. JMP6 software (SAS Institute Inc., Cary, NC, USA) was used to run the hierarchical cluster and to analyze the flow cytometry data. Further details are available in the Online Supplementary Appendix.
Results
Production of conditional Pig-a-deficient mice
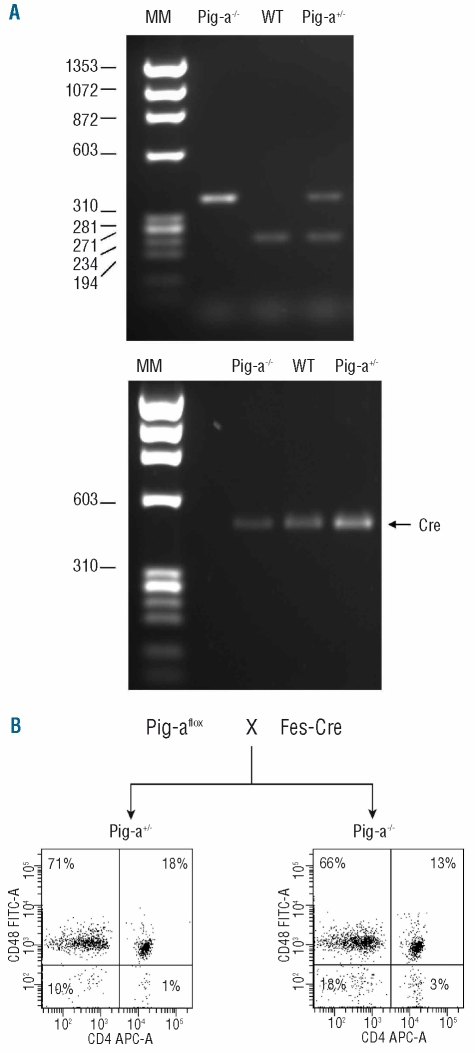
Cross-breeding of Fes-Cre and Pig-aflox mice produced Pig-a knock-out (Pig-a−/−), heterozygous (Pig-a+/−) and WT (Pig-a+/+) females as well as knock-out (Pig-a−/0) and WT (Pig-a+/0) hemizygous males. Genotype was confirmed by PCR analysis: Pig-a−/− (or Pig-a−/0) knock-out mice carried a 350 bp knock-out band, WT Pig-a+/+ (or Pig-a+/0) mice showed a 230 WT bp band, and Pig-a+/− mice displayed both bands,23 while all mice carried the Fes-Cre transgene, detectable as a 470 bp band20 (Figure 1A). Flow cytometry of peripheral blood using CD24, Gr-1 and CD48 antibodies to detect GPI-linked proteins and TER-119, CD11b, CD45R and CD4 as lineage-specific markers was used to measure the proportion of circulating GPI− cells in all animals; Figure 1B shows representative FACS dot plots for a female Pig-a−/− and a Pig-a+/− animal at 4 months of age (Figure 1B). We performed serial assessments of 66 Pig-a−/− (or Pig-a−/0) mice from 2 to 11 months of age; the proportion of GPI− cells increased over time to about 40% in T cells, 8% in B cells and 15% in red blood cells (data not shown).
Figure 1.
Generation of Pig-a−/− mice. Mice carrying the Pig-aflox construct were mated with mice carrying the Cre recombinase transgene driven by the Fes promoter to produce WT (Pig-a+/+), heterozygous (Pig-a+/−), homozygous Pig-a−/− and hemizygous (Pig-a−/0) mice. (A) PCR analysis identified a 230 bp band for WT mice and a 350 bp band for the knock-out (Pig-a−/− Pig-a, −/0) mice while heterozygous (Pig-a+/−) mice had both PCR fragments. All mice were positive for the Cre recombinase. (B) A representative Facs plot shows the comparison between a Pig-a+/− and a female Pig-a−/− animal highlighting that the leukocytes of Pig-a−/− mice contain a fraction of cells that are negative for the CD48 antigen.
Up-regulation of immune-related gene expression in glycosyl phosphatidylinositol-deficient bone marrow cells
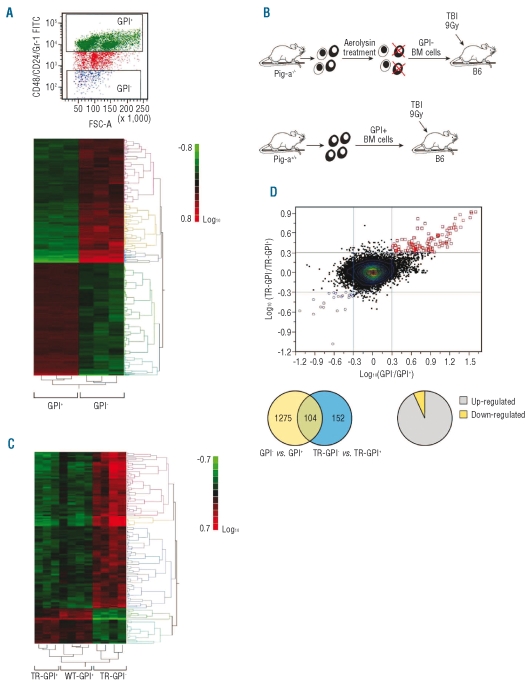
To define molecular differences between GPI− and GPI+ cells, we sorted GPI− and GPI+ bone marrow cells from assorted female Pig-a−/− and male Pig-a−/0 mice and analyzed gene expression on three GPI− and three GPI+ cell pools through microarray analysis. We found distinctive gene expression patterns with clusters of genes showing significant differential expression between the two cell types (Figure 2A). Of the 45,101 transcripts potentially available for screening, 669 genes (1275 probesets) were selected with 2-fold change in expression, 10% false discovery rate, and present calls. Using Ingenuity Software, we found that GPI-cells showed up-regulation of the expression of genes involved in major canonical pathways such as immune responses, cell proliferation, and hematologic development (data not shown).
Figure 2.
Differences between GPI− and GPI+ bone marrow (BM) cells were analyzed by microarray. (A) BM cells from Pig-a−/− mice were stained with CD24/CD48/Gr1 antibodies and sorted into [CD24/CD48/Gr1] − (GPI−) and [CD24/CD48/Gr1]+ (GPI+) cell fractions in order to apply microarray analysis. Three GPI− and GPI+ cell pools, each pool derived from three to six female Pig-a−/− and Pig-a−/0 mice, were used for RNA extraction and 500 ng RNA were used in the Mouse Genome 430 2.0 arrays as described in the Online Supplementary Data. Gene expression profile showed clusters of genes up-regulated in GPI− cells. (B) Aerolysin-treated, GPI− BM cells from Pig-a−/− and normal BM cells from Pig-a+/+ donors were transplanted into lethally irradiated (9 Gy) B6 recipients. (C) Four months later, BM cells from four recipients of GPI− donor cells (TR-GPI−), three recipients of normal GPI+ donor cells (TR-GPI+), and four B6 mice (WT-GPI+) were sorted and analyzed for gene expression by microarray. Distinctive gene expression profiles were detected between TR-GPI−, TR-GPI+ and WT-GPI+ cells showing 296 probesets (225 genes) in the comparison between TR-GPI− and WT-GPI+. (D) Overall, 152 probesets (117 genes) were selected between TR-GPI− and TR-GPI+. No essential gene expression difference was observed between TR-GPI+ and WT-GPI+ cells: 104 genes with more than 2-fold change were found to be common to the two microarray experiments.
We then questioned whether the molecular differences between GPI− and GPI+ cells would be preserved when GPI− cells achieve dominance. For this purpose, we transplanted GPI− bone marrow cells (after aerolysin treatment of bone marrow from Pig-a−/− donors) into lethally irradiated normal B6 recipients (Figure 2B). The blood cell counts of recipients of GPI− bone marrow cells were the same as those of recipients of GPI+ bone marrow cells (data not shown). Using the criteria of 2-fold change, 10% false discovery rate and present call, 296 probesets (225 genes) were selected in the comparison between TR-GPI− and WT-GPI+ of which 145 probesets were up-regulated in the TR-GPI− group. Comparison between TR-GPI− and TR-GPI+ cells found 152 probesets (117 genes) of which 124 were up-regulated in TR-GPI− (Figure 2C). The gene-expression profile for TR-GPI− cells was similar to that of untransplanted GPI− cells, showing a high level of expression of T-cell-related and immune-functional genes (Online Supplementary Table S1).
We further compared the consistency of some probe-sets/genes with more than 2-fold change from the two microarray experiments. Gene expression differences between GPI− and GPI+ cells showed very high concurrence with gene expression differences between TR-GPI− and TR-GPI+ cells (upper right and low left corners of the dot plot; Figure 2D, upper panel).
Between the two lists of differentially-expressed genes, 104 genes with more than 2-fold change were common between GPI− and TR-GPI− cells. Only six genes were down-regulated in GPI− and TR-GPI− cells (Online Supplementary Table S1 and Figure 2D, lower panel).
Enrichment of inactive CD8 T cells in glycosyl phosphatidylinositol-deficient cells
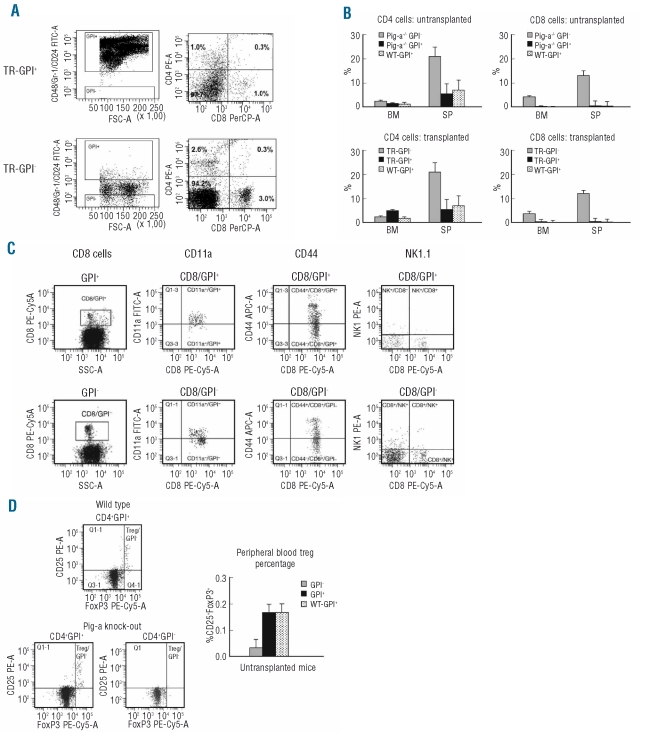
Up-regulated immune-function gene expression in GPI-cells led us to analyze CD4+ and CD8+ T-cell proportions in GPI− and GPI+ cells from female Pig-a−/− and male Pig-a−/0 mice, as well as in TR-GPI− and TR-GPI+ cells from transplant recipients, since previous studies had reported higher numbers of T lymphocytes in the GPI− cell population.19,20 FACS plots represent the difference in the proportion of CD4 and CD8 cells in transplanted mice (Figure 3A). The proportions of CD8 T cells were indeed significantly higher (P<0.01) in GPI− than in GPI+ and WT-GPI+ cells in both the bone marrow and spleen, with or without transplantation. The proportion of CD4 T cells was significantly higher (P<0.05) in GPI− cells from the spleen but not from the bone marrow when compared to their respective GPI+ and WT-GPI+ cell fractions (Figure 3B).
Figure 3.
T-cell enrichment was characterized in untransplanted and transplanted GPI-deficient bone marrow (BM) cells. (A) Facs plot presents percentage of CD4 and CD8 in peripheral blood (PB) of TR-GPI− compared to TR-GPI+ animals. (B) In the BM and splenocytes (SP), both GPI− and TR-GPI− cells contained higher proportions of CD8 T cells than did GPI+ and TR-GPI+ cells. Results are the mean ± SEM derived from the comparison of three TR-GPI− and TR-GPI+ mice. (C) Expression of CD11a, CD44 and NK 1.1 was also analyzed in GPI−CD8+ T cells and GPI+CD8+T cells in BM and PB comparing three Pig-a−/− and Pig-a−/0 with three Pig-a+/− mice. Results are represented as mean ± SEM. (D) GPI− and GPI+ cells were also analyzed for the presence of CD4+CD25+FoxP3+ Treg cells. Data shown are representative or means±SD from six animals used for the measurements.
We measured CD11a expression as a marker of T-cell activation.29 In the bone marrow, CD4+ and CD8+ T cells from GPI− and TR-GPI− cells had lower levels of CD11a expression relative to the expression on CD4+ and CD8+ T cells from GPI+ and TR-GPI+ cells. In the spleen, the proportion of CD11a+ CD8 T cells was much lower (P<0.006) in GPI− and TR-GPI− cells (7.4±3.2% and 29.5±4.0%, respectively) than in GPI+ and TR-GPI+ cells (52.8±3.2% and 55.5±4.0%, respectively), indicating that the enlarged pool of GPI− CD8 T cells was less activated than the normal T cells. We also analyzed CD44 expression on T cells and found no significant difference in the proportion of CD44+ cells in CD4 and CD8 T cells from either GPI− or GPI+ cells or WT-GPI+ cells, in either untransplanted or transplanted animals. Similarly, there was no difference in the proportions of natural killer and natural killer T cells between GPI− and GPI+ cells in peripheral blood (Figure 3C).
Another cellular change was that the proportion of CD4+CD25+FoxP3+ regulatory T (Treg) cells was significantly lower (P<0.01) in peripheral blood GPI− (0.03±3%) than in peripheral blood GPI+ (0.17±3%; P<0.04) cells (Figure 3D). The decline in Treg presence was also detected in TR-GPI− cells when compared to TR-GPI+ cells (data not shown).
Vβ clonality in glycosyl phosphatidylinositol-deficient CD8 T cells
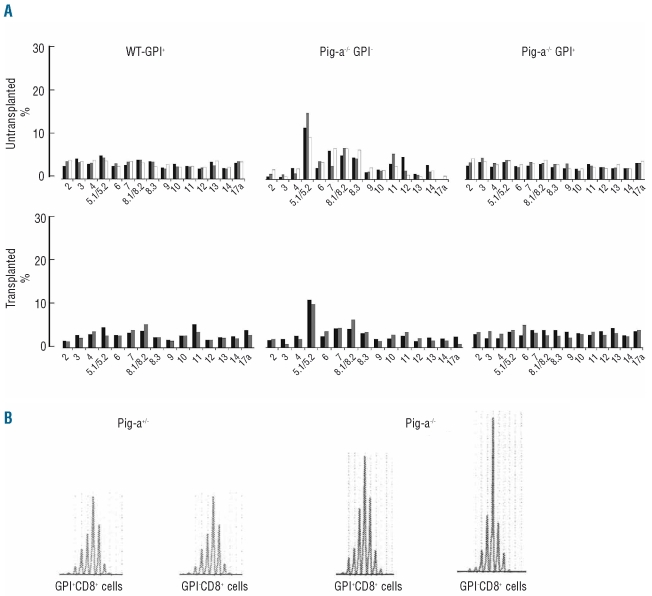
To determine T-cell clonality, we analyzed the usage of TCR β variable region in bone marrow CD8 T cells from Pig-a−/− mice, in comparison to littermate Pig-a+/+ controls. Of the 15 Vβ subfamilies analyzed, 5–6 subfamilies were over-represented in GPI− CD8 T cells. In particular, Vβ 5.1/5.2 was over-represented among GPI− CD8 T and TR-GPI− CD8 T cells, constituting an average of 22–23 ± 5%, a 2.8- and 2.5-fold increase from the 8–9.1 ± 0.5% Vβ 5.1/5.2 cells in GPI+ CD8 T cells and TR-GPI+ CD8 T cells, respectively (Figure 4A). Thus, expansion of Vβ 5.1/5.2 CD8 T cells was a shared phenotype in GPI− and TR-GPI− cells. In order to confirm clonal expansion, we isolated GPI− and GPI+ CD8 cells from male Pig-a−/0 and Pig-a+/− mice for spectratyping, which showed that the Vβ 5.1 was the specific skewed clone present in GPI− CD8 T cells of Pig-a-deficient mice (Figure 4B). We also performed sequencing of PCR products amplified using specific oligonucleotides for Vβ 5.2; absence of skewing (data not shown) confirmed the specificity for Vβ 5.1.
Figure 4.
GPI-deficient cells show T cell clonality. (A) Usage of TCR β variable region by CD8 T cells was analyzed in three WT-GPI+ from Pig-a+/+, three Pig-a−/− GPI−, two T-WT-GPI+, two TR-GPI−, and two TR-GPI+ bone marrow samples. The average of Vβ 5.1/5.2 in the GPI−CD8 T cells from Pig-a−/− mice and TR-GPI− mice was 22–23±5% compared to an average of 8–9.1±0.5% in GPI+ CD8 T cells and TR-GPI+ CD8 T cells. (B) Vβ skewing was confirmed by CDR3 analysis using specific primers for Vβ5.1 on sorted GPI−CD8+ and GPI+CD8+ cells from splenocytes of, respectively, Pig-a+/− and Pig-a−/0. The Vβ5.1 displayed 47% of total peak area in GPI−CD8+ T cells compared to 38% GPI+CD8+ from the Pig-a−/0 animal.
Functional responses in glycosyl phosphatidylinositol-deficient T cells
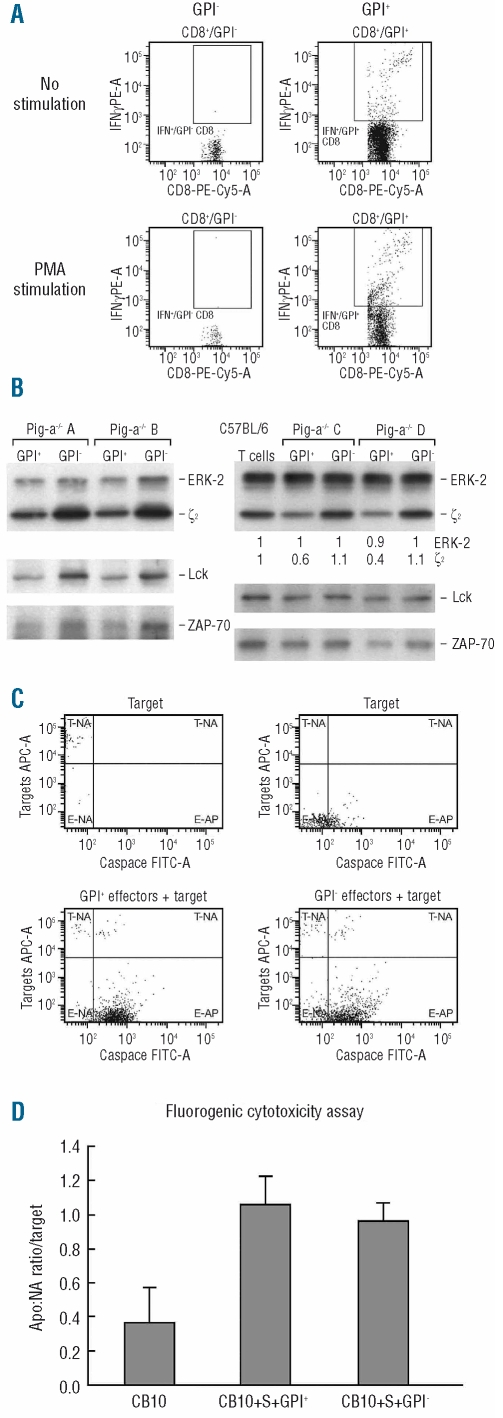
We next examined interferon-γ expression in CD4 and CD8 T cells from GPI− and TR-GPI− cell fractions from female Pig-a−/− and male Pig-a−/0 mice (Figure 5A). After 5 h of phorbol myristate acetate and ionomycin stimulation, the proportion of interferon-γ-expressing CD4 T cells was much lower in GPI− (5.9±5.1%) than in GPI+ (75±5.1%) and WT-GPI+ (49±3.2%) cells (P<0.0001). Similarly, the proportion of interferon-γ-expressing CD8 T cells was also much lower in GPI− (8.0±7.3%) than in GPI+ (76±7.3%) and WT-GPI+ (56±4.7%) cells (P<0.0001) (Figure 5A). Similar differences were observed in TR-GPI− compared to TR-GPI+ cells (data not shown).
Figure 5.
Functionality of GPI-deficient T cells was tested by flow cytometry. (A) Intracellular expression of interferon-γ was measured in CD4 and CD8 T cells from GPI− and GPI+ cells following stimulation with phorbol myristate acetate + ionomycin. (B) Sorted GPI+ T cells and GPI− T cells from Pig-a−/− mice and Pig-a−/0, as well as GPI+ T cells from WT B6 mice, were analyzed by immunoblotting to detect ζ-chain, Lck and ZAP-70 protein levels. (C) Target cell apoptosis was assessed with an in vitro CytoxiLux assay using sorted GPI− T cells and GPI+ T cells as effectors and minor-H mismatched C.B10 bone marrow cells as targets followed by co-incubation at 37°C with 5% CO2 for 60 min. (D) At a 20:1 effector: target cell ratio, both GPI− T cells and GPI+ T cells induced target cell apoptosis detected by caspase 8 activity, shown by representative dot plots and means with standard errors using cells from two donor animals each with triplicate measurements.
To study GPI− T-cell signaling characteristics and to confirm the gene expression profile data, we sorted GPI+ T cells and GPI− T cells from female Pig-a−/− and male Pig-a−/0 mice, as well as WT-GPI+ T cells from B6 mice, and analyzed the levels of TCR-associated signaling proteins by immunoblotting.30 Signals of TCR-associated ζ-chain, Lck and ZAP-70 were stronger in GPI− T cells than in GPI+ T cells from the same Pig-a−/− mice (Figure 5B, left panel). Further analysis revealed that the levels of TCR-associated ζ-chain, Lck and ZAP-70 proteins were similar between WT-GPI+ T cells and GPI− T cells, and were reduced in GPI+ T cells from Pig-a−/− mice. We detected a loss of ζ-chain without a decrease in expression of Lck and ZAP-70, suggesting that the protein loss was sequential from the ζ-chain, an active member of TCR signaling (Figure 5B, right panel).
Finally, we tested GPI− T-cell function as immune effectors by co-incubating sorted GPI− T cells and GPI+ T cells (effectors) from female Pig-a−/− mice with minor-H antigen mismatched C.B10 bone marrow cells (targets) in an in vitro CytoToxiLux assay. Both GPI− and GPI+ T cells induced the same levels of target cell apoptosis, indicating that GPI− T cells are functionally competent (Figure 5, C and D).
Discussion
Conditional deletion of the exon 6 portion of the Pig-a gene in murine hematopoietic cells resulted in an increase of GPI− cells in lymphohematopoietic tissues. The GPI− cell clone was small in size, enlarged gradually over time, but did not become dominant. Among the different cell lineages, the percentage of GPI− cells was lower in erythrocytes, granulocytes, and B cells but much higher in T lymphocytes. The presence of GPI− cells was not associated with obvious hematologic abnormalities since blood and bone marrow cell counts were normal, as in other Pig-a gene deletion models studied under different conditions in which the exon 2 portion of the Pig-a gene was deleted.15,17,19,20
Murakami et al. reported that, in their Pig-a knock-out model, GPI− cells dominated lymphoid organs and the proportions of GPI− cells were similarly high among CD4−CD8− double negative cells, CD44+CD25− cells, and in later stages of T-cell development,19 from which the authors inferred a role for GPI-anchored proteins in lymphocyte ontogeny, possibly leading to escape from negative selection in the thymus.20 Our results are consistent with other reports indicating that deletion of either exon 2 or 6 of the Pig-a gene causes expansion of T cells in the GPI− cell population. This finding, however, differs from observations in PNH patients showing higher numbers of GPI− granulocytes and red blood cells but lower and variable numbers of GPI− T cells. We speculate that other factors in addition to Pig-a deficiency may contribute to the manifestation of clinical PNH in humans, factors that may not be present in the Pig-a deletion mouse models, resulting in the accumulation of GPI− T cells instead of GPI− granulocytes and red cells. Unlike PNH patients, Pig-a deletion mouse models do not, of course, manifest clinical PNH.
We further found from our study that the enlarged population of GPI−CD8 T cells was naive and inactive, based on the expression of CD44 and CD11a. These GPI− T cells were not natural killer T cells, since they did not express NK1.1. Unlike mice carrying a FoxP3 gene deletion which develop a fatal autoimmune lymphoproliferative syndrome,31 the conditional Pig-a knock-out mice, despite having much reduced CD4+CD25+FoxP3+ Treg cells in the GPI− T-cell population, developed no obvious signs of autoimmunity. The role of regulatory T cells in the development of PNH is unknown. The levels of regulatory T cells are low in aplastic anemia.32 A decline in regulatory T cells might contribute to promote the selective damage to normal GPI+, but not GPI−, hematopoietic cells potentially mediated by an autoimmune attack.33 However, in our Pig-a deficient mouse model, inadequate effector T-cell activation may account for the lack of autoimmunity and bone marrow failure despite reduced presence of regulatory T cells.
The expression of T-cell-related immune response genes, including genes important for TCR signaling, was up-regulated in GPI− cells. This characteristic was retained when Pig-a knock-out bone marrow cells were treated with aerolysin to enrich for GPI− cells34,25 and then engrafted into lethally irradiated recipients. There was no consistent difference in gene expression between TR-GPI+ and WT-GPI+ bone marrow cells, suggesting that the transplantation procedure itself caused no alteration in immune function gene expression. Immunoblotting also revealed that the protein levels of ζ-chain, Lck, and ZAP-70 were lower in GPI+ T cells than in GPI− T cells from the same Pig-a knock-out mice. In human GPI-deficient T cells derived from PNH patients, the expression of TCR signaling proteins was not different when GPI− and GPI+ cells were stimulated in vitro.35–37 This discrepancy might be due to the different experimental systems used in the studies, as we analyzed gene expression on primary cells without stimulation. In our study, the protein levels of ζ-chain, Lck and ZAP-70 were normal in GPI− T cells and comparable to those in WT-GPI+ T cells from normal B6 mice, but they were reduced in GPI+ T cells from Pig-a knock-out mice. Expanded GPI− CD8 T cells were naive and inactive, in agreement with the findings of human studies that demonstrated impairment in TCR engagements leading to failure to phosphorylate tyrosine kinases35 and a reduced response to lectin stimulation in GPI− T cells from patients with PNH.37 The pattern of changes in ζ-chain, Lck, and ZAP-70 in our model was similar to that of the alterations observed in T cells isolated from individuals infected by human immunodeficiency virus.38 While the mechanism of human immunodeficiency virus-induced modulation of signaling proteins is not known, the extent of changes/number of T cells affected suggests that the effect is indirect. Perturbation of protein expression detected in GPI+ T cells isolated from Pig-a knock-out mice suggested a possible “bystander effect” mediated by activities of GPI− T cells to induce down-regulation of some of the TCR proximal signaling molecules in GPI+ T cells.
Of particular interest was the oligoclonality of GPI− CD8 T cells from both untransplanted and transplanted animals. Approximately 5–6 Vβ subfamilies were over-represented in GPI− CD8+ T cells, with a marked increase in Vβ 5.1/5.2. Immunoscope analysis confirmed that the Vβ 5.1 clone had a single prominent peak. Skewing was restricted to GPI−CD8 T cells and not present in either GPI+ CD8 T cells derived from the same Pig-a knock-out mice or in GPI+ and GPI− cells from Pig-a+/− animals. In humans, oligoclonal expansion of cytotoxic T cells with highly homologous TCR-β molecules suggests an autoimmune process linked to the pathogenesis of PNH.33 The nature of the target antigens for autoreactive T cells is unknown, but presumably these antigens are expressed on phenotypically normal GPI+ hematopoietic stem cells.33,39–41 We postulate that reduced T-cell activation and decreased response to stimulation-related interferon-γ in GPI− cells would explain why the enlarged pool of clonally-skewed GPI− CD8 T cells did not cause clinical bone marrow failure and PNH in Pig-a deficient mice, even when the proportion of immunosuppressive Treg cells was much reduced. GPI− CD8 T cells are functionally competent and might express their killing activity under some conditions. In most patients with PNH, bone marrow failure is believed to be caused by immune-mediated injury of hematopoietic progenitor cells, as observed in aplastic anemia.34 The bone marrow failure environment is also thought to be conducive to the selective expansion of PIG-A mutant hematopoietic stem cell clones. If stress or injury signals produced in the damaged tissues are needed to activate the primary immune response,42 the lack of danger signals from tissue damage in Pig-a knock-out mice might also explain the inactive status of expanded oligoclonal CD8 T cells.
The mechanism of PNH clonal expansion remains unclear. One theory postulates that PNH cells might escape an immune attack because one or more of the missing GPI-anchor proteins could be the immune target. The association of expanded PNH clones with HLA-DR213 and of PNH clones with an increased level of auto-antibodies14 serving as predictors of responsiveness to immunosuppressive therapies43 are compatible with such a selective clonal expansion mechanism. The immunoselection of PIG-A mutant cells was demonstrated in the emergence of PNH lymphocytes in patients with non-Hodgkin’s lymphoma who had been treated with antibodies against the GPI-linked lymphocyte antigen CD52.44,45 However, a more recent clinical study found no evidence of decreased sensitivity to T-cell-mediated immune attack in PNH cells after non-myeloablative allogeneic hematopoietic stem cell transplantation.46 A comparative survival advantage of the PNH cells might be due to the decreased survival of phenotypically normal cells, since GPI-anchored protein-deficient CD34+ cells from PNH patients had a similar rate of proliferation when compared to normal CD34+ progenitors.47 Gene expression analysis of CD34+ cells also revealed that PNH cells from PNH patients shared a similar gene expression profile with CD34+ cells from normal volunteers, while the normal cells from PNH patients showed down-regulation of anti-apoptotic genes.48 PIG-A mutation in leukemic cells appeared to confer insensitivity to killing by natural killer cells because of the absence of stress inducible membrane proteins, such as UL 16 binding proteins (ULBP).49,50 In cells lines deficient in GPI-anchored proteins, lipid rafts in GPI− cells contained important anti-apoptotic proteins not present in the lipid rafts of GPI+ cells, enabling GPI− cells to avert cell death.51 An alternative theory to explain PNH clonal expansion is that the PIG-A mutation causes intrinsic defects that confer resistance to apoptosis. In vitro, GPI-deficient cell lines exhibited resistance to apoptosis when effectors were raised against GPI-deficient cells, arguing against a GPI-anchored protein being the possible target of the immune attack in PNH.52
In our mouse model, conditional Pig-a gene deletion in hematopoietic cells led to enrichment of T cells that lacked GPI and GPI-anchored proteins, especially oligoclonal CD8 T cells, in hematopoietic tissues. The reduced activation, lower interferon-γ production, and absence of danger signals might explain why these skewed GPI−CD8 T cells did not cause detectable marrow failure, autoimmunity, PNH or other abnormalities, despite reduced immunosuppressive Treg cells.
Acknowledgments
we thank Irena Stefanova of the National Institute of Allergy and Infection Disease, National Institutes of Health (NIH) for performing the protein analysis and providing helpful comments; Rodrigo T. Calado of the Hematology Branch, National Heart, Lung, and Blood Institute, NIH, for technical help in the cell transplantation study and for valuable discussions; and Regis P. de Latour of the Hematology Branch, National Heart, Lung, and Blood Institute, NIH, for providing important advice and helpful comments. We also thank F.M. Ellison of the Food and Drug Administration for expert technical assistance.
Footnotes
Funding: this work was supported by the Intramural Research Program of the National Institutes of Health.
The online version of this paper has a supplementary appendix.
Authorship and Disclosures
VV designed the study, executed the experiments, collected and analyzed the data, and drafted the manuscript; NR performed the microarray gene expression experiments; DL analyzed the array data; KK sorted cells; MJD maintained the animal colony and performed experiments; JC and NSY designed the study and edited the paper.
The authors reported no potential conflicts of interest.
References
- 1.Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108(8):2509–19. doi: 10.1182/blood-2006-03-010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luzzatto L. Paroxysmal nocturnal hemoglobinuria: an acquired X-linked genetic disease with somatic-cell mosaicism. Curr Opin Genet Dev. 2006;16(3):317–22. doi: 10.1016/j.gde.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Takeda J, Miyata T, Kawagoe K, Iida Y, Endo Y, Fujita T, et al. Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell. 1993;73(4):703–11. doi: 10.1016/0092-8674(93)90250-t. [DOI] [PubMed] [Google Scholar]
- 4.Parker CJ. The pathophysiology of paroxysmal nocturnal hemoglobinuria. Exp Hematol. 2007;35(4):523–33. doi: 10.1016/j.exphem.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Ware RE, Howard TA, Kamitani T, Change HM, Yeh ET, Seldin MF. Chromosomal assignment of genes involved in glycosylphosphatidylinositol anchor biosynthesis: implications for the pathogenesis of paroxysmal nocturnal hemoglobinuria. Blood. 1994;83(12):3753–7. [PubMed] [Google Scholar]
- 6.Kinoshita T, Inoue N, Takeda J. Role of phosphatidylinositol-linked proteins in paroxysmal nocturnal hemoglobinuria pathogenesis. Annu Rev Med. 1996;47:1–10. doi: 10.1146/annurev.med.47.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Davitz MA, Low MG, Nussenzweig V. Release of decay-accelerating factor (DAF) from the cell membrane by phosphatidylinositol-specific phospholipase C (PIPLC). Selective modification of a complement regulatory protein. J Exp Med. 1986;163(5):1150–61. doi: 10.1084/jem.163.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessler M, Mason PJ, Hillmen P, Miyata T, Yamada N, Takeda J, et al. Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO J. 1994;13(1):110–7. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosse WF, Ware RE. The molecular basis of paroxysmal nocturnal hemoglobinuria. Blood. 1995;86(9):3277–86. [PubMed] [Google Scholar]
- 10.Rotoli B, Robledo R, Luzzatto L. Decreased number of circulating BFU-Es in paroxysmal nocturnal hemoglobinuria. Blood. 1982;60(1):157–9. [PubMed] [Google Scholar]
- 11.Moore JG, Humphries RK, Frank MM, Young N. Characterization of the hematopoietic defect in paroxysmal nocturnal hemoglobinuria. Exp Hematol. 1986;14(3):222–9. [PubMed] [Google Scholar]
- 12.Dunn DE, Tanawattanacharoen P, Boccuni P, Nagakura S, Green SW, Kirby MR, et al. Paroxysmal nocturnal hemoglobinuria cells in patients with bone marrow failure syndromes. Ann Intern Med. 1999;131(6):401–8. doi: 10.7326/0003-4819-131-6-199909210-00002. [DOI] [PubMed] [Google Scholar]
- 13.Maciejewski JP, Follmann D, Nakamura R, Saunthararajah Y, Rivera CE, Simonis T, et al. Increased frequency of HLA-DR2 in patients with paroxysmal nocturnal hemoglobinuria and the PNH/aplastic anemia syndrome. Blood. 2001;98(13):3513–9. doi: 10.1182/blood.v98.13.3513. [DOI] [PubMed] [Google Scholar]
- 14.Sugimori C, Chuhjo T, Feng X, Yamazaki H, Takami A, Teramura M, et al. Minor population of CD55-CD59- blood cells predicts response to immunosuppressive therapy and prognosis in patients with aplastic anemia. Blood. 2006;107(4):1308–14. doi: 10.1182/blood-2005-06-2485. [DOI] [PubMed] [Google Scholar]
- 15.Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, et al. Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood. 1996;87(9):3600–6. [PubMed] [Google Scholar]
- 16.Dunn DE, Yu J, Nagarajan S, Devetten M, Weichold FF, Medof ME, et al. A knock-out model of paroxysmal nocturnal hemoglobinuria: Pig-a(−) hematopoiesis is reconstituted following intercellular transfer of GPI-anchored proteins. Proc Natl Acad Sci USA. 1996;93(15):7938–43. doi: 10.1073/pnas.93.15.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosti V, Tremml G, Soares V, Pandolfi PP, Luzzatto L, Bessler M. Murine embryonic stem cells without pig-a gene activity are competent for hematopoiesis with the PNH phenotype but not for clonal expansion. J Clin Invest. 1997;100(5):1028–36. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luzzatto L. Paroxysmal murine hemoglobinuria(¿): a model for human PNH. Blood. 1999;94(9):2941–4. [PubMed] [Google Scholar]
- 19.Murakami Y, Kinoshita T, Maeda Y, Nakano T, Kosaka H, Takeda J. Different roles of glycosylphosphatidylinositol in various hematopoietic cells as revealed by a mouse model of paroxysmal nocturnal hemoglobinuria. Blood. 1999;94(9):2963–70. [PubMed] [Google Scholar]
- 20.Tremml G, Dominguez C, Rosti V, Zhang Z, Pandolfi PP, Keller P, et al. Increased sensitivity to complement and a decreased red blood cell life span in mice mosaic for a nonfunctional Piga gene. Blood. 1999;94(9):2945–54. [PubMed] [Google Scholar]
- 21.Feldman RA, Gabrilove JL, Tam JP, Moore MA, Hanafusa H. Specific expression of the human cellular fps/fes-encoded protein NCP92 in normal and leukemic myeloid cells. Proc Natl Acad Sci USA. 1985;82(8):2379–83. doi: 10.1073/pnas.82.8.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller P, Payne JL, Tremml G, Greer PA, Gaboli M, Pandolfi PP, et al. FES-Cre targets phosphatidylinositol glycan class A (PIGA) inactivation to hematopoietic stem cells in the bone marrow. J Exp Med. 2001;194(5):581–9. doi: 10.1084/jem.194.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarutani M, Itami S, Okabe M, Ikawa M, Tezuka T, Yoshikawa K, et al. Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc Natl Acad Sci USA. 1997;94:7400–5. doi: 10.1073/pnas.94.14.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller P, Tremml G, Rosti V, Bessler M. X inactivation and somatic cell selection rescue female mice carrying a Piga-null mutation. Proc Natl Acad Sci USA. 1999;96(13):7479–83. doi: 10.1073/pnas.96.13.7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodsky RA, Mukhina GL, Nelson KL, Lawrence TS, Jones RJ, Buckley JT. Resistance of paroxysmal nocturnal hemoglobinuria cells to the glycosylphosphatidylinositol-binding toxin aerolysin. Blood. 1999;93(5):1749–56. [PubMed] [Google Scholar]
- 26.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci USA. 1993;90(9):4319–23. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JA, Rao P, Graor H, Rothchild K, O’Keefe C, Maciejewski JP. CDR3 spectratyping identifies clonal expansion within T-cell subpopulations that demonstrate therapeutic antitumor activity. Surgery. 2004;136(2):295–302. doi: 10.1016/j.surg.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Risitano AM, Kook H, Zeng W, Chen G, Young NS, Maciejewski JP. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V beta CDR3 spectratyping and flow cytometry. Blood. 2002;100(1):178–83. doi: 10.1182/blood-2002-01-0236. [DOI] [PubMed] [Google Scholar]
- 29.Choi EY, Christianson GJ, Yoshimura Y, Sproule TJ, Jung N, Joyce S, et al. Immunodominance of H60 is caused by an abnormally high precursor T cell pool directed against its unique minor histocompatibility antigen peptide. Immunity. 2002;17(5):593–603. doi: 10.1016/s1074-7613(02)00428-4. [DOI] [PubMed] [Google Scholar]
- 30.Burkhardt AL, Stealey B, Rowley RB, Mahajan S, Prendergast M, Fargnoli J, et al. Temporal regulation of non-transmembrane protein tyrosine kinase enzyme activity following T cell antigen receptor engagement. J Biol Chem. 1994;269(38):23642–7. [PubMed] [Google Scholar]
- 31.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–7. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 32.Solomou EE, Rezvani K, Mielke S, Malide D, Keyvanfar K, Visconte V, et al. Deficient CD4+ CD25+ FOXP3+ T regulatory cells in acquired aplastic anemia. Blood. 2007;110(5):1603–6. doi: 10.1182/blood-2007-01-066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gargiulo L, Lastraioli S, Cerruti G, Serra M, Loiacono F, Zupo S, et al. Highly homologous T-cell receptor beta sequences support a common target for autoreactive T cells in most patients with paroxysmal nocturnal hemoglobinuria. Blood. 2007;109(11):5036–42. doi: 10.1182/blood-2006-10-052381. [DOI] [PubMed] [Google Scholar]
- 34.Brodsky RA. Advances in the diagnosis and therapy of paroxysmal nocturnal hemoglobinuria. Blood Rev. 2008;22(2):65–74. doi: 10.1016/j.blre.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romagnoli P, Bron C. Phosphatidylinositol-based glycolipid-anchored proteins enhance proximal TCR signaling events. J Immunol. 1997;158(12):5757–64. [PubMed] [Google Scholar]
- 36.Romagnoli P, Bron C. Defective TCR signaling events in glycosylphosphatidylinositol-deficient T cells derived from paroxysmal nocturnal hemoglobinuria patients. Int Immunol. 1999;11(9):1411–22. doi: 10.1093/intimm/11.9.1411. [DOI] [PubMed] [Google Scholar]
- 37.Schubert J, Uciechowski P, Zielinska-Skowronek M, Tietjen C, Leo R, Schmidt RE. Differences in activation of normal and glycosylphosphatidylinositol-negative lymphocytes derived from patients with paroxysmal nocturnal hemoglobinuria. J Immunol. 1992;148(12):3814–9. [PubMed] [Google Scholar]
- 38.Stefanova I, Saville MW, Peters C, Cleghorn FR, Schwartz D, Venzon DJ, et al. HIV infection--induced posttranslational modification of T cell signaling molecules associated with disease progression. J Clin Invest. 1996;98(6):1290–7. doi: 10.1172/JCI118915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plasilova M, Risitano A, Maciejewski JP. Application of the molecular analysis of the T-cell receptor repertoire in the study of immune-mediated hematologic diseases. Hematology. 2003;8(3):173–81. doi: 10.1080/1024533031000107505. [DOI] [PubMed] [Google Scholar]
- 40.Zeng W, Maciejewski JP, Chen G, Young NS. Limited heterogeneity of T cell receptor BV usage in aplastic anemia. J Clin Invest. 2001;108(5):765–73. doi: 10.1172/JCI12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young NS. The problem of clonality in aplastic anemia: Dr Dameshek’s riddle, restated. Blood. 1992;79(6):1385–92. [PubMed] [Google Scholar]
- 42.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296(5566):301–5. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 43.Feng X, Chuhjo T, Sugimori C, Kotani T, Lu X, Takami A, et al. Diazepam-binding inhibitor-related protein 1: a candidate autoantigen in acquired aplastic anemia patients harboring a minor population of paroxysmal nocturnal hemoglobinuria-type cells. Blood. 2004;104(8):2425–31. doi: 10.1182/blood-2004-05-1839. [DOI] [PubMed] [Google Scholar]
- 44.Rawstron A, Barrans S, Blythe D, Davies F, English A, Pratt G, et al. Distribution of myeloma plasma cells in peripheral blood and bone marrow correlates with CD56 expression. Br J Haematol. 1999;104(1):138–43. doi: 10.1046/j.1365-2141.1999.01134.x. [DOI] [PubMed] [Google Scholar]
- 45.Hertenstein B, Wagner B, Bunjes D, Duncker C, Raghavachar A, Arnold R, et al. Emergence of CD52-, phosphatidylinositolglycan-anchor-deficient T lymphocytes after in vivo application of Campath-1H for refractory B-cell non-Hodgkin lymphoma. Blood. 1995;86(4):1487–92. [PubMed] [Google Scholar]
- 46.Takahashi Y, McCoy JP, Jr, Carvallo C, Rivera C, Igarashi T, Srinivasan R, et al. In vitro and in vivo evidence of PNH cell sensitivity to immune attack after nonmyeloablative allogeneic hematopoietic cell transplantation. Blood. 2004;103(4):1383–90. doi: 10.1182/blood-2003-04-1281. [DOI] [PubMed] [Google Scholar]
- 47.Chen G, Kirby M, Zeng W, Young NS, Maciejewski JP. Superior growth of glycophosphatidy linositol-anchored protein-deficient progenitor cells in vitro is due to the higher apoptotic rate of progenitors with normal phenotype in vivo. Exp Hematol. 2002;30(7):774–82. doi: 10.1016/s0301-472x(02)00811-1. [DOI] [PubMed] [Google Scholar]
- 48.Chen G, Zeng W, Maciejewski JP, Kcyvanfar K, Billings EM, Young NS. Differential gene expression in hematopoietic progenitors from paroxysmal nocturnal hemoglobinuria patients reveals an apoptosis/immune response in ‘normal’ phenotype cells. Leukemia. 2005;19(5):862–8. doi: 10.1038/sj.leu.2403678. [DOI] [PubMed] [Google Scholar]
- 49.Nagakura S, Ishihara S, Dunn DE, Nishimura J, Kawaguchi T, Horikawa K, et al. Decreased susceptibility of leukemic cells with PIG-A mutation to natural killer cells in vitro. Blood. 2002;100(3):1031–7. doi: 10.1182/blood.v100.3.1031. [DOI] [PubMed] [Google Scholar]
- 50.Hanaoka N, Kawaguchi T, Horikawa K, Nagakura S, Mitsuya H, Nakakuma H. Immunoselection by natural killer cells of PIGA mutant cells missing stress-inducible ULBP. Blood. 2006;107(3):1184–91. doi: 10.1182/blood-2005-03-1337. [DOI] [PubMed] [Google Scholar]
- 51.Szpurka H, Schade AE, Jankowska AM, Maciejewski JP. Altered lipid raft composition and defective cell death signal transduction in glycosylphosphatidylinositol anchor-deficient PIG-A mutant cells. Br J Haematol. 2008;142(3):413–22. doi: 10.1111/j.1365-2141.2008.07203.x. [DOI] [PubMed] [Google Scholar]
- 52.Savage WJ, Barber JP, Mukhina GL, Hu R, Chen G, Matsui W, et al. Glycosylphosphatidylinositol-anchored protein deficiency confers resistance to apoptosis in PNH. Exp Hematol. 2009;37(1):42–51. doi: 10.1016/j.exphem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]