Abstract
Background
The histone deacetylase inhibitor ITF2357 has potent cytotoxic activity in multiple myeloma in vitro and has entered clinical trials for this disease.
Design and Methods
In order to gain an overall view of the activity of ITF2357 and identify specific pathways that may be modulated by the drug, we performed gene expression profiling of the KMS18 multiple myeloma cell line treated with the drug. The modulation of several genes and their biological consequence were verified in a panel of multiple myeloma cell lines and cells freshly isolated from patients by using polymerase chain reaction analysis and western blotting.
Results
Out of 38,500 human genes, we identified 140 and 574 up-regulated genes and 102 and 556 down-modulated genes at 2 and 6 h, respectively, with a significant presence of genes related to transcription regulation at 2 h and to cell cycling and apoptosis at 6 h. Several of the identified genes are particularly relevant to the biology of multiple myeloma and it was confirmed that ITF2357 also modulated their encoded proteins in different multiple myeloma cell lines. In particular, ITF2357 down-modulated the interleukin-6 receptor α (CD126) transcript and protein in both cell lines and freshly isolated patients’ cells, whereas it did not significantly modify interleukin-6 receptor β (CD130) expression. The decrease in CD126 expression was accompanied by decreased signaling by interleukin-6 receptor, as measured by STAT3 phosphorylation in the presence and absence of inter-leukin-6. Finally, the drug significantly down-modulated the MIRHG1 transcript and its associated microRNA, miR-19a and miR-19b, known to have oncogenic activity in multiple myeloma.
Conclusions
ITF2357 inhibits several signaling pathways involved in myeloma cell growth and survival.
Keywords: pleiotropic anti-myeloma, ITF2357, interleukin-6 receptor, miR-19a, miR-19b
Introduction
Multiple myeloma (MM) is an incurable B-cell malignancy caused by a clonal proliferation of terminally differentiated plasma cells. Over the last years the search for new agents that specifically target pathways relevant to plasma cell survival and proliferation has gained impulse from novel insights into the pathogenesis of MM, including the role of specific growth factors such as interleukin (IL)-6 and insulin-like growth factor 1 (IGF-1), of growth factor receptors such as fibroblast growth factor receptor 3 (FGFR3), the influence of the microenvironment and the dysregulation of various signal transduction pathways and cell cycling.1 More recently, an important role for specific microRNA (miRNA), in particular the miR-17~92 cluster, in B-cell neoplasms including MM, has gained much support.2–5
Gene regulation has been recognized to depend not only on transcription factor activation but also on the specific pattern of chromatin modifications, including histone acetylation, methylation, phosphorylation and sumoylation, present in the genome of each cell.6 These patterns regulate complex biological processes, including cell growth, apoptosis and immune responses. Histone deacetylases (HDAC) are a family of at least 18 enzymes involved in the regulation of the acetylation of histones as well as other cellular proteins. HDAC inhibitors have emerged as promising molecules for the treatment of cancer, including MM, as well inflammatory diseases.7–10
ITF2357 is a recently identified HDAC inhibitor of the hydroxamate family, which inhibits both class I and class II HDAC.11 ITF2357 induces apoptosis of MM and acute myelogenous leukemia (AML) cells and this apoptosis takes place following induction of p21 and down-modulation of Bcl-2 and Mcl-1 proteins.12 ITF2357 has also shown anti-tumor activity against hepatoma cells in vitro.13 Finally, in vitro ITF2357 is a particularly potent inhibitor of hematopoietic colony formation by JAK2V617F-bearing progenitor cells from patients with chronic myeloproliferative neoplasms.14 When tested on normal cells, ITF2357 inhibits the production of pro-inflammatory cytokines such as IL-1, IL-6, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ by peripheral blood mononuclear cells11 and of IL-6 and vascular endothelium growth factor (VEGF) by mesenchymal stromal cells.12 The pro-apoptotic activities of the drug, as well as its capacity to inhibit IL-6 and VEGF production, suggest that it could be a useful compound for the treatment of MM and leukemias. Indeed, it has entered phase II clinical trials for the treatment of MM, AML, Hodgkin’s lymphoma, as well as JAK2V617F-positive myeloproliferative neoplasms (www.clinicaltrials.gov). On the other hand, the drug’s capacity to inhibit pro-inflammatory cytokines has led to it being tested in clinical trials on inflammatory diseases such as idiopathic arthritis and Crohn’s disease.
Although some of the effects of ITF2357 at the molecular level have already been identified, such as p21 induction and down-modulation of Bcl2 and Mcl1 expression,12 an overall analysis of the genes whose expression is modified by ITF2357 has not yet been performed. We, therefore, planned genome-wide screening of transcripts that are induced or repressed by ITF2357 treatment. We chose a human MM cell line as the target and relatively early time points, 2 and 6 h, in order to identify the first gene subsets induced or repressed by the drug.
Design and Methods
Reagent and cells
ITF2357 (diethyl-[6-(4-hydroxycarbamoyl-phenyl carbamoy-loxymethyl)-naphthalen-2-yl methyl]-ammonium chloride; monohydrate; Italfarmaco, patent WO 97/43251, US 6034096) was synthesized by Italfarmaco (Milan, Italy) and its purity confirmed by high performance liquid chromatography.
All MM cell lines were grown in Iscove’s modified Dulbecco’s medium (Cambrex Bio Science, Verviers, Belgium) with 10% fetal calf serum (Euroclone, Wetherby, West Yorkshire, UK), 2 mM glutamine (Euroclone) and 110 μM gentamycin (PHT Pharma, Milan, Italy). In the case of the IL-6-dependent line CMA-03, 10 ng/mL recombinant human IL-6 (Biosource International) was added. Mononuclear cells were isolated after informed consent from the bone marrow or peripheral blood of MM or plasmacytoma patients, respectively, and plasma cells purified (≥90% purity) by positive selection with anti-CD138 magnetic beads (Miltenyi Biotec, San Diego, CA, USA).
Gene expression profiling and microarray data analysis
Exponentially growing KMS18 cells were cultured in the presence or absence of 0.25 μM ITF2357 in triplicate flasks for 2 or 6 h. After washing, cell pellets were resuspended in TRIzol Reagent (Life Technologies, Inc., Rockville, MD, USA). Total RNA was then purified using the RNeasy® total RNA Isolation Kit (Qiagen, Valencia, CA, USA). Preparation of biotin-labeled cRNA, hybridization to GeneChip® Human Genome U133 Plus 2.0 Arrays (Affymetrix Inc., Santa Clara, CA) and scanning of the chips (7G Scanner, Affymetrix Inc.) were carried out according to the manufacturers’ protocols. The images were processed and array normalization was performed using Affymetrix® GeneChip Operating Software (GCOS) 1.4. The supervised gene expression analyses were performed using the Gene@Work software platform as previously described.15 Full support and a δ value of 0.0001 were chosen for the analysis. Average fold change (FC) and zg score thresholds were fixed at 2 and 2.5, respectively, and were expressed using the untreated group as baseline. The selected probe list was visualized by means of DNAChip Analyzer (dChip) software.16 The functional analysis of the selected lists was conducted using NetAffx (Affymetrix at https://www.affymetrix.com/analysis/netaffx/) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) Tool 2008 (U.S. National Institutes of Health at http://david.abcc.ncifcrf.gov/), using the functional annotation clustering tool. Only Gene Ontology, Biological Process, and Molecular Function terms were selected as annotation categories and high classification stringency was set for the analysis.
Quantitative real-time polymerase chain reaction analysis
For measurements of p21, CD126 and SDC1, cDNA was synthesized using 500 ng of purified RNA and the Superscript Reverse Transcriptase II (Invitrogen) followed by RNase H treatment. Real-time polymerase chain reaction (PCR) analysis was performed using the standard primer sets from SABiosciences (Frederick, MD, USA). Amplification was performed on an iCycler iQ system using the iQ SYBR Green PCR Supermix (Bio-Rad). The relative expression of genes in treated and untreated samples was calculated using 18S rRNA as the normalization gene.
To measure the expression of miR-19a and miR-19b, total RNA was extracted with TRIzol Reagent (Life Technologies) and not further purified. Total RNA (50 ng) was reverse-transcribed and then subjected to real-time PCR using reagents from the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and TaqMan® MicroRNA Assays. RNA samples were normalized on the basis of the Z30 TaqMan® MicroRNA Assays-Control. MIRHG1 expression was analyzed as described elsewhere,17 using 50 ng of cDNA equivalents and gene-specific primers and probe: forward 5′-TTTTTACTAATTTTGTGTACTTTTATTGTGTCGATGT-3′, reverse 5′-CACTTTAGTGCTACAGAAGCTGTCA-3′, probe FAM-ACCAGGCAGATTCT-NFQ (Custom TaqMan® Gene Expression Assay, Applied Biosystems). A GAPDH-specific Pre-Developed Assay Reagent (Applied Biosystems) was used as the internal control. Quantitative real time PCR (Q-RT-PCR) amplifications were performed on an Applied Biosystems 7900 Sequence Detection System, and run in duplicate from each reverse transcriptase reaction. miRNA and gene expression were quantified relatively using the 2−ΔΔCt method (Applied Biosystems User Bulletin No. 2), and expressed as the amount of target transcript, normalized to the housekeeping gene and relative to the untreated sample.
Immunofluorescence by flow cytometry
Cells were plated at 0.5×106/mL (cell lines) or 1×106/mL (patients’ samples) in the presence or absence of 0.1–0.5 μM ITF2357. After 24 h of culture at 37 °C, the cells were collected, washed and stained with phycoerythrin (PE)- or fluorescein isothiocyanate (FITC)-conjugated antibodies: CD126-PE, CD130-PE, CD28-PE, CD40-FITC and CD138-FITC (BD Biosciences, San José, CA, USA) or appropriate isotype controls. For MHC class I chain-related protein A (MICA), unconjugated mouse monoclonal antibody was used (R&D Systems, Minneapolis, MN, USA) with FITC labeled secondary antibody (BD Biosciences). Stained cells were analyzed using a FACScan Instrument, collecting at least 5000 events/sample using Cell-Quest software (BD Biosciences). Dead cells were excluded from all the analyses from the forward and side scatter plots.
Western blotting
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting were performed according to standard procedures using total cellular extracts. The following antibodies were used after blocking in 5% non-fat milk: polyclonal anti-pSTAT3 (Tyr705), STAT3 (Cell Signaling Technology Inc., Danvers, MA, USA), FGFR3 and actin (Santa Cruz, Heidelberg, Germany). Signals were detected by chemilumiscence using ECL (Amersham-Pharmacia Biotech, Cologno Monzese, Italy).
Statistical analyses
Statistical significances were calculated using the Student’s t test.
Results
Genome wide analysis of genes modulated by ITF2357 in the KMS18 multiple myeloma cell line
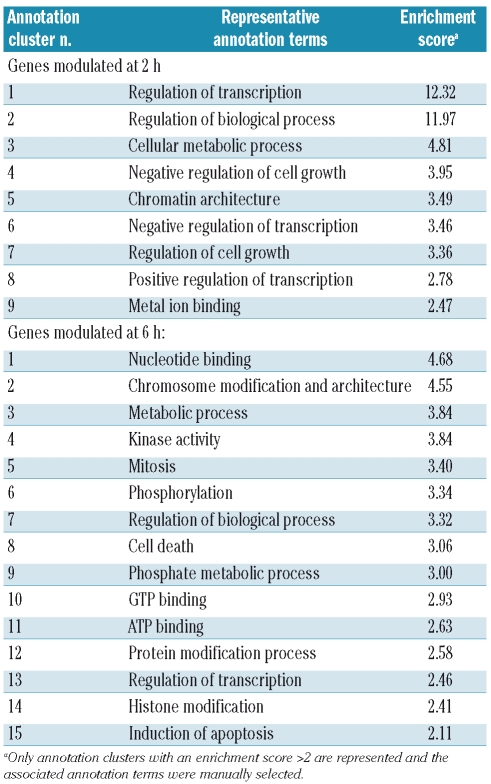
In order to identify genes whose expression was modulated early by ITF2357 in MM cells, we performed gene expression profiling on the KMS18 cell line treated for 2 and 6 h with 0.25 μM ITF2357. A total of 410 probe sets, corresponding to 140 up-regulated (Online Supplementary Table S1) and 102 down-regulated genes (Online Supplementary Table S2), and 1,612 probe sets, corresponding to 574 up-regulated (Online Supplementary Table S3) and 556 down-regulated genes (Online Supplementary Table S4), were identified by supervised analysis at 2 and 6 h, respectively. In view of the relatively high number of genes that were found to be modulated by ITF2357 treatment, particularly at 6 h, we used the DAVID Functional Annotation Clustering Tool in order to identify the functional categories that were most significantly modulated. The analysis identified 48 annotation clusters of genes modulated at 2 h and 103 clusters modulated at 6 h. Table 1 shows the most significant annotation clusters, with enrichment scores greater than 2 (9 clusters at 2 h and 15 clusters at 6 h).
Table 1.
Functional annotation clustering analysis of genes modulated by ITF2357 at 2 and 6 h.
At 2 h, involvement of genes related to transcription regulation was most evident (about 30% of genes, present in clusters 1, 5, 6 and 8, Table 1). The individual genes composing cluster 1 (transcription regulation) and modulated at 2 h are shown in more detail in Figure 1. Their pattern of expression at 6 h is also shown in the same figure. These genes included a relevant number of zinc finger transcription factors, most of them carrying the Kruppel motif C2H2. We also detected modulated genes related to chromatin modification processes such as ARID4A, JARID1B, which interact with RB in transcriptional repression,18,19 and down-regulation of three subunits of the histone acetyltransferase complex NuA4 (EPC1, EPC2 and ING3).20 Interestingly, a significant number of genes involved in transcription regulation modulated by ITF2357 encode proteins known to regulate cell growth: in particular, the MYC oncogene was down-regulated about 3-fold. The tumor suppressor genes ING1 and ING2 were up-regulated whereas the transcriptional repressors E2F6 and E2F8 were repressed (Figure 1).
Figure 1.
Effect of ITF2357 on genes related to transcription regulation included in the annotation cluster 1 after 2 h of treatment. The average fold change in expression level of treated versus untreated control at 2 and 6 h is shown for each gene. Genes are ranked according to their fold change at 2 h of treatment. The probe set with the highest zg score was selected whenever more than one recognized the same gene. * indicates genes that were also significantly modulated at 6 h.
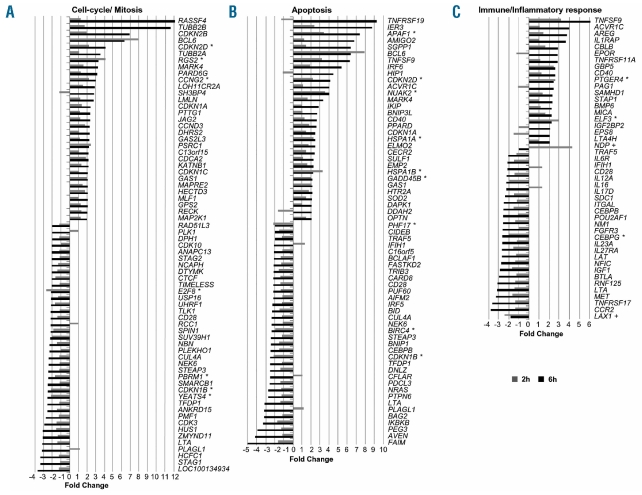
At 6 h we found 15 annotation clusters with an enrichment score greater than 2. The clusters at this time point were more diversified than at 2 h and included categories related to cell cycling/mitosis (cluster 5), apoptosis (clusters 8 and 15), protein kinase activity (clusters 4 and 6), as well as transcription regulation (clusters 13 and 14) and chromatin structure (clusters 2 and 14) (Table 1). Given the known biological effects of ITF2357, the modulation of single genes modulated at 6 h and related to cell cycle/mitosis and apoptosis are presented in more detail in Figure 2A–B. Among the genes involved in cell cycling, we found a number of genes related to cyclins. The cyclin inhibitors CDKN1A (p21), CDKN1C (p57), CDKN2B (p15) and CDKN2D (p19) as well as cyclins CCNG2 and CCND3 were up-regulated by ITF2357, whereas CDKN1B (p27) and the cyclin-dependent kinases CDK10, CDK3 were decreased. Other modulations of note were the strong induction (12-fold) of RASSF4, a gene for a RAS-associated protein that induces cell cycle arrest and apoptosis,21 of the BCL6 proto-oncogene that acts as a transcriptional repressor recruiting a histone deacetylase-repressing complex,22 and of GAS1, a gene associated with growth arrest23 (Figure 2A).
Figure 2.
Effect of ITF2357 on genes involved in cell-cycling, apoptosis and immunity. The average fold change in expression level of treated versus untreated control at 2 and 6 h is shown for each gene. Genes related to cell cycling (A), apoptosis (B) and immunity/inflammation (C) are ranked according to their fold change at 6 h of treatment. The probe set with the highest zg score was selected whenever more than one recognized the same gene.* indicates genes that were also significantly modulated at 2 h. +in panel C indicates genes specifically modulated at 2 h.
The cluster of modulated genes that control the apoptotic process includes APAF1, responsible for activation of pro-caspase-9,24 DAPK1 a positive mediator of cell death induced by IFN-γ25 and several genes belonging to the TNF pathway (CD40, TRAF5, TNFRSF19, TNFSF9 and GADD45B) (Figure 2B).
Analysis of cytokine and immune-related genes
Given the known role of ITF2357 in regulating the expression of cytokines and growth factors important for cancer growth and its clinical activity in inflammatory conditions, we carried out a more detailed analysis of genes related to immunity and inflammation and modulated by ITF2357. The majority of these were modulated at 6 h (19 up- and 25 down-regulated), three of them were also modulated at 2 h (CEBPG, PTGER4 and ELF3), while NPD and LAX1 were significantly up- and down-regulated, respectively, only at 2 h (Figure 2C). Among the genes down-modulated at 6 h we noted those encoding for several pro-inflammatory cytokines (LTA/TNF-β, IL-12A, IL-16, IL-17D and IL-23A), growth factors (IGF1) and receptors (CD28, IL-6R, IL27RA, TNFRSF17, BTLA, CCR2, FGFR3, MET). Most other modulated genes related to immunity were transcription factors (CEBPG, CEBPB, NM1, POU2AF1 and NFIC). Several of the modulated genes have been implicated in MM biology, as discussed below.
Validation of the gene expression profile data
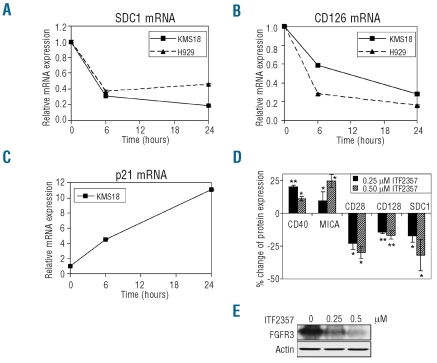
We next validated selected modulated genes by quantitative PCR. The mRNA for SDC1 (Syndecan 1, CD138) was down-modulated by 70% and 85% in the presence of 0.25 μM ITF2357 in the KMS18 cell line at 6 and 24 h, respectively, as expected from the gene chip data. Similar data were obtained in the H929 MM cell line (Figure 3A). CD126 (IL-6Rα) mRNA was decreased by 42 and 70% at 6 and 24 h respectively in KMS18 cells and by 73 and 83% in the H929 cell line at the same time points (Figure 3B). In contrast, p21 mRNA was induced by ITF2357, with 4- and 11-fold increases in KMS18 cells at 6 and 24 h, respectively, also in agreement with gene chip analysis (Figure 3C).
Figure 3.
Validation of gene chip results with selected genes. (A–C) KMS18 or H929 cells were treated with 0.25 μM ITF2357 for the indicated times and the mRNA levels for SDC1 (A) CD126 (B) or p21 (C) relative to untreated controls was analyzed by quantitative PCR. The results are the mean ± SD of triplicate wells. (D) KMS18 cells treated for 24 h with either 0.25 μM (black bars) or 0.5 μM ITF2357 (striped bars). Expression of CD40, MICA, CD28, CD126 and SDC1 proteins was analyzed by immunofluorescence on the FACS. The results are the mean ± SD of at least three independent experiments and show the percentage expression obtained after subtracting untreated control values. *P<0.05, **P<0.01 with respect to untreated control. (E) KMS18 cells treated for 24 h with ITF2357 and expression of FGFR3 and actin analyzed by western blot. The data shown are representative of two independent experiments.
Since biologically relevant changes are likely to require protein activity, we also validated ITF2357-mediated modulation of several immune-related genes at the protein level in KMS18 cells. In particular, we observed a reproducible induction by ITF2357 of CD40 positive cells (11–20% over control) and MICA (10–25%), consistent with gene chip data (Figure 3D). As expected, we also found reproducible decreases in CD28 (23–30%), CD126 (15–17%), SDC1 (17–32%) (Figure 3D) and FGFR3 protein expression (46–78%) (Figure 3E) in KMS18 cells treated for 24 h with the drug.
ITF2357 down-modulates CD126-mediated signaling
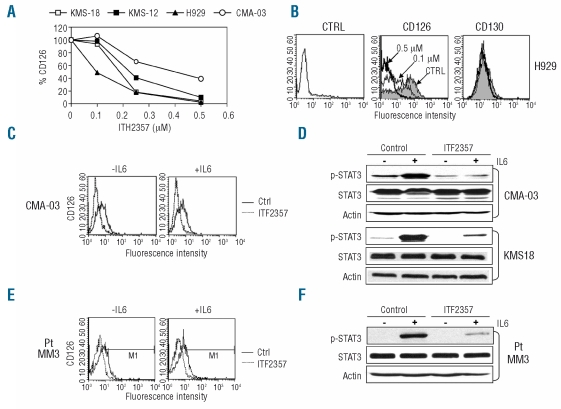
Given the well established importance of IL-6 signaling in MM, we further analyzed the role of ITF2357 on IL-6R expression in a panel of cell lines. Of seven MM cell lines analyzed, all stained positive for CD126 in the absence of ITF2357 treatment, with percentages of expression ranging from 8.1 to 98%. Only in one case (KMS11) was CD126 expression very weak (1.6%) (Online Supplementary Table S5). Similarly the IL-6R β chain (CD130) involved in signal transduction was expressed in all MM cell lines, although in two of them, KMS18 and CMA-03, expression was rather weak (8.7 and 4.2%, respectively) (Online Supplementary Table S5). We then analyzed the effect of ITF2357 on CD126 and CD130 expression in several MM cell lines. As shown in Figure 4A, ITF2357 induced a marked decrease in CD126 expression in all cell lines examined (P<0.001), with an ED50 varying from 0.1 to 0.3 μM. The FACS histograms of treated and control H929 cell line are shown in Figure 4B as an example. The effect of ITF2357 was specific for CD126 since other markers, in particular CD130 and CD54, were not modified by the drug at concentrations of up to 0.5 μM in any of the cell lines analyzed (Figure 4B and data not shown). Furthermore down-modulation of CD126 in the IL-6-dependent MM cell line CMA-03 occurred in both the presence and the absence of exogenous IL-6 (Figure 4C).
Figure 4.
Effect of ITF2357 on CD126 expression and function in MM cells. (A) MM cell lines were treated with increasing concentrations of ITF2357 for 24 h and stained with CD126-PE or control antibody. The percentage CD126 stained cells relative to untreated control are shown. All cell lines were analyzed in two independent experiments. (B) The H929 MM cell line was treated with medium (gray profile), 0.1 μM (thin lines) or 0.5 μM ITF2357 (thick lines) for 24 h. Cells were then stained with CD126-PE, CD130-PE or control antibodies, as indicated, and analyzed on the FACS. (C–F) The indicated MM cell lines (C–D) or purified CD138+ cells from a MM patient (Pt MM3, E–F) were treated for 24 h in the absence (continuous line) or presence (broken line) of 0.5 μM ITF2357 and then recombinant human IL-6 was added during the last 30 min of culture. Cells were then stained with CD126-PE antibody and analyzed on the FACS (C,E). Total cell extracts were analyzed in western blots for the indicated proteins (D,F). The data are representative of six different cell lines and freshly isolated cells from three patients. Most cell lines were analyzed in two independent experiments.
We then determined whether CD126 down-modulation in MM was accompanied by decreased STAT3 signaling in response to exogenous IL-6. MM cell lines were cultured in the presence or absence of 0.5 μM ITF2357 for 24 h and then stimulated for 30 min with recombinant human IL-6. Extracted proteins were analyzed by western blotting for total and phosphorylated STAT3 (Tyr705). The results of two representative cells lines, CMA-03 and KMS18, of six are shown in Figure 4D. A variable but detectable level of basal phosphorylated STAT3 (p-STAT3) could be detected in the absence of IL-6 in all cell lines and this was, as expected, up-regulated by IL-6 treatment, by a mean of 15-fold (P<0.001). Interestingly ITF2357 treatment for 24 h reduced basal p-STAT3 in all cases. Indeed, in four of the six cell lines, basal p-STAT3 became undetectable after ITF2357 treatment. ITF2357 also reduced IL-6-induced p-STAT3 by a mean of 82% (±16%) in the six cell lines (Figure 4D and data not shown). This reduction was highly significant (P<0.001). In contrast, total STAT3 was not significantly modified in any of the cell lines (Figure 4D and data not shown).
We then examined whether the drug also affected freshly isolated MM samples. In all three cases analyzed, MM cells expressed CD126 and more weakly CD130 (Online Supplementary Table S5). Purified CD138+ myeloma cells were treated for 24 h with 0.5 μM ITF2357. The drug down-modulated CD126 expression both in the presence and absence of exogenous IL-6 (Figure 4E) and diminished the STAT3 response to IL-6 by a mean of 87% (Figure 4F and data not shown). Also in this case inhibition was observed in all three cases and was statistically significant (P<0.01).
These data demonstrate that the down-modulation of CD126 expression by ITF2357 has functional consequences, since it markedly diminishes the response to exogenous IL-6, measured as STAT3 phosphorylation, in both MM cell lines and patients’ cells.
ITF2357 down-modulates expression of MIRHG1 and associated miR-19a and miR-19b
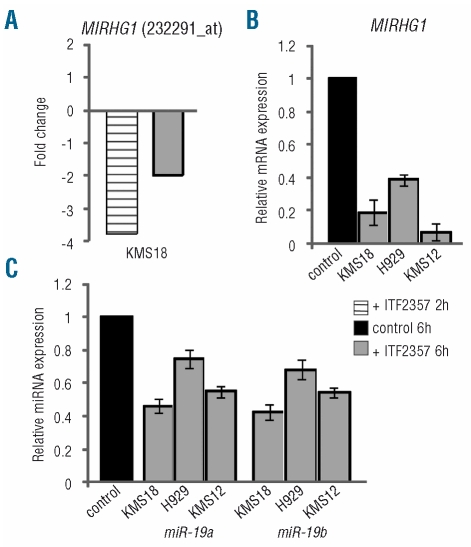
In the course of gene chip analysis, we observed that one of the modulated transcripts was C13orf25 (also called MIRHG1), located at locus 13q31.3, and known to host the miR-17~92 cluster, a cluster of microRNA that has oncogenic activity in MM cells.5 The MIRHG1 transcript was modulated at both 2 and 6 h in the gene chip analysis (Figure 5A). Given the potential importance of this locus, we validated these data by quantitative PCR using a MIRHG1-specific probe in KMS18, H929 and KMS12 cell lines treated for 6 h with 0.5 μM ITF2357. As shown in Figure 5B, there was strong down-modulation (range, 61% to 93%) of the MIRHG1 transcript in all three cell lines. We also analyzed expression of two selected miRNA, the miR-19a and miR-19b transcripts, belonging to the miR-17~92 cluster. ITF2357 down-modulated these two miRNA specifically, in all cell lines tested, with levels of inhibition ranging from 25 to 57% (Figure 5C, P<0.05 for miRNA-19a in KMS18 and KMS12 cell lines and P<0.01 for miRNA-19b in all cell lines).
Figure 5.
Effect of ITF2357 on expression of MIRHG1 and miR-19a and miR-19b. (A) Average fold change of MIRHG1 expression in the KMS18 cell line, 2 and 6 h after treatment with 0.25 μM ITF2357, as determined by microarray analysis. (B,C) Relative expression of MIRHG1 (B) and miR-19a and miR-19b (C) in KMS18, H929 and KMS12 cell lines as determined by Q-RT-PCR 6 h after treatment with 0.5 μM ITF2357. Data shown are the means and standard errors of at least two experiments performed in duplicate.
We conclude that ITF2357 down-modulates the MIRHG1 mRNA transcript as well as miR-19a and miR-19b located within this locus.
Discussion
In order to identify the possible mechanism of action of ITF2357 in MM, we analyzed the global gene expression profile of the KMS18 cell line treated with the drug. We identified 242 and 1,130 genes modulated by ITF2357 at 2 and 6 h, respectively. Several genes were validated at the RNA and protein level. Interestingly, genes modulated at 2 h mostly belong to the transcription regulation family (about 30% of modulated genes), whereas those regulated at 6 h are involved in a wider range of functional categories, including cell growth and apoptosis. This suggests that a primary effect of ITF2357 is to modulate transcription regulators, which in turn directs expression of other genes leading, for example, to cell cycle arrest and cell death.
ITF2357 regulated numerous genes involved in the cell cycle and apoptosis. More specifically, the drug repressed MYC and several CDK, and up-regulated the tumor suppressors ING1 and ING2,26 as well as several cyclin kinase inhibitors, including p21. ITF2357 also up-regulated several pro-apototic proteins, most notably APAF1, which was induced by more than 7-fold. Although APAF-1 was not further analyzed in MM cells, we and others have observed its induction by HDAC inhibitors in other cell types27 (LL and JG, unpublished data). Most notably, APAF-1 has been shown to play a role in the mitochondrial apoptosis pathway and caspase 9 activation induced by several HDAC inhibitors.28 Thus APAF-1 up-regulation may also be involved in the previously demonstrated ITF2357-mediated caspase 9 cleavage.12 Although a genome-wide comparison is not possible, the gene pattern modulated by ITF2357 is in line with that previously observed in MM1 cells treated with other HDAC inhibitors.29,30 Indeed several genes shown here, such as CD138, CD126, IGF1, TNFRSF1, MYB, CDK3, CDK1B, CDK1A (p21), CDKN1C (p57), CDKN2D (p19) and APAF1, have been previously identified to be modulated in MM cells by the pan-HDAC inhibitor SAHA.29 Interestingly, CDK1A, CDKN1C, CDKN1B (p27), MYC, BID and CD138 have also been described to be modulated in MM cells by the class I specific HDAC inhibitor valproic acid.30 A full understanding of the role of the various HDAC isoforms in regulating specific sets of genes does, however, await further analyses with isoform-specific inhibitors using identical cellular targets and time points.
Gene arrays have long been used to identify genes that are altered in MM cells and have contributed significantly to the understanding of MM pathogenesis, prognosis and differential response to drugs.31 Although a detailed comparison of our results with those of previous authors is beyond the scope of this work, we identified several genes modulated by ITF2357 which are specifically related to plasma cell and MM biology. The expression of CD28 on malignant plasma cells correlates with a poor prognosis and disease progression in MM32 and the direct activation of the receptor by anti-CD28 monoclonal antibody suppresses plasma cell proliferation.33 FGFR3 is a target gene of the t(4;14) chromosomal translocation occurring in 15% of MM and it, too, is related to a poor prognosis.34–36 Furthermore FGFR3 expression promotes MM proliferation in cooperation with IL-6.37,38 Thus it is interesting to note that we found that both FGFR3 and CD126 were down-modulated by ITF2357 and the down-regulation was verified at the protein level. Other modulated genes of interest are the hepatocyte growth factor receptor MET, a potent myeloma growth and survival factor,39 TNFRS17 (BCMA) involved in B-cell lineage maturation and its ligand BAFF that protects MM cells from apoptosis.40 CEBPB is an IL-6-dependent protein, critical for cell cycling, apoptosis and induction of metastasis in some tumors and is thought to play a role in regulation of genes whose transcription is affected by cyclin D1 overexpression.41 POU2AF1, a B-cell specific transcriptional co-activator associated with OCT1 and OCT2, is involved in the control of expansion and maintenance of mature B cells and promotes growth of MM plasma cells through the transactivation of BCMA.42,43 The down-modulation of IGF-1, a molecule that has been implicated as an autocrine growth factor in MM, is also of note.44,45 We also found modulation of several genes involved in TNF pathways, including CD40. CD40 is expressed in a large fraction of MM patients and its activation promotes growth/survival of MM cells and their adhesion to both fibronectin and bone marrow stem cells, with the consequent secretion of IL-6 and other cytokines.46 Anti-CD40 monoclonal antibody has significant anti-tumor activity against MM cells, providing a specific antibody-mediated therapy for MM cells expressing CD40,46 and suggesting the possibility of combined therapies, since CD40 protein expression was found to be up-regulated by ITF2357. Similarly, we observed that the expression of the MICA gene and protein are up-regulated in the presence of ITF2357. MICA is expressed on bone marrow plasma cells from patients with MM and monoclonal gammopathy of undetermined significant, making the MM cells more sensitive to the cytotoxic effect of natural killer cells.47
Exposure of the neoplastic B-cell line studied here to ITF2357 also resulted in repression of a set of ‘inflammatory’ cytokines and receptors. This is of interest since HDAC inhibitors have been known for several years to have anti-inflammatory properties8,11,48,49 and ITF2357 inhibits production of several pro-inflammatory cytokines by lipopolysaccharide-stimulated peripheral blood cells in vitro and in vivo.11 The inhibition by ITF2357 of inflammatory gene expression is in agreement with the recently suggested role of chromatin modification, including acetylation of histones and transcription factors, in regulating inflammatory genes.50–54 More specifically, current models suggest that histone acetyltransferase remodeling complexes such as NuA4, SWI/SNF and Mi-2b may determine the chromatin structure of these genes and their responsiveness to external inflammatory stimuli.50–52,55 It is, therefore, intriguing to note that ITF2357 modulated several genes belonging to such complexes, particularly those for NuA4 (EPC1 and EPC2 and ING3).
Perhaps the most important modulated gene related to both inflammation and MM pathogenesis is CD126, the IL-6 receptor. IL-6 is an important MM growth factor and CD126 membrane expression has prognostic significance in the disease.56–58 We demonstrated here the functional significance of CD126 down-modulation by ITF2357, with decreased CD126 expression at the protein level in the MM cell lines and freshly isolated plasma cells from patients. The effect on CD126 protein expression was specific, and other surface proteins, such as the signaling molecule CD130 (gp130) or the adhesion molecule CD54, were not affected by ITF2357. More importantly, down-modulation of CD126 led to a decreased response to IL-6 signaling, assessed by STAT3 phosphorylation. This would be of particular importance for MM cells dependent upon IL-6 for growth. In addition, in all MM cell lines analyzed, exposure to ITF2357 led to decreased basal expression of p-STAT3 in the absence of exogenous IL-6. This could be due to inhibition by ITF2357 of an autocrine IL-6 loop, such as in U266 cells which secrete IL-6. In non-IL-6 producers, ITF2357 may inhibit other stimulatory signals that induce STAT3 phosphorylation. Interestingly we have observed that many AML cell lines and freshly isolated AML samples express CD126 and that ITF2357 down-modulated this protein and diminished the STAT3 response to exogenous IL-6 also in AML cells (JG and VB, unpublished data). Such data suggest that the phenomenon described here may be common to several cancer cell types. This is of interest given the growing evidence for a role of IL-6 and its receptor in neoplastic progression in a wide variety of cancers.59
A very novel mechanism described here by which ITF2357 may affect MM growth is the inhibition of expression of miR-19a and miR-19b. These two microRNA belong to the miR-17~92 cluster, which is localized within the MIRHG1 locus (also called C13orf25) on chromosome 13q31.3 and has been strongly implicated in tumorigenesis in B-cell neoplasms, including MM, as well as other cancer cell types.2,4,60 Indeed deletion of miR-17~92 leads to a block in B-cell proliferation and development, whereas its over-expression induces B-cell hyperproliferation and autoimmune disease.3 This miRNA cluster is frequently amplified in diffuse large cell lymphomas of the germinal center subtype.4,60 More recently, miR-19a and miR-19b have been directly implicated in myeloma pathogenesis.5 Indeed these miRNA have been shown to be up-regulated in MM patients’ cells and cell lines compared to normal plasma cells. Furthermore miR-19a and miR-19b antagonists suppressed MM tumor growth in nude mice.5 We found in this study that ITF2357 down-modulated both the non-protein coding host gene MIRHG1, and its associated miR-19a and miR-19b transcripts. In B cells, miR-19a and 19b have been shown to repress SOCS1 protein, an inhibitor of the JAK-STAT pathway, the pro-apoptotic protein BIM, the cell cycle regulators E2F1 and PTEN, as well as the p21 protein.2
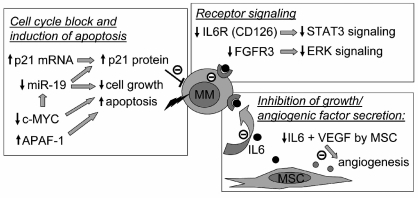
Collectively our data demonstrate that ITF2357 has pleiotropic activities, several of which may contribute to its anti-myeloma activity, as summarized in Figure 6. ITF2357 inhibits secretion of growth and angiogenic factors, in particular IL-6 and VEGF, by bone marrow stromal cells.12 An additional mechanism proposed here is the down-modulation of CD126 leading to decreased response to exogenous or endogenous IL-6. This combined action on the cytokine and its receptor should inhibit IL-6 signaling most effectively. In addition to the IL-6 axis, ITF2357 modulates several genes implicated in MM growth and apoptosis, including c-MYC, FGFR3, APAF-1, p21, miR-19a and miR-19b.2,61 Current data from the literature suggest that some of these effects may be linked. For example c-MYC is known to modulate miR-17~92. Thus MYC down-modulation may mediate miR-19 repression and this in turn may enhance p21 protein expression.62 These effects are likely to explain, at least in part, the cell cycle block and induction of apoptosis by the drug. In conclusion, although it is likely that ITF2357 will be best combined with other drugs for a fully effective treatment of MM, extensive characterization of the pleiotropic effects of the drug, as described here, should help in designing the best combination strategies in the context of MM.
Figure 6.
ITF2357 has pleiotropic effects on several signaling pathways relevant to MM biology The different mechanisms by which ITF2357 may affect MM growth, as deduced from this and previous reports,12,2 are indicated.
Footnotes
Funding: this work was supported in part by grants from the Italian Association for Cancer Research (AIRC) to JG (IG 2008-10) and AN (IG 2007-09) and by the Associazione Italiana Contro le Leucemia, Linfomi e Mieloma (AIL)-Sezione Paolo Belli and Sezione Milano. KT was supported by a fellowship from Fondazione Italiana Ricerca sul Cancro (FIRC).
Authorship and Disclosures
KT, VB, OP, ML and GF performed the experiments and prepared the figures and tables. JG, LL, MI, AR and AN participated in the design and funding of the project. JG and LL wrote the manuscript. AR and AN provided patients’ material.
GL and PM are employees of Italfarmaco, which produces ITF2357. PM is co-inventor of patent of drug under study; AR have research funding by Italfarmaco; JG received research grant from Italfarmaco. The other authors reported no potential conflicts of interest.
The online version of this article has a supplementary appendix.
References
- 1.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–98. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64(9):3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 5.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105(35):12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Kelly WK, Marks PA. Drug insight: histone deacetylase inhibitors–development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat Clin Pract Oncol. 2005;2(3):150–7. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 8.Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol. 2006;209(3):611–6. doi: 10.1002/jcp.20781. [DOI] [PubMed] [Google Scholar]
- 9.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 10.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269(1):7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11(1–12):1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golay J, Cuppini L, Leoni F, Micò C, Barbui V, Domenghini M, et al. The histone deacetylase inhibitor ITF2357 has anti-leukemic activity in vitro and in vivo and inhibits IL-6 and VEGF production by stromal cells. Leukemia. 2007;21(9):1892–900. doi: 10.1038/sj.leu.2404860. [DOI] [PubMed] [Google Scholar]
- 13.Armeanu S, Pathil A, Venturelli S, Mascagni P, Weiss TS, Göttlicher M, et al. Apoptosis on hepatoma cells but not on primary hepatocytes by histone deacetylase inhibitors valproate and ITF2357. J Hepatol. 2005;42(2):210–7. doi: 10.1016/j.jhep.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Guerini V, Barbui V, Spinelli O, Salvi A, Dellacasa C, Carobbio A, et al. The histone deacetylase inhibitor ITF2357 selectively targets cells bearing mutated JAK2(V617F) Leukemia. 2008;22(4):740–7. doi: 10.1038/sj.leu.2405049. [DOI] [PubMed] [Google Scholar]
- 15.Mattioli M, Agnelli L, Fabris S, Baldini L, Morabito F, Bicciato S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24(15):2461–73. doi: 10.1038/sj.onc.1208447. [DOI] [PubMed] [Google Scholar]
- 16.Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;(Suppl 37):120–5. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 17.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6(10):986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 18.Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21(8):2918–32. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashuba V, Protopopov A, Podowski R, Gizatullin R, Li J, Klein G, et al. Isolation and chromosomal localization of a new human retinoblastoma binding protein 2 homologue 1a (RBBP2H1A) Eur J Hum Genet. 2000;8(6):407–13. doi: 10.1038/sj.ejhg.5200474. [DOI] [PubMed] [Google Scholar]
- 20.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14(2):147–54. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64(23):8688–93. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- 22.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32(4):606–13. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 23.Del Sal G, Ruaro ME, Philipson L, Schneider C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell. 1992;70(4):595–607. doi: 10.1016/0092-8674(92)90429-g. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1(7):949–57. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 25.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9(1):15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 26.Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer. 2008;123(7):1483–90. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]
- 27.Wallace DM, Cotter TG. Histone deacetylase activity in conjunction with E2F-1 and p53 regulates Apaf-1 expression in 661W cells and the retina. J Neurosci Res. 2009;87(4):887–905. doi: 10.1002/jnr.21910. [DOI] [PubMed] [Google Scholar]
- 28.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101(52):18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101(2):540–5. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neri P, Tagliaferri P, Di Martino MT, Calimeri T, Amodio N, Bulotta A, et al. In vivo anti-myeloma activity and modulation of gene expression profile induced by valproic acid, a histone deacetylase inhibitor. Br J Haematol. 2008;143(4):520–31. doi: 10.1111/j.1365-2141.2008.07387.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahtouk K, Hose D, De Vos J, Moreaux J, Jourdan M, Rossi JF, et al. Input of DNA microarrays to identify novel mechanisms in multiple myeloma biology and therapeutic applications. Clin Cancer Res. 2007;13(24):7289–95. doi: 10.1158/1078-0432.CCR-07-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robillard N, Jego G, Pellat-Deceunynck C, Pineau D, Puthier D, Mellerin MP, et al. CD28, a marker associated with tumoral expansion in multiple myeloma. Clin Cancer Res. 1998;4(6):1521–6. [PubMed] [Google Scholar]
- 33.Bahlis NJ, King AM, Kolonias D, Carlson LM, Liu HY, Hussein MA, et al. CD28-mediated regulation of multiple myeloma cell proliferation and survival. Blood. 2007;109(11):5002–10. doi: 10.1182/blood-2006-03-012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chesi M, Nardini E, Brents LA, Schröck E, Ried T, Kuehl WM, et al. Frequent translocation t(4;14)(p16.3; q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16(3):260–4. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23(26):6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 36.Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, et al. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90(10):4062–70. [PubMed] [Google Scholar]
- 37.Plowright EE, Li Z, Bergsagel PL, Chesi M, Barber DL, Branch DR, et al. Ectopic expression of fibroblast growth factor receptor 3 promotes myeloma cell proliferation and prevents apoptosis. Blood. 2000;95(3):992–8. [PubMed] [Google Scholar]
- 38.Ishikawa H, Tsuyama N, Liu S, Abroun S, Li FJ, Otsuyama K, et al. Accelerated proliferation of myeloma cells by interleukin-6 cooperating with fibroblast growth factor receptor 3-mediated signals. Oncogene. 2005;24(41):6328–32. doi: 10.1038/sj.onc.1208782. [DOI] [PubMed] [Google Scholar]
- 39.Hov H, Holt RU, Rø TB, Fagerli UM, Hjorth-Hansen H, Baykov V, et al. A selective c-met inhibitor blocks an autocrine hepatocyte growth factor growth loop in ANBL-6 cells and prevents migration and adhesion of myeloma cells. Clin Cancer Res. 2004;10(19):6686–94. doi: 10.1158/1078-0432.CCR-04-0874. [DOI] [PubMed] [Google Scholar]
- 40.Moreaux J, Legouffe E, Jourdan E, Quittet P, Rème T, Lugagne C, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103(8):3148–57. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114(3):323–34. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 42.Schubart K, Massa S, Schubart D, Corcoran LM, Rolink AG, Matthias P. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat Immunol. 2001;2(1):69–74. doi: 10.1038/83190. [DOI] [PubMed] [Google Scholar]
- 43.Zhao C, Inoue J, Imoto I, Otsuki T, Lida S, Ueda R, et al. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene. 2008;27(1):63–75. doi: 10.1038/sj.onc.1210637. [DOI] [PubMed] [Google Scholar]
- 44.Ménoret E, Maiga S, Descamps G, Pellat-Deceunynck C, Fraslon C, Cappellano M, et al. IL-21 stimulates human myeloma cell growth through an autocrine IGF-1 loop. J Immunol. 2008;181(10):6837–42. doi: 10.4049/jimmunol.181.10.6837. [DOI] [PubMed] [Google Scholar]
- 45.Maiso P, Ocio EM, Garayoa M, Montero JC, Hofmann F, Garcia-Echeverria C, et al. The insulin-like growth factor-I receptor inhibitor NVP-AEW541 provokes cell cycle arrest and apoptosis in multiple myeloma cells. Br J Haematol. 2008;141(4):470–82. doi: 10.1111/j.1365-2141.2008.07049.x. [DOI] [PubMed] [Google Scholar]
- 46.Tai YT, Li X, Tong X, Santos D, Otsuki T, Catley L, et al. Human anti-CD40 antagonist antibody triggers significant antitumor activity against human multiple myeloma. Cancer Res. 2005;65(13):5898–906. doi: 10.1158/0008-5472.CAN-04-4125. [DOI] [PubMed] [Google Scholar]
- 47.Carbone E, Neri P, Mesuraca M, Fulciniti MT, Otsuki T, Pende D, et al. HLA class I, NKG2D, and natural cytotoxicity receptors regulate multiple myeloma cell recognition by natural killer cells. Blood. 2005;105(1):251–8. doi: 10.1182/blood-2004-04-1422. [DOI] [PubMed] [Google Scholar]
- 48.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99(5):2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, et al. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122(4):596–606. doi: 10.1111/j.1365-2567.2007.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193(12):1351–9. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20(3):282–96. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130(1):7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447(7147):972–8. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 54.Calao M, Burny A, Quivy V, Dekoninck A, Van Lint C. A pervasive role of histone acetyltransferases and deacetylases in an NF-kappaB-signaling code. Trends Biochem Sci. 2008;33(7):339–49. doi: 10.1016/j.tibs.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10(1):92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dankbar B, Padró T, Leo R, Feldmann B, Kropff M, Mesters RM, et al. Vascular endothelial growth factor and interleukin-6 in paracrine tumor-stromal cell interactions in multiple myeloma. Blood. 2000;95(8):2630–6. [PubMed] [Google Scholar]
- 57.Hideshima T, Richardson P, Anderson KC. Novel therapeutic approaches for multiple myeloma. Immunol Rev. 2003;194:164–76. doi: 10.1034/j.1600-065x.2003.00053.x. [DOI] [PubMed] [Google Scholar]
- 58.Perez-Andres M, Almeida J, Martin-Ayuso M, De Las Heras N, Moro MJ, Martin-Nuñez G, et al. Soluble and membrane levels of molecules involved in the interaction between clonal plasma cells and the immunological microenvironment in multiple myeloma and their association with the characteristics of the disease. Int J Cancer. 2009;124(2):367–75. doi: 10.1002/ijc.23941. [DOI] [PubMed] [Google Scholar]
- 59.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2(3):175–87. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 60.Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105(36):13520–5. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sirohi B, Powles R. Multiple myeloma. Lancet. 2004;363(9412):875–87. doi: 10.1016/S0140-6736(04)15736-X. [DOI] [PubMed] [Google Scholar]
- 62.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]