Abstract
A clear picture of animal relationships is a prerequisite to understand how the morphological and ecological diversity of animals evolved over time. Among others, the placement of the acoelomorph flatworms, Acoela and Nemertodermatida, has fundamental implications for the origin and evolution of various animal organ systems. Their position, however, has been inconsistent in phylogenetic studies using one or several genes. Furthermore, Acoela has been among the least stable taxa in recent animal phylogenomic analyses, which simultaneously examine many genes from many species, while Nemertodermatida has not been sampled in any phylogenomic study. New sequence data are presented here from organisms targeted for their instability or lack of representation in prior analyses, and are analysed in combination with other publicly available data. We also designed new automated explicit methods for identifying and selecting common genes across different species, and developed highly optimized supercomputing tools to reconstruct relationships from gene sequences. The results of the work corroborate several recently established findings about animal relationships and provide new support for the placement of other groups. These new data and methods strongly uphold previous suggestions that Acoelomorpha is sister clade to all other bilaterian animals, find diminishing evidence for the placement of the enigmatic Xenoturbella within Deuterostomia, and place Cycliophora with Entoprocta and Ectoprocta. The work highlights the implications that these arrangements have for metazoan evolution and permits a clearer picture of ancestral morphologies and life histories in the deep past.
Keywords: phylogenomics, Acoelomorpha, Nemertodermatida, Cycliophora, Xenoturbella, Ctenophora
1. Introduction
(a). Scalability in phylogenomic analyses
As the cost of sequencing DNA has fallen, broad-scale phylogenetic studies have begun to shift away from pre-selected gene fragments isolated by directed PCR—the traditional target gene approach—to high-throughput sequencing strategies that collect data from many genes at random. These sequencing methods, which include expressed sequence tags (ESTs) and whole-genome shotgun sequencing, theoretically allow gene selection to be part of the data analysis rather than project design since gene selection does not affect, and could be informed by, data acquisition. Existing phylogenetic studies already vary in size by at least four orders of magnitude and are anticipated to grow much larger, so scalable gene selection methods (i.e. tools that are able to accommodate datasets of very different sizes) based on explicit criteria will become increasingly important. In addition to facilitating larger analyses, such tools would make it possible to evaluate the specific effects of gene selection criteria on phylogenetic results. The development of methods and criteria for matrix assembly, rather than the manual curation of gene lists, would also facilitate the construction of matrices for a wide diversity of phylogenetic problems, including matrices optimized for subclades, superclades or entirely different groups of organisms.
The last several years have seen a proliferation of tools for identifying homologous sequences and evaluating orthology (Chen et al. 2007), but fully automated phylogenomic matrix construction based on explicit criteria is still in its infancy. A recent study that included new EST data for 29 broadly sampled animals applied a largely automated method for gene selection (Dunn et al. 2008) that relied on phenetic Markov clustering (MCL; van Dongen 2000) followed by phylogenetic evaluation of clusters. User intervention was required to evaluate some cases of paralogy. That study supported previous views that broad taxon sampling is critical for improving the phylogenetic resolution of the metazoan tree of life. Some important relationships still remained unresolved, however, and other critical taxa were unsampled.
(b). The base of Bilateria
The existence and placement of Acoelomorpha, a group hypothesized to consist of Acoela and Nemertodermatida, have been particularly problematic. Resolving the placement of acoelomorphs is essential for rooting the bilaterian tree and understanding the early phylogeny of bilaterian animals, particularly for the reconstruction of the evolution of animal organ systems (Baguñà & Riutort 2004; Hejnol & Martindale 2008b; Bourlat & Hejnol 2009). This is therefore one of the most important remaining problems in animal phylogenetics. Acoela has been recovered as the sister group to all other bilaterian animals in direct sequencing analyses, though their placement with respect to Nemertodermatida has been inconsistent (Ruiz-Trillo et al. 1999, 2002; Jondelius et al. 2002; Wallberg et al. 2007; Paps et al. 2009). The position of acoels has not been resolved satisfactorily in previous EST-based analyses (Philippe et al. 2007; Dunn et al. 2008; Egger et al. 2009). In fact, they were the most unstable taxon in the Dunn et al. (2008) study. In a more recent phylogenomic study, Egger et al. (2009) found an acoel to be the sister group to the rest of Bilateria, but questioned the result based on data on stem cell distribution and proliferation, as well as the mode of epidermal replacement, and suggested that acoels could alternatively be part of Platyhelminthes. Critically, the second major acoelomorph group, Nemertodermatida, has yet to be included in any phylogenomic analysis.
Here we simultaneously address new analytical challenges of building phylogenomic matrices using entirely explicit criteria, investigate central questions in animal phylogenetics regarding Acoelomorpha and several other important taxa, and explore the effects of subsampling this matrix. We do this by collecting new data from relevant animals, developing new orthology evaluation methods that enable the construction of much larger data matrices and applying optimized tools for high-performance computing architectures. The new data we generated (see electronic supplementary material, table S1) focus on the putative group Acoelomorpha, including two species of the previously unsampled Nemertodermatida. We also added new EST or whole-genome data for additional taxa of special interest. Publicly available data were incorporated, largely derived from the same taxa considered in a previous analysis (Dunn et al. 2008), but also including additional key taxa such as the placozoan Trichoplax adhaerens and the gastropod mollusc Lottia gigantea. Our new gene selection strategy relies exclusively on explicit criteria, allowing it to be fully automated, and it is scalable across projects of very different sizes. This new method improves the ability to build large matrices, though at a trade-off of shallower gene extraction from poorly sampled EST libraries.
2. Material and methods
(a). Data acquisition and matrix assembly
New data were generated for seven taxa (electronic supplementary material, table S1) that were selected to address several key questions in animal phylogeny, and a total of 94 taxa were included in the present analyses (electronic supplementary material, table S2). Sequencing and assembly of ESTs were performed as previously described (Dunn et al. 2008). The new ESTs were strategically collected from species in groups that were unstable (according to leaf stability metrics; see below and Dunn et al. 2008), undersampled or unrepresented in previous studies. These include a sponge, two acoels, two nemertodermatids, an entoproct and a cycliophoran. All new data, not just the sequences used for phylogenetic inference, have been deposited in the National Center for Biotechnology Information (NCBI) Trace Archive. Publicly available data for a variety of other taxa were incorporated into the analysis (see electronic supplementary material, table S2).
(b). Homology assignment and paralogue pruning
Phenetic sequence clustering was similar to that of Dunn et al. (2008), though taxon sampling criteria were relaxed considerably as described below. Unless specified otherwise, all software versions and settings are the same as in that study. Amino acid sequences were used at all stages of analysis. Sequence similarity was assessed with the previously described BLAST strategy (Dunn et al. 2008) and then grouped with MCL (van Dongen 2000). An MCL inflation parameter of 2.2 was used (see electronic supplementary material). Clusters were required to (i) include at least four taxa, (ii) include at least one taxon from which data were collected in this or the previous study, (iii) include at least one of the taxa used as BLAST subjects, (iv) have a mean of less than five sequences per taxon, (v) have a median of less than two sequences per taxon and (vi) have no representatives of a HomoloGene group that had sequences in more than one MCL cluster. Clusters that failed any of these criteria were not considered further. Sequences for each cluster that passed these criteria were aligned with Muscle (Edgar 2004), trimmed with GBlocks (Castresana 2000) and a maximum likelihood (ML) tree was inferred by RAxML.
The assessment of cluster phylogenies herein differs markedly from Dunn et al. (2008). In the first step, monophyly masking, all but one sequence were deleted in clades of sequences derived from the same taxon. The retained sequence was chosen at random. Paralogue pruning, the next step, consisted of identifying the maximally inclusive subtree that has no more than one sequence per taxon. This tree is then pruned away for further analysis, and the remaining tree is used as a substrate for another round of pruning. The process is repeated until the remaining tree has no more than one sequence per taxon. If there were multiple maximally inclusive subtrees of the same size in a given round, then they were all pruned away at the same time.
Subtrees produced by paralogue pruning were then filtered to include only those with (i) four or more taxa and (ii) 80 per cent of the taxa present in the original cluster from which they were derived (see electronic supplementary material). Fasta-format files with sequences corresponding to each terminal in the final subtrees were then generated, aligned and concatenated into a supermatrix.
(c). Phylogenetic inference
Phylogenetic analyses were conducted on an IBM BlueGene/L system at the San Diego Supercomputer Center, comprising three racks of 1024 nodes each, with two processors per node. The total analysis time was 2.25 million processor hours. The relatively low amount of per node RAM on the IBM BlueGene/L (BG/L) means that the likelihood computations for a single tree topology need to be conducted concurrently on several nodes, essentially by distributing the alignment columns across processors. The dedicated parallel version of RAxML for the current analysis is based on RAxML v. 7.0.4. A significant software engineering effort was undertaken to transform the initial proof-of-concept parallelization on an IBM BG/L into production-level code that covers the full functionality of RAxML. Among other things, the performance of the code was improved by 30 per cent (compared with the original BG/L version) via optimization of the compute-intensive loops in the phylogenetic likelihood kernel. In general, the fine-grained parallelization strategy deployed here at the level of the phylogenetic likelihood kernel needs to be applied on all state-of-the-art supercomputer architectures to better accommodate the immense memory requirements of current phylogenomic studies (Stamatakis & Ott 2008). The ability to now split the likelihood calculation for a single matrix across multiple nodes, rather than just dividing bootstrap replicates across nodes, overcomes hurdles from memory limitations per node that are encountered with large alignments, allows for a short response time for a single tree search and enables the convenient exploitation of thousands of CPUs. The adaptation of RAxML to the IBM BG/L also required the development of solutions to avoid memory fragmentation.
Models of molecular evolution were evaluated using the Perl script available from the RAxML website. ML searches and bootstrap analyses were executed under the Gamma model of rate heterogeneity. Tree sets were summarized with Phyutility (Smith & Dunn 2008), which was used to map bootstrap support onto the most likely trees, calculate leaf stability and prune taxa.
3. Results
(a). Data matrix assembly
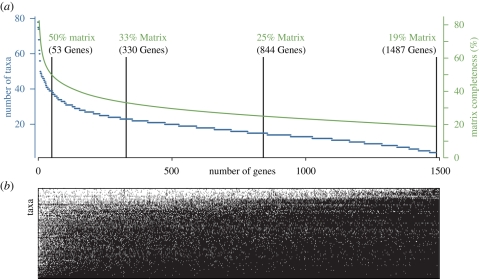
MCL generated 7445 clusters, of which 2455 passed the taxon sampling and other phenetic criteria described above. Paralogue pruning, the phylogenetic evaluation and pruning of these clusters to generate sets of orthologues with no more than one sequence per taxon resulted in 4732 subtrees with four or more taxa (the minimum size of a phylogenetically informative tree), of which 1487 passed the additional criteria described in the methods. This process is robust to noisy data, even when two haplotypes are included for nearly every gene in the Branchiostoma floridae genome (see electronic supplementary material on the robustness of matrix assembly). The final 1487-gene, 94-taxon matrix (figure 1) was 270 580 amino acids long, and had 19 per cent occupancy (i.e. on average 19% of the genes were sampled for each taxon) and 251 152 distinct column patterns. Of the 150 genes from the previous study (Dunn et al. 2008), 56 corresponded to genes in the new 1487-gene matrix. The omission here of genes considered in that previous analysis, or other such studies, does not necessarily indicate that they were unfit for phylogenetic inference, only that they were not accepted according to the different set of criteria used here that are optimized for other purposes.
Figure 1.
Genes are ranked by decreasing taxon sampling. (a) The number of taxa sampled for each gene is shown along the left vertical axis and indicated by blue data points, while the cumulative matrix completeness is shown on the right vertical axis indicated by a green continuous line. Vertical lines indicate the gene cutoffs for the four matrices that were analysed. (b) ‘Bird's eye’ view of the matrix. A white cell indicates a sampled gene. Taxa are sorted from the best sampled at the top to the least sampled at bottom (gene ordering is the same as in (a)).
Relative to the previous study (Dunn et al. 2008), the number of gene sequences in the new matrix was greatly increased for taxa with many sequenced genes (i.e. the number of unique protein predictions following EST assembly and translation), but was reduced for taxa with the smallest numbers of sequenced genes (electronic supplementary material, table S2), despite there being nearly ten times as many genes in the total matrix (1487 versus 150). The reasons for this are explored in greater detail in the electronic supplementary material, along with comparisons to the 150-gene matrix supplemented to include all 94 taxa considered here (electronic supplementary material, fig. S1). The best-sampled taxon, Homo sapiens, had 1351 (90.9%) of the 1487 genes, whereas the most poorly sampled taxon, Phoronis vancouverensis, had only 2 (0.14%; yellow circles in figure 2). Positions of taxa with the least data were not well resolved. The new matrix construction strategy was therefore disproportionally beneficial for well-sampled taxa. Poorly sampled taxa such as P. vancouverensis were not excluded from analyses a priori because heterogeneous sampling success is common in EST datasets, and is therefore of analytical interest. Also, the later application of leaf stability indices allows for the evaluation of support between stable taxa, even when poorly sampled, unstable taxa are included in the analysis.
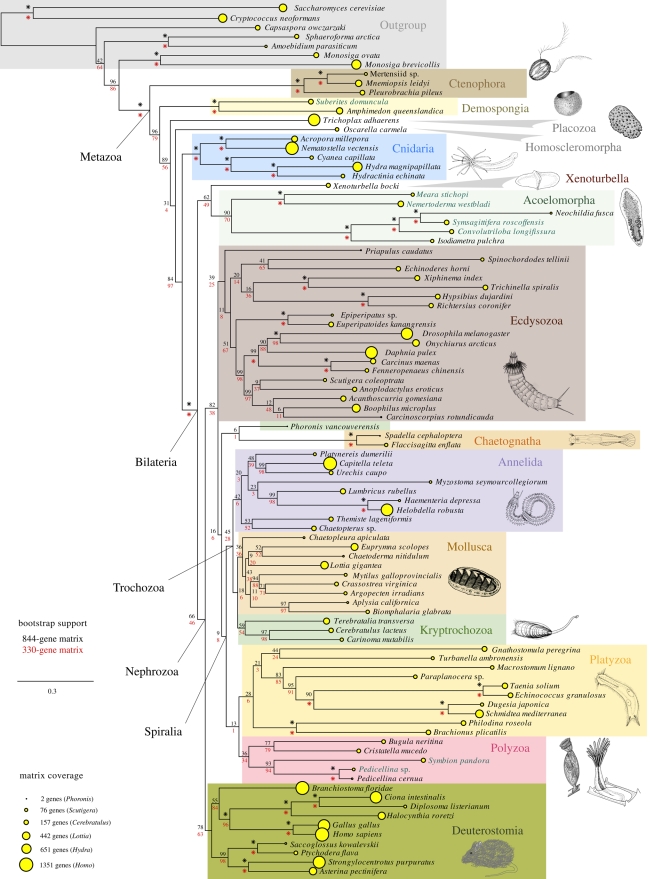
Figure 2.
Phylogram of the most likely tree found in ML searches of the 1487-gene matrix (37 searches, log likelihood = −6 124 157.6). The area of the yellow circle at each tip is proportional to the number of genes present in the 1487-gene matrix for the indicated species (see table S2 in the electronic supplementary material for values). Bootstrap support from analyses of the 844-gene (black values above nodes, 201 bootstrap replicates) and 330-gene (red values below nodes, 210 bootstrap replicates) subsamples of the 1487-gene matrix are also shown at each node. Asterisk indicates 100 per cent bootstrap support. Species for which new EST data are produced are highlighted with green species names.
Our analyses address the potential impact of missing data in several ways (see electronic supplementary material; §4). We found no indication that missing data have resulted in systematic error, though the analyses we were able to conduct were necessarily constrained by the size of the large matrix and the subject in general still requires greater attention.
(b). Gene subsampling comparisons: large, sparse matrices versus smaller, denser matrices
We analysed the complete 1487-gene matrix with 19 per cent gene occupancy, and three nested subsamples with 25, 33 and 50 per cent occupancy (figure 1). These subsets were constructed from the best-sampled genes and had 844, 330 and 53 genes, respectively. The RTREV model, with empirically estimated amino acid frequencies (F option; for details, see RAxML manual) was selected for all four matrices and used in all analyses. Partitioned analyses that apply a different model to each gene were not possible owing to load balancing problems in the likelihood kernel that resulted in severely decreased computational efficiency. The load balance problem is due to a strong variation in per-partition pattern numbers.
The optimal trees across analyses (figures 2–4) are in broad agreement with most recent phylogenomic and targeted-gene analyses in depicting, for example, monophyly of Protostomia and Deuterostomia as the fundamental bilaterian clades, and the division of protostomes into Ecdysozoa and Spiralia (the latter sometimes referred to as Lophotrochozoa; but see Giribet et al. 2009). The analyses consistently resolve Spiralia into two major clades: Trochozoa, which unites Mollusca and Annelida with a nemertean–brachiopod group recently named Kryptrochozoa (Giribet et al. 2009); and a grouping of Platyzoa together with an ectoproct–entoproct–cycliophoran clade that we discuss below under the name Polyzoa, introduced by Cavalier-Smith (1998). A more contentious issue is the base of the metazoan tree, and, after the addition of new ctenophore and sponge ESTs (compared with Dunn et al. 2008), and the complete genome of T. adhaerens, our most inclusive datasets support ctenophores as sister to all other metazoans. The positions of sponges and T. adhaerens relative to each other varied across matrix subsamples as described in the electronic supplementary material.
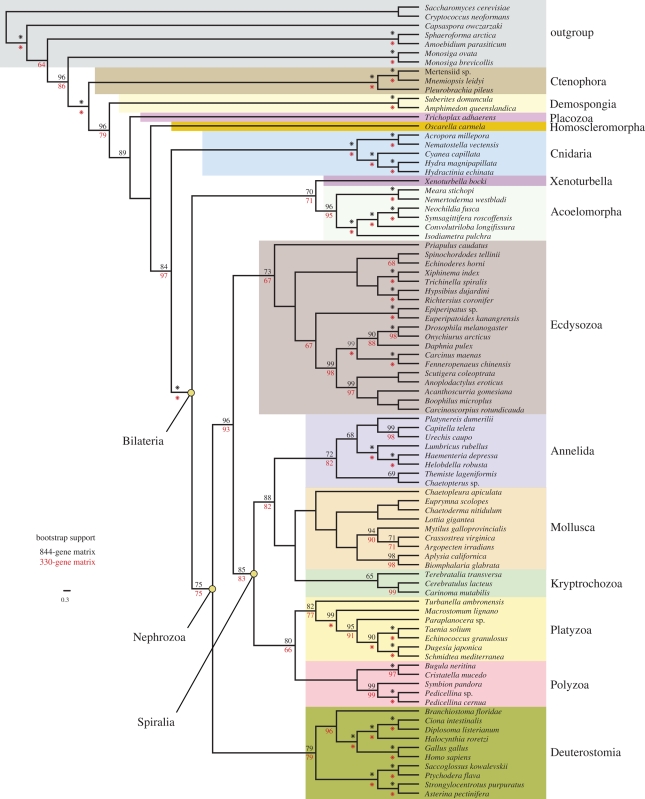
Figure 3.
Cladogram showing bootstrap support for relationships between taxa from figure 2 with a leaf stability of 87 per cent or higher. This criterion was met by 87 taxa, though only bilaterian taxa are shown (other relationships were not impacted by the removed taxa). The 844-gene (black values above nodes) and 330-gene (red values below nodes) subsamples are also shown at each node. Asterisk indicates 100 per cent bootstrap support.
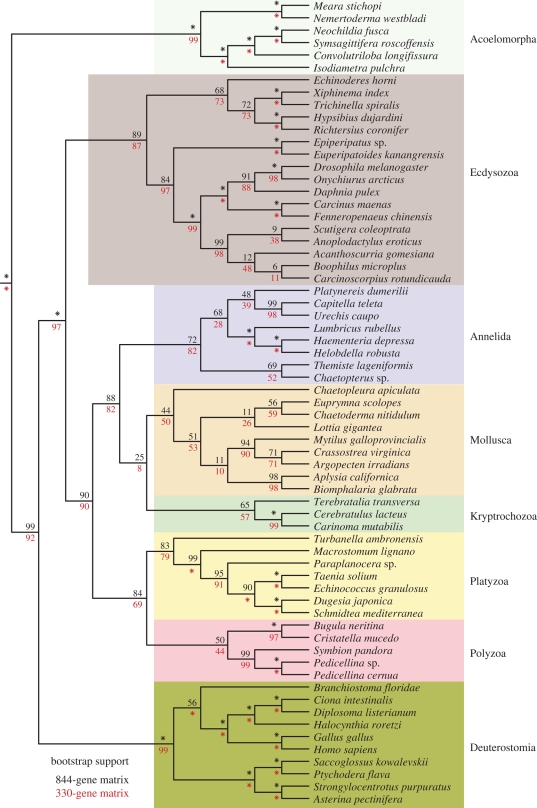
Figure 4.
Cladogram showing bootstrap support for relationships between taxa from figure 2 with a leaf stability of 90 per cent or higher. This criterion was met by 84 taxa, though only bilaterian taxa are shown (other relationships were not impacted by the removed taxa). The 844-gene (black values above nodes) and 330-gene (red values below nodes) subsamples are also shown at each node. Asterisk indicates 100 per cent bootstrap support. The taxa included in figure 3, but not here, are Xenoturbella bocki, Spinochordodes tellinii and Priapulus caudatus.
Analyses of the 53-gene subset were largely unresolved, with little convergence even between ML replicates (not shown) and poor bootstrap support at almost all deep nodes (electronic supplementary material, fig. S2). Differences between ML analyses of the 1487-, 844- and 330-gene matrices were restricted to the placement of a small number of taxa (see electronic supplementary material for details). Analyses of the 330-gene matrix recovered most of the relationships found from the 844-gene and 1487-gene matrices, many of which were not recovered in the 150-gene matrix from a previous study (electronic supplementary material, fig. S1) or the 53-gene matrix (electronic supplementary material, fig. S2). Bootstrap support values for many relationships were similar in the 330-gene and 844-gene analyses (figures 2–4; electronic supplementary material, fig. S3). Bootstrap support for the 1487-gene matrix was not evaluated owing to computational limitations.
(c). Taxon subsampling: stability and the visualization of phylogenetic relationships
Different taxa within the same phylogenetic analysis can have widely disparate stability (Thorley & Wilkinson 1999). In the present analyses most taxa are quite stable (leaf stability; electronic supplementary material, table S2)—their relationships with each other are consistent and well supported across bootstrap replicates. Other taxa, however, have inconsistent relationships across and within analyses. These unstable taxa tend to be poorly sampled in the matrix generated here, as for Phoronis and some molluscs.
A small number of unstable taxa can obscure strongly supported relationships between stable taxa, even if they have no effect on those relationships. Unless visualization tools are used that can identify stable relationships that are not affected by unstable taxa and assess support for these relationships directly, strong signals present in the data may not be discernible. We have addressed this issue by looking at support for relationships between nested subsamples of the most stable taxa, as assessed by leaf stability indices (Thorley & Wilkinson 1999; Smith & Dunn 2008). Three different leaf stability cutoffs were used (see electronic supplementary material for details on cutoff selection): 0 per cent (figure 2, i.e. no threshold), 87 per cent (figure 3) and 90 per cent (figure 4).
There were minimal differences in support values between analyses where taxa were removed prior to phylogenetic analysis versus after phylogenetic analysis (electronic supplementary material, fig. S4), indicating that unstable taxa had very little impact on the inference of the relationships between stable taxa. This indicates that taxa that are unstable do not negatively impact the ability of large-scale phylogenetic analyses to infer relationships between other taxa, though they do increase the computational burden of the studies.
4. Discussion
(a). Acoelomorpha as sister group to other Bilateria
The hypothesis that acoels (and subsequently nemertodermatids) were outside of Nephrozoa (all other bilaterian animals, i.e. protostomes and deuterostomes) has been one of the biggest challenges generated from molecular sequence data (Ruiz-Trillo et al. 1999, 2002; Jondelius et al. 2002) to the traditional view of animal phylogeny. Acoels have been difficult to place using molecular data in part due to rapid sequence evolution of the species examined, and two recent phylogenomic efforts have failed to place them with confidence (Philippe et al. 2007; Dunn et al. 2008), though Egger et al. (2009) show a similar result to ours. Notably, no EST or genomic data have been previously available for Nemertodermatida, the other major group of acoelomorphs, leaving their position unresolved. Here we find up to 100 per cent bootstrap support for the sister-group relationship of Acoela and Nemertodermatida (figure 4), together forming Acoelomorpha, and our analyses place this group as sister to Nephrozoa. This provides strong evidence that the deepest split within Bilateria is between Acoelomorpha and Nephrozoa. This result is evident only in analyses of the new large matrices and is not recovered when taxon sampling alone is improved (electronic supplementary material, fig. S1). The signal for this placement is therefore dependent on widespread gene sampling, although a similar result is obtained by Egger et al. (2009) using only 43 genes.
The morphological analysis by Ehlers (1985) listed several apomorphies for Acoelomorpha. The strongest morphological argument for this relationship is the complex epidermal ciliary root system with an intercalated network of one anterior and two lateral rootlets that is present in both acoels and nemertodermatids (Ehlers 1985; note that Ehlers regarded Acoelomorpha as a clade of Platyhelminthes). As seen here, Egger et al. (2009) found the acoel Isodiametra pulchra to be the sister to Nephrozoa. However, they questioned the result based on morphological grounds and noted similarities among acoels and rhabditophoran platyhelminths in epidermal cell replacement via mesodermally placed stem cells, and expression of a piwi-like gene in somatic and gonadal stem cells, concluding that the conflict between the phylogenomic and morphological data meant placement of acoels could not presently be resolved. This argument does not take into account other morphological data (e.g. sac-like body, non-ganglionated nervous system, absence of excretory organs, etc.), which have been used by Haszprunar (1996) to argue for a basal position of acoelomorphs in Bilateria. Furthermore, arguments regarding gene content (only three Hox genes, limited number of bilaterian microRNAs, etc.), is consistent with placement for Acoelomorpha as sister to the rest of Bilateria. The stem cell and expression data presented by Egger et al. (2009) can reasonably be interpreted as convergence or symplesiomorphy across Bilateria.
Except for one study based on myosin heavy chain type II (Ruiz-Trillo et al. 2002), molecular analyses have consistently shown a paraphyly of Acoelomorpha, with Nemertodermatida as sister to Nephrozoa and Acoela as sister to this assemblage (Jondelius et al. 2002; Ruiz-Trillo et al. 2002; Wallberg et al. 2007; Paps et al. 2009). This resulted in the previous dismissal of Acoelomorpha. Instead, our results indicate that Acoelomorpha is a clade and forms the most relevant outgroup for comparisons between protostomes and deuterostomes, providing critical insight into the origin, evolution and development of metazoan organ systems (Hejnol & Martindale 2008b; Bourlat & Hejnol 2009). Acoelomorphs possess an orthogonal nervous system (consisting of multiple longitudinal dorsal and ventral cords) and an anterior ring-shaped centralization (absent in some species; Raikova et al. 2001). The placement of Acoelomorpha as sister to Nephrozoa is therefore consistent with older hypotheses that this orthogonal nerve organization is ancestral for Bilateria (Reisinger 1972).
In both nemertodermatids and acoels, there is a single opening to the digestive system, as in cnidarians and ctenophores. A recent study shows that this opening is homologous to the bilaterian mouth and suggests that the anus might have evolved multiple times independently in Bilateria by a connection between the gonoduct and the endoderm of the gut (Hejnol & Martindale 2008a). These data strongly reject old hypotheses about the transition of a cnidarian polyp-like ancestor to a coelomate ancestor of protostomes and deuterostomes (the ‘Enterocoely hypothesis’; Remane 1950).
(b). Diminishing support for the placement of Xenoturbella in Deuterostomia
After an odyssey through the animal tree of life, the enigmatic Xenoturbella bocki seemed to have settled down as part of Deuterostomia; either as a sister group to Ambulacraria (Echinodermata + Hemichordata; Bourlat et al. 2003, 2006; Dunn et al. 2008) or as a sister group to all deuterostomes (Perseke et al. 2007). None of the analyses presented here find strong support for the placement of Xenoturbella with Deuterostomia. Instead, analyses of the new gene matrix (figures 2 and 3) place Xenoturbella with Acoelomorpha (70–71% bootstrap support). This is consistent with falling support for the placement of Xenoturbella within Deuterostomia as data have been added in other studies (Philippe et al. 2007; Dunn et al. 2008), though these previous studies failed to place it with other specific taxa.
The placement of Xenoturbella with Acoelomorpha is not surprising from a morphological point of view and morphological arguments were used by Haszprunar (1996) to include Xenoturbella in Acoelomorpha. In the original description of Xenoturbella (Westblad 1949) it was already regarded as close relative to acoels. The gross anatomy of Xenoturbella—a completely ciliated worm with only a ventral mouth opening to its digestive system and a basiepidermal nervous system—is similar to that of acoelomorphs. Several ultrastructural features, such as the epidermal ciliary rootlets including the unique ciliary tips (Franzén & Afzelius 1987; Lundin 1998), and specific degenerating epidermal cells that get resorbed into the gastrodermal tissue (Lundin 2001), are also found in Acoelomorpha (Lundin & Hendelberg 1996). The simplicity of its nervous system, especially the lack of a stomatogastric system and its basiepidermal localization, is also consistent with a close relationship to Acoelomorpha (Raikova et al. 2000). In contrast, strong morphological support for the placement of Xenoturbella as a deuterostome has not been forthcoming. A detailed ultrastructural study of its epidermis describes the previously noted morphological similarities to the epidermis of hemichordates as superficial and points out the differences in the organization of the ciliary apparatus and the junctional structures (Pedersen & Pedersen 1988).
(c). Cycliophorans, ectoprocts, entoprocts and their relatives
This is the first inclusion of Cycliophora in a phylogenomic study. The new data and analyses place the cycliophoran Symbion pandora with strong support as sister to entoprocts, consistent with a series of anatomical similarities in ultrastructure and developmental features (Funch & Kristensen 1995). The cycliophoran/entoproct grouping is a result recently recovered with molecular sequence data (Passamaneck & Halanych 2006; Paps et al. 2009).
In most of our analyses, the clade composed of Entoprocta and Cycliophora is placed as sister to Ectoprocta (=Bryozoa to some authors), although with low bootstrap support (figures 2–4). This relationship was suggested by Funch & Kristensen (1995) and a recent phylogenomic analysis found evidence for a clade of entoprocts and ectoprocts, which they referred to as Bryozoa (Hausdorf et al. 2007), but cycliophorans were not sampled. For many years, Ectoprocta and Entoprocta were treated as not being closely related, though Nielsen (2001, and references therein) has long argued for uniting the two groups as Bryozoa. Cavalier-Smith (1998) resurrected the name Polyzoa (originally coined for what is now accepted as Bryozoa) as a taxon to include bryozoans, entoprocts and cycliophorans. Our molecular analyses find evidence for this group, to which we also apply the name Polyzoa. The 844-gene analysis provides more than 80 per cent bootstrap support (figures 3 and 4) for Polyzoa being sister to Platyzoa within Spiralia, and this topology is widely recovered across analyses, though with varying support. Certain features of one polyzoan group, Entoprocta, support the placement of the clade within Spiralia. Two entoprocts that have been studied show spiral cleavage (Marcus 1939; Malakhov 1990), though further detailed embryological analyses are needed.
(d). Ctenophores and the base of the animal tree
Dunn et al. (2008) found strong support for the placement of ctenophores, rather than sponges, as the sister group to all other animals, although it was cautioned that this result should be treated provisionally until taxon sampling was improved. The present paper considers further ctenophore and sponge EST data, as well as Trichoplax genome data (Srivastava et al. 2008), and still gets the same result in analyses of the 1487-, 844- and 330-gene matrices (figure 2). Since the completion of the analyses presented here, an EST study with sampling from all major groups of sponges has been published (Philippe et al. 2009). This study placed Porifera as sister to other metazoans, but bootstrap support was low (62% for other animals, Eumetazoa, to the exclusion of sponges). A recent analysis of a manually curated set of mitochondrial and nuclear genes, together with a small morphological matrix, concluded that ‘Diploblastica’ (including Porifera), not Ctenophora or Porifera, is sister to all other animals (Schierwater et al. 2009). This topology, however, was statistically indistinguishable from a tree that placed Ctenophora as sister to all other animals (see table 1 in Schierwater et al. 2009). Analyses of the deepest splits in the animal tree of life clearly require further taxon sampling, with both new EST and genome projects for Porifera and Ctenophora in particular, before they can be rigorously evaluated.
(e). Phylogenetic inference
This study demonstrates the feasibility of a scalable, fully automated phylogenomic matrix construction method that requires little a priori knowledge for gene selection and is therefore portable to any group of organisms and any scale of phylogenetic problem. Such tools are critical if phylogenomic analyses are to leverage the new high-throughput sequencing technologies. Priorities for future development include improvement of the representation in the final analyses of taxa with relatively few available sequences.
Acknowledgements
This work is supported by the National Science Foundation under the AToL programme to G.G. (EF05-31757), M.Q.M. (EF05-31558) and W.C.W. (EF05-31677), by NASA to M.Q.M. and through IBM Blue Gene/L time provided by the San Diego Supercomputer Center. Additional support to individual project members was also provided from multiple sources. S. roscoffensis ESTs have been sequenced by Genoscope, France. Thanks to S. Smith for implementing monophyly masking in a developmental version of Phyutility and John Bishop for contributing with Pedicellina cultures.
References
- Baguñà J., Riutort M.2004The dawn of bilaterian animals: the case of acoelomorph flatworms. Bioessays 26, 1046–1057 (doi:10.1002/bies.20113) [DOI] [PubMed] [Google Scholar]
- Bourlat S. J., Hejnol A.2009Acoels. Curr. Biol. 19, R279–R280 [DOI] [PubMed] [Google Scholar]
- Bourlat S. J., Nielsen C., Lockyer A. E., Littlewood D. T. J., Telford M. J.2003Xenoturbella is a deuterostome that eats molluscs. Nature 424, 925–928 (doi:10.1038/nature01851) [DOI] [PubMed] [Google Scholar]
- Bourlat S. J., et al. 2006Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444, 85–88 (doi:10.1038/nature05241) [DOI] [PubMed] [Google Scholar]
- Castresana J.2000Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.1998A revised six-kingdom system of life. Biol. Rev. 73, 203–266 (doi:10.1017/S0006323198005167) [DOI] [PubMed] [Google Scholar]
- Chen F., Mackey A. J., Vermunt J. K., Roos D. S.2007Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2, e383 (doi:10.1371/journal.pone.0000383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C. W., et al. 2008Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 (doi:10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- Edgar R. C.2004MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (doi:10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., et al. 2009To be or not to be a flatworm: the acoel controversy. PLoS ONE 4, e5502 (doi:10.1371/journal.pone.0005502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers U.1985Das phylogenetische System der Plathelminthes Stuttgart, Germany: Gustav Fischer [Google Scholar]
- Franzén A., Afzelius B.1987The ciliated epidermis of Xenoturbella bocki (Platyhelminthes, Xenoturbellida) with some phylogenetic considerations. Zool. Scripta 16, 9–17 (doi:10.1111/j.1463-6409.1987.tb00046.x) [Google Scholar]
- Funch P., Kristensen R. M.1995Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature 378, 711–714 (doi:10.1038/378711a0) [Google Scholar]
- Giribet G., Dunn C. W., Edgecombe G. D., Hejnol A., Martindale M. Q., Rouse G. W.2009Assembling the spiralian tree of life. In Animal evolution: genes, genomes, fossils and trees (eds Telford M. J., Littlewood D. T. J.), pp. 52–64 Oxford, UK: Oxford University Press [Google Scholar]
- Haszprunar G.1996Plathelminthes and Plathelminthomorpha—paraphyletic taxa. J. Zool. Syst. Evol. Res. 34, 41–48 [Google Scholar]
- Hausdorf B., Helmkampf M., Meyer A., Witek A., Herlyn H., Bruchhaus I., Hankeln T., Struck T. H., Lieb B.2007Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol. Biol. Evol. 24, 2723–2729 (doi:10.1093/molbev/msm214) [DOI] [PubMed] [Google Scholar]
- Hejnol A., Martindale M. Q.2008aAcoel development indicates the independent evolution of the bilaterian mouth and anus. Nature 456, 382–386 (doi:10.1038/nature07309) [DOI] [PubMed] [Google Scholar]
- Hejnol A., Martindale M. Q.2008bAcoel development supports a simple planula-like urbilaterian. Phil. Trans. R. Soc. B 363, 1493–1501 (doi:10.1098/rstb.2007.2239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jondelius U., Ruiz-Trillo I., Baguñà J., Riutort M.2002The Nemertodermatida are basal bilaterians and not members of the Platyhelminthes. Zool. Scripta 31, 201–215 (doi:10.1046/j.1463-6409.2002.00090.x) [Google Scholar]
- Lundin K.1998The epidermal ciliary rootlets of Xenoturbella bocki (Xenoturbellida) revisited: new support for a possible kinship with the Acoelomorpha (Platyhelminthes). Zool. Scripta 27, 263–270 (doi:10.1111/j.1463-6409.1998.tb00440.x) [Google Scholar]
- Lundin K.2001Degenerating epidermal cells in Xenoturbella bocki (phylum uncertain). Nemertodermatida and Acoela (Platyhelminthes). Belgian J. Zool. 131, 153–157 [Google Scholar]
- Lundin K., Hendelberg J.1996Degenerating epidermal bodies (‘pulsatile bodies’) in Meara stichopi (Plathelminthes, Nemertodermatida). Zoomorphology 116, 1–5 (doi:10.1007/BF02526924) [Google Scholar]
- Malakhov V. V.1990Description of the development of Ascopodaria discreta (Coloniales, Barentsiidae) and discussion of the Kamptozoa status in the animal kingdom. Zool. Zh. 69, 20–30 [Google Scholar]
- Marcus E.1939Bryozoarios marinhos brasileiros III. Boletim da Faculdade de filosofia, ciências e letras, Universidade di Sao Paolo, Zoologia 3, 113–299 [Google Scholar]
- Nielsen C.2001Animal evolution New York, NY: Oxford University Press [Google Scholar]
- Paps J., Baguñà J., Riutort M.2009Lophotrochozoa internal phylogeny: new insights from an up-to-date analysis of nuclear ribosomal genes. Proc. R. Soc. B 276, 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck Y., Halanych K. M.2006Lophotrochozoan phylogeny assessed with LSU and SSU data: evidence of lophophorate polyphyly. Mol. Phylogenet. Evol. 40, 20–28 (doi:10.1016/j.ympev.2006.02.001) [DOI] [PubMed] [Google Scholar]
- Pedersen K., Pedersen L.1988Ultrastructural observations on the epidermis of Xenoturbella bocki Westblad, 1949, with a discusion of epidermal cytoplasmic filament systems of Invertebrates. Acta Zool. 69, 231–246 [Google Scholar]
- Perseke M., Hankeln T., Weich B., Fritzsch G., Stadler P. F., Israelsson O., Bernhard D., Schlegel M.2007The mitochondrial DNA of Xenoturbella bocki: genomic architecture and phylogenetic analysis. Theory Biosci. 126, 35–42 (doi:10.1007/s12064-007-0007-7) [DOI] [PubMed] [Google Scholar]
- Philippe H., Brinkmann H., Martinez P., Riutort M., Baguñà J.2007Acoel flatworms are not Platyhelminthes: evidence from phylogenomics. PLoS ONE 2, e717 (doi:10.1371/journal.pone.0000717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., et al. 2009Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 (doi:10.1016/j.cub.2009.02.052) [DOI] [PubMed] [Google Scholar]
- Raikova O. I., Reuter M., Jondelius U., Gustafsson M. K. S.2000An immunocytochemical and ultrastructural study of the nervous and muscular systems of Xenoturbella westbladi (Bilateria inc. sed.). Zoomorphology 120, 107–118 (doi:10.1007/s004350000028) [Google Scholar]
- Raikova O. I., Reuter M., Justine L.2001Contributions to the phylogeny and systematics of the Acoelomorpha. In Interrelationships of the Platyhelminthes (eds Littlewood D. T. J., Bray R. A.), pp. 13–23 London, UK: Taylor & Francis [Google Scholar]
- Reisinger E.1972Die Evolution des Orthogons der Spiralier und das Archicölomatenproblem. Z. Zool. Syst. Evolutionsforsch. 10, 1–43 (doi:10.1111/j.1439-0469.1972.tb00783.x) [Google Scholar]
- Remane A.1950Die Entstehung der Metamerie der Wirbellosen. Zool. Anz. Suppl. 14, 18–23 [Google Scholar]
- Ruiz-Trillo I., Riutort M., Littlewood D. T. J., Herniou E. A., Baguñà J.1999Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science 283, 1919–1923 (doi:10.1126/science.283.5409.1919) [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I., Paps J., Loukota M., Ribera C., Jondelius U., Baguñà J., Riutort M.2002A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proc. Natl Acad. Sci. USA 99, 11 246–11 251 (doi:10.1073/pnas.172390199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B., Eitel M., Jakob W., Osigus H. J., Hadrys H., Dellaporta S. L., Kolokotronis S. O., Desalle R.2009Concatenated analysis sheds light on early metazoan evolution and fuels a modern ‘urmetazoon’ hypothesis. PLoS Biol. 7, e20 (doi:10.1371/journal.pbio.1000020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. A., Dunn C. W.2008Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716 (doi:10.1093/bioinformatics/btm619) [DOI] [PubMed] [Google Scholar]
- Srivastava M., et al. 2008The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 (doi:10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Ott M.2008Efficient computation of the phylogenetic likelihood function on multi-gene alignments and multi-core architectures. Phil. Trans. R. Soc. B 363, 3977–3984 (doi:10.1098/rstb.2008.0163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley J. L., Wilkinson M.1999Testing the phylogenetic stability of early tetrapods. J. Theor. Biol. 200, 343–344 (doi:10.1006/jtbi.1999.0999) [DOI] [PubMed] [Google Scholar]
- van Dongen S.2000Graph clustering by flow simulation. PhD thesis, University of Utrecht, Holland [Google Scholar]
- Wallberg A., Curini-Galletti M., Ahmadzadeh A., Jondelius U.2007Dismissal of Acoelomorpha: Acoela and Nemertodermatida are separate early bilaterian clades. Zoolog. Scripta 36, 509–523 (doi:10.1111/j.1463-6409.2007.00295.x) [Google Scholar]
- Westblad E.1949Xenoturbella bocki n. g., n. sp., a peculiar, primitive Turbellarian type. Arkiv för Zool. 1, 3–29 [Google Scholar]