Abstract
Across angiosperms, variable rates of molecular substitution are linked with life-history attributes associated with woody and herbaceous growth forms. As the number of generations per unit time is correlated with molecular substitution rates, it is expected that rates of phenotypic evolution would also be influenced by differences in generation times. Here, we make the first broad-scale comparison of growth-form-dependent rates of niche evolution. We examined the climatic niches of species on large time-calibrated phylogenies of five angiosperm clades and found that woody lineages have accumulated fewer changes per million years in climatic niche space than related herbaceous lineages. Also, climate space explored by woody lineages is consistently smaller than sister lineages composed mainly of herbaceous taxa. This pattern is probably linked to differences in the rate of climatic niche evolution. These results have implications for niche conservatism; in particular, the role of niche conservatism in the distribution of plant biodiversity. The consistent differences in the rate of climatic niche evolution also emphasize the need to incorporate models of phenotypic evolution that allow for rate heterogeneity when examining large datasets.
Keywords: life history, rates of evolution, phylogenetics, climatic niche
1. Introduction
Heterogeneity in the rate of molecular evolution has been widely documented (Laird et al. 1969; Kohne 1970; Sibley & Ahlquist 1984; Wu & Li 1985; Bousquet et al. 1992; Gaut et al. 1992, 1996, 1997; Andreasen & Baldwin 2001; Bromham et al. 2002; Smith & Donoghue 2008; Ho 2009) and has been attributed to a variety of causes, including differences in body size (Martin & Palumbi 1993; Bromham et al. 1996), metabolic rate (Martin & Palumbi 1993; Gillooly et al. 2005), DNA repair (Sarich & Wilson 1973; Britten 1986) and generation time (Mooers & Harvey 1994; Bromham et al. 1996), with correlations among these variables also being observed (Martin & Palumbi 1993). In plants, there is strong evidence for heterogeneity in the rate of molecular evolution between woody and herbaceous species (Gaut et al. 1992, 1996; Laroche et al. 1997; Kay et al. 2006; Smith & Donoghue 2008), and to a lesser extent between annuals and perennials (Andreasen & Baldwin 2001). Both the differences in rate between woody and herbaceous species, and between annual and perennial plants, probably reflect a difference in generation time (Sturtevant 1965; Wu & Li 1985; Li et al. 1987; Gaut et al. 1992, 1996; Smith & Donoghue 2008). This evidence suggests that plant lineages with longer generation times (i.e. the time to first flower) tend to accumulate nuclear substitutions at a slower rate per unit time than lineages with shorter generation times.
While the number of generations per unit time has been shown to be correlated with molecular substitution rates (e.g. Gaut et al. 1992, 1996; Kay et al. 2006; Smith & Donoghue 2008), it remains unclear whether life-history attributes such as generation time influence the tempo of phenotypic evolution. Early studies suggested that molecular and phenotypic rates were largely decoupled (Kimura 1983; Ohta 1992; Bromham et al. 2002). More recently, Davies & Savolainen (2006) found evidence for such a correlation in plants, with molecular change accounting for 2 to 11 per cent of the variance in the rate of morphological evolution (see also Xiang et al. 2008 for similar results within Cornus). Though evidence for a correlation between rates of molecular and phenotypic evolution is clearly mounting, to date there remains no clear consensus on this issue. A correlation between rates of molecular and phenotypic evolution would be remarkable and would provide evidence for the far-reaching consequences of life history across both genotypic and phenotypic scales.
Before molecular data were available, generation time was thought to exert a profound influence on the rates of phenotypic evolution (e.g. Sinnott 1916; Simpson 1944). The number of generations per unit time can influence the potential for adaptive phenotypic change by modulating the rate at which advantageous mutations arise in a population (Sinnott 1916). However, Simpson (1944, pp. 62–64) postulated that although generation time could cause dramatic differences in phenotypic evolution, this had not been widely observed. Yet phylogenetic information has not specifically been brought to bear on these questions relating generation time, morphological evolution and molecular evolution. Recent advances in large-scale phylogeny construction (Smith & Donoghue 2008; Smith et al. 2009) combined with sophisticated model-based phylogenetic comparative methods provide an ideal framework in which to adequately address this longstanding question.
Here, we demonstrate the correlation between growth form and rates of phenotypic evolution as measured by climate tolerance in flowering plants. We examine this pattern across previously published phylogenies representing a diverse set of flowering plant species. Because generation time is correlated with plant habit, designations such as ‘woody’ and ‘herbaceous’ provide convenient distinctions for testing this pattern of rate heterogeneity. In this study, we focus on climate tolerance, which includes a particularly interesting set of traits to examine. This is not only because abundant spatial data allow for the compilation of very large datasets, but also results pertaining to climate tolerance evolution have broad implications for the evolution of flowering plants and the ability of plants to respond to climate fluctuations.
2. Material and methods
(a). Phylogenetic trees
Smith & Donoghue (2008) previously examined molecular rate heterogeneity and plant habit in five large angiosperm phylogenies: Apiales, Commelinidae (Cantino et al. 2007), Dipsacales, ‘Primulales’ and Moraceae + Urticaceae (Rosales). From this study, we relied on both the time-calibrated phylogenies and the information on plant habit. We also use the uncalibrated branch lengths from these phylogenies to assess the adequacy of molecular branch lengths in properly scaling phenotypic change according to life history (see below).
(b). Geographical and climate data
To estimate a climate tolerance for a given species, we retrieved all GPS coordinates provided by the Global Biodiversity Information Facility (GBIF; http://gbif.org). This resulted in 889 species for Apiales, 3174 species for Commelinidae (commelinids; Cantino et al. 2007), 290 species for Dipsacales, 301 species for Primulales (Ericales, APG II) and 351 species for Moraceae + Urticaceae (Rosales). We then extracted the mean climate data for each of the 19 BIOCLIM variables at 2.5 arc-minutes resolution developed by Hijmans et al. (2005), which describe the major temperature and precipitation dimensions of a given species. The BIOCLIM variables are biologically meaningful layers derived from monthly rainfall and temperature values. Because our datasets involved taxonomically broad samples spanning a wide range in geographical and climatic spaces, we used all 19 BIOCLIM variables in our analyses.
(c). Testing for differences in climatic niche dimensions among growth form
To determine the climatic niche for each species in each clade, we conducted a principal component analysis across all 19 BIOCLIM variables. Because the data represent multicollinear species values, we used a principal component analysis that specifically incorporates phylogenetic information while calculating eigenvectors, eigenvalues, loadings and species scores (Revell in press). While this method takes into account phylogenetic information when estimating the eigenstructure of the data, the resulting PC scores remain in the original species space and therefore require a comparative approach for any subsequent analysis due to shared ancestry. The first two principal axes (PC1 and PC2) were chosen as these represented the major axes of temperature and precipitation dimensions, and accounted for a majority of the shared variation among all species of a given clade. These analyses were performed in R (R Development Core Team 2009) using code generously provided by L. J. Revell.
To assess whether there were mean differences in climatic niche space, we used a phylogenetic MANOVA (multivariate analysis of variance) to test whether significant differences in cross-species trait means among growth form were larger than expected based on a random model of Brownian motion (BM) evolution (sensu Garland et al. 1993). We used the R (R Development Core Team 2009) package GEIGER (Harmon et al. 2008) to generate 1000 Monte Carlo simulations using our input tree topology and time-calibrated branch lengths. All simulations were carried out under a gradual model of BM evolution and we relied on Wilks's λ as our multivariate test statistic. Wilks's λ measures the proportion of variance in multiple dependent variables (i.e. niche axes) that remains unexplained by the independent variable (i.e. growth form). The null distribution of Wilks's λ was compared against the observed Wilks's λ calculated from a conventional MANOVA. If the observed Wilks's λ was less than 95 per cent of the null distribution, then trait differences were greater than expected based on a model of BM evolution. We carried out this analysis separately for each clade.
To determine whether there were differences in the variance of climatic niche space between sister lineages that differ only in predominate growth form, we examined the observed variance in PC1 and PC2, separately. To do so, we calculated a sample variance (mean-squared error; MSE) that is the average squared difference between a tip value and the phylogenetic mean (â), or the maximum-likelihood ancestral trait value at the root node of a given tree (Schluter et al. 1997; Blomberg et al. 2003). In this case, the phylogenetic mean represented the ancestral trait value for a particular PC axis, analysed separately, at the root of a subtree consisting of sister lineages. To estimate a phylogenetic mean, we used a phylogenetic generalized least-squares analysis that incorporated the BM covariance structure of a phylogeny into the calculation of the mean (Martins & Hansen 1997; Garland & Ives 2000). Because we analysed each PC axis separately, the independent variable was treated as fixed to 1 for all values of the dependent variable (the PC axis). In this way, the phylogenetic mean is equivalent to the estimate of the grand mean of the dependent variable, which is also the slope intercept of the regression model (Garland & Ives 2000; Rohlf 2001). We calculated a disparity ratio from the observed disparity in the herb lineage to the disparity observed by the woody lineage. Therefore, a disparity ratio greater than 1 would indicate that the herbaceous lineage exhibits greater tip disparity than the woody lineage. It is important to note that while this is somewhat analogous to the rate analysis of O'Meara et al. (2006), we view this analysis as a more detailed description of the observed climate space occupied by a given lineage.
We also calculated a disparity ratio based on the expected tip variances predicted by the tree structure underlying each growth form lineage comparison. The expected disparity was calculated from eqn (1) in O'Meara et al. (2006) and is a function of three factors: (i) the rate of BM evolution (which, in this case, is irrelevant as it factors out of the numerator and denominator, so we arbitrarily set it to one); (ii) the path length of a tip to the root of the clade of interest; and (iii) the average entry in the covariance matrix (O'Meara et al. 2006). Larger values of factors (i) and (ii) increase disparity while larger values of factor (iii) decrease disparity. Our particular interest was the influence that speciation had on our observed disparity ratio. Speciation events occurring more recently necessarily increase the average entry in the covariance matrix and, consequently, reduce the expected disparity among tip taxa. Thus, our use of the expected disparity was meant to examine whether the observed asymmetry in the ratio of tip variance between herbaceous and woody lineages was greater than the disparity expected based solely on their respective underlying tree topology and branch lengths. Both the observed and expected disparity ratio analyses were carried out using code written by J.M.B. for R (R Development Core Team 2009). It is important to note that while this is somewhat analogous to the evolutionary rate analysis of O'Meara et al. (2006), our method should be viewed as a more detailed description of the observed climate space occupied by a given lineage, whereas the evolutionary rate analysis describes the tempo of filling that space.
(d). Estimating rates of climatic niche evolution
To assess whether there are differences in the rate of climatic niche evolution among woody and herbaceous lineages, we compared the fit of a single-rate model of BM evolution with that of a multiple-rate model. The single-rate model assumes that all lineages accumulated evolutionary changes in climate tolerance at the same rate—σ2, or the variance of phenotypic evolution—while the multiple-rate model assigns a separate rate to lineages that differ in growth form (e.g. σ2HERB and σ2WOODY). We carried out model comparisons using the ‘non-censored’ approach in BROWNIE v. 2.1 (O'Meara et al. 2006). Because the non-censored approach includes information about internal branches when estimating rate parameters, we used the likeliest state reconstructed along each edge, a procedure implemented in BROWNIE to obtain an estimate of the ancestral growth form state (e.g. woody or herbaceous) across all branches in a given tree. The best-fitting model was chosen based on the second-order information criterion, AICc (Akaike 1974; Sugiura 1978), owing to low sample size in some cases.
In phylogenies with branch lengths proportional to substitutions per site (i.e. a non-ultrametric phylogeny), branch lengths may represent estimates of evolutionary rate times time, instead of just time. If phenotypic evolution were proportional to rates of molecular evolution, the phylogenies with branch lengths proportional to substitutions per site would be a better model for variances and covariances of species than the ultrametric trees. Because molecular branch lengths apparently reflect the bias of generation time on accumulated molecular rates of evolution (Smith & Donoghue 2008), we also conducted these rate analyses to assess whether molecular branch lengths better accommodate for generation time by generally supporting a single-rate model.
3. Results
(a). The major axes of climate tolerance
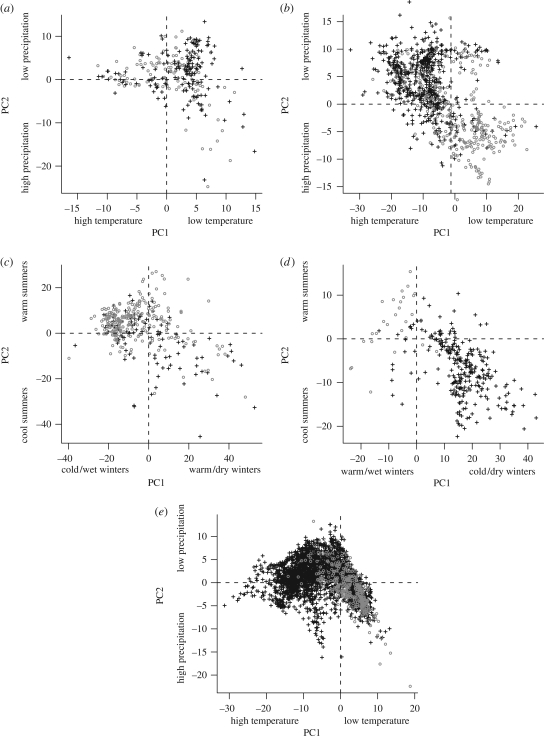
Principal component analysis of the 19 BIOCLIM variables was used to uncover the primary axes of climate variation exhibited among species contained within each of the five angiosperm clades we examined. The first (PC1) and second (PC2) components explained 34.6–43.2 and 23.1–29.7 per cent of the variation, respectively, accounting for a majority of the total variation (electronic supplementary material, tables S1–S5). These axes mainly reflected temperature and precipitation seasonality gradients (figure 1).
Figure 1.
The primary axes (PC1 and PC2) of climate variation exhibited among woody (grey circles) and herbaceous (black crosses) species contained within (a) Dipsacales, (b) Apiales, (c) Moraceae + Urticaceae, (d) Primulales and (e) Commelinidae. Interpretations of the variable loadings for each axis are indicated as axis subheadings.
The variable loadings for PC1 calculated for Dipsacales, Apiales and the commelinids indicated that species separate along a gradient of temperature seasonality (figure 1). In all three clades, annual mean temperature was negatively associated with mean daily temperature range, temperature seasonality, and annual temperature range. For Moraceae + Urticaceae and Primulales, PC1 was associated with both temperature and precipitation patterns in the winter months (figure 1).
For Dipsacales, Apiales and the commelinids, PC2 indicated that species separate along precipitation seasonality. The loadings for these clades showed strong associations with annual precipitation, especially variables associated with precipitation during the warmest months. In contrast, PC2 for Moraceae + Urticaceae and Primulales was associated mainly with temperature conditions during the warmest months. In both cases, PC2 loaded strongest with maximum temperature in the warmest month and mean temperature in the warmest quarter.
(b). Growth form separation in climate tolerance
In all but one clade, there were significant differences in mean climate tolerance among extant woody and herbaceous species, but no more different than would be expected given a model of BM evolution (table 1). That is, mean differences in climate tolerance observed among growth form could have arisen by chance. Primulales was the only clade to vary significantly (Wilks's λ = 0.598, F2,298, p = 0.005; table 1), even after incorporating both chance and phylogeny. However, this is probably due to sampling, as nearly all the woody species sampled for Primulales were contained within Theophrastaceae sensu stricto, a clade of trees and shrubs restricted to the northern regions of South America. The sister families, Myrisinaceae and Primulaceae, are much more widely distributed around the northern Hemisphere. Thus, the appearance of significant niche differentiation among woody and herbaceous species of Primulales may simply reflect geography.
Table 1.
Results from a conventional MANOVA and Monte Carlo simulations to test for significant niche differences between herbaceous and woody species relative to those expected based on stochastic BM evolution. The critical value is the 95th percentile obtained from a distribution of 1000 Monte Carlo simulated Wilks's λ-statistics assuming a gradual model of BM evolution. p-values from Monte Carlo simulations are the proportion of simulated Wilks's λ that are greater than the observed test statistic obtained from a conventional MANOVA. Italics indicate p < 0.05.
| clade | Wilks' λ | F-ratio | Monte Carlo p-value |
|---|---|---|---|
| Dipscales | |||
| growth form | 0.963 | 5.46 | 0.932 |
| Apiales | |||
| growth form | 0.621 | 269.80 | 0.090 |
| Moraceae + Urticaceae | |||
| growth form | 0.850 | 30.70 | 0.740 |
| Primulales | |||
| growth form | 0.598 | 100.20 | 0.005 |
| commelinids | |||
| growth form | 0.836 | 312.00 | 0.337 |
Although in general we detected no meaningful differences in overall climate tolerance among growth form, we did observe a general asymmetry in the amount of climatic niche space occupied by extant woody and herbaceous species. Lineages composed of predominately herbaceous extant species consistently occupied a greater amount of climatic niche space described by PC1 than sister lineages composed of predominately woody extant species (table 2). Moreover, the observed asymmetry in the herb-to-woody tip disparity ratio was greater than that predicted based on the underlying tree topology and branch lengths. These patterns were similar for PC2, with the exception of sister lineage comparisons within Apiales and commelinids (table 2). In the case of Apiales, species contained within Pittosporaceae + Myodocarpaceae exhibited nearly twice the variation in precipitation tolerance than the strictly herbaceous Apiaceae (table 2).
Table 2.
Disparity ratios depicting asymmetry in the observed tip variance (MSE) observed between sister lineages that differ only in predominate growth form exhibited by tip taxa (herb versus woody). The expected ratio is the expected disparity ratio between the sister lineages based solely on the underlying tree topology and branch lengths. Observed tip variance was calculated as the MSE of the tip data measured from the phylogenetically correct mean (â) or the estimated trait value at the root node of the tree. Expected tip variance was calculated from eqn (1) in O'Meara et al. (2006). Capri, Caprifoliaceae + Heptacodium; rest D, rest of the Dipsacales; Pitt, Pittosporaceae; Myodo, Myodocarpaceae; Api, Apiaceae; Mora, Moraceae; Urtica, Urticaceae; Theo, Theophrastaceae; Prim, Primulaceae; Myrs, Myrsinaceae; Areca, Arecaceae; rest C, rest of the commelinids.
| clade | herb : woody |
||
|---|---|---|---|
| PC1 | PC2 | expected | |
| Dipsacales | |||
| Capri versus rest D | 1.72 | 1.530 | 0.633 |
| Apiales | |||
| Pitt + Myodo versus Api | 1.17 | 0.582 | 0.942 |
| Moraceae + Urticaceae | |||
| Mora versus Urtica | 1.28 | 1.690 | 0.930 |
| Primulales | |||
| Theo versus Prim + Myrs | 2.85 | 1.610 | 0.939 |
| commelinids | |||
| Areca versus rest C | 3.67 | 0.883 | 1.510 |
(c). Rates of niche evolution
We assessed the fit of two BM models that differed in the number of evolutionary rate parameters. For both PC1 and PC2 in each clade, using time-calibrated branch lengths, a two-rate model that inferred separate rates for woody and herbaceous species was strongly favoured over a single-rate model of BM (table 3). Herbs consistently accumulated changes in climate tolerance at much higher rates than related woody lineages; the estimated rate parameters ranged from approximately 2 to 12 times as high in herbs as in trees/shrubs. In only one case did we estimate tree/shrubs to have a higher rate. Changes in precipitation seasonality (PC2) for herbs within Apiales accumulated at a rate that was nearly three times slower than related woody lineages (table 3). Interestingly, in each clade, results obtained when using molecular branch lengths also supported a two-rate model based on growth form for PC1, with varying results for PC2 (table 4).
Table 3.
Parameter estimates and fit of single- and multiple-rate models of BM to the major axes of climate tolerance using phylogenies with branch lengths in units of millions of years. Bold indicates the favoured model based on the sample-size-corrected AICc. Positive ΔAICc denotes significant contribution of the multiple-rate BM. σ2, estimated single-rate BM parameter; σ2HERB, rate parameter for herbaceous lineages; σ2WOODY, rate parameter for woody lineages.
| clade | single-rate BM, σ2 | multiple-rate BM |
||
|---|---|---|---|---|
| ΔAICc | σ2HERB | σ2WOODY | ||
| Dipsacales | ||||
| PC1 | 7.35 | 135.9 | 12.40 | 1.43 |
| PC2 | 4.36 | 33.8 | 6.21 | 2.23 |
| Apiales | ||||
| PC1 | 7.97 | 36.0 | 9.43 | 4.90 |
| PC2 | 5.63 | 115.1 | 3.52 | 10.10 |
| Moraceae + Urticaceae | ||||
| PC1 | 8.18 | 44.5 | 15.10 | 5.20 |
| PC2 | 4.98 | 68.4 | 10.30 | 2.76 |
| Primulales | ||||
| PC1 | 6.55 | 5.2 | 6.95 | 2.81 |
| PC2 | 4.71 | 20.6 | 5.09 | 0.955 |
| commelinidsa | ||||
| PC1 | 4.10 | 61.0 | 4.46 | 1.42 |
| PC2 | 2.77 | 21.5 | 2.94 | 1.52 |
aOne of five randomly sampled subsets of 1200 commelinid species.
Table 4.
Parameter estimates and fit of single- and multiple-rate models of BM to the major axes of climate tolerance. Analyses were performed on phylogenies with branch lengths in units of substitutions per site (rates multiplied by 10−4 to allow for easier comparison). Bold indicates the favoured model based on the sample-size-corrected AICc. Positive ΔAICc denotes significant contribution of the multiple-rate BM. σ2, estimated single-rate BM parameter; σ2HERB, rate parameter for herbaceous lineages; σ2WOODY, rate parameter for woody lineages.
| clade | single-rate BM, σ2 | multiple-rate BM |
||
|---|---|---|---|---|
| ΔAICc | σ2HERB | σ2WOODY | ||
| Dipsacales | ||||
| PC1 | 110.84 | 155.90 | 189.93 | 18.21 |
| PC2 | 3.61 | 1.00 | 32.98 | 39.91 |
| Apiales | ||||
| PC1 | 98.94 | 8.50 | 105.36 | 83.49 |
| PC2 | 1.09 | 93.00 | 40.14 | 148.35 |
| Moraceae + Urticaceae | ||||
| PC1 | 52.99 | 6.50 | 82.94 | 38.99 |
| PC2 | 42.67 | 1.90 | 442.45 | 42.01 |
| Primulales | ||||
| PC1 | 41.81 | 77.50 | 45.69 | 0.86 |
| PC2 | 21.35 | 80.20 | 23.38 | 0.39 |
| commelinidsa | ||||
| PC1 | 6.79 | 185.60 | 7.64 | 0.23 |
| PC2 | 3.92 | 260.70 | 4.46 | 0.26 |
aOne of five randomly sampled subsets of 1200 commelinid species.
4. Discussion
Our results demonstrate that the rate of climatic niche evolution is growth-form-dependent. We hypothesize that, because growth form in plants is correlated to generation time, these differences in rates of climatic niche evolution are associated with differences in generation time. With the exception of Primulales, these results did not reflect differences in the average climate space occupied by woody and herbaceous species. Instead, the growth form dependency reflected differences in the overall amount of occupied climatic niche space. Across both climate axes (PC1 and PC2), woody species generally exhibited less interspecific variation in occupied climate space than herbaceous species of similar age (table 2). This pattern is clearly linked to differences in the rate of climatic niche evolution. We found that, in all but one of the ten comparisons, the rate of climatic niche evolution in woody lineages ranged from nearly 2 to 12 times slower than related herbaceous lineages. Even molecular branch lengths—which, if phenotypic evolution were proportional to rates of molecular evolution, would be a better model for variances and covariances of species—also strongly favoured a two-rate model for the primary climatic niche axis (table 4). Taken together, our results strongly suggest that life attributes such as generation time can impose limits to climate tolerance evolution in flowering plants.
Recently, there has been renewed interest in the extent to which niches evolve (e.g. Wiens & Donoghue 2004; Wiens & Graham 2005; Donoghue 2008). The theory of niche conservatism suggests that closely related species tend to exhibit very similar habitat requirements, implying that speciation does not necessarily result in evolutionary shifts across niche dimensions (Wiens 2004; Wiens & Graham 2005; Kozak & Wiens 2006). Donoghue (2008) suggests that, when faced with a changing environment, plant movements along corridors may be favoured over evolving key adaptations in place. Our results reveal an additional layer of complexity in understanding plant responses to changing environmental conditions. Because of the slower rates of both phenotypic and molecular evolution in woody lineages, there is an apparent constraint imposed by growth form and, by correlation, possibly generation time. Therefore, woody lineages may disperse more rapidly than accumulated mutation rates can allow for adaptation. Hence, instead of adapting to new climate tolerances, woody species may not diverge far from ancestral climate tolerances before they migrate to available niche space. This is evidenced by the clear reduction in observed climate space occupied by woody species (figure 1; table 2), while herbaceous lineages, lacking molecular and phenotypic constraints imposed by generation time, may be better able to accommodate themselves to new, emerging environments.
The tendency for woody plants to be less labile with regard to niche evolution also has implications for the evolution of the earliest angiosperms. The climatic conditions under which the first angiosperms have evolved have been the subject of considerable study, as these conditions would have implications for the evolution of a suite of morphological traits that characterize angiosperms (e.g. Stebbins 1965; Cronquist 1988; Donoghue & Doyle 1989; Taylor & Hickey 1992; Sun et al. 2002; Field et al. 2004). Hypotheses for the early angiosperm habit have ranged from shrubs (Stebbins 1965; Cronquist 1988) to herbs (Donoghue & Doyle 1989; Taylor & Hickey 1992) to aquatic plants (Sun et al. 2002; but see Friis et al. 2003). More recently, Field et al. (2004), using phylogenetic reconstructions, inferred a woody growth form as the ancestral state for early angiosperms. From our results, a woody growth habit would limit the degree of adaptation of early angiosperms to changing or fluctuating climates. In many studies, the phylogenetic branch leading to angiosperms is long, measured either by molecular substitutions or time, suggesting a low net diversification rate (Magallón & Sanderson 2001). The findings in this paper may help further our understanding of the low net diversification rate characterizing early angiosperm lineages by attributing lower rates of habitat evolution to growth habit.
Although we demonstrate the correlation of growth form and rates of climatic niche evolution, we cannot rule out other factors that would also influence the rates of climate evolution. Specifically, population size may play a significant role, as it can influence the rate at which species adapt (Lynch 2007). For example, if certain lineages exhibit larger population sizes, as some have proposed for both woody (e.g. Petit & Hampe 2006) and herbaceous (e.g. Hamrick & Godt 1996) lineages, strong stabilizing selection may inherently reduce the rate of phenotypic evolution. Alternatively, large population sizes can allow for faster responses to changing selection coefficients. However, large datasets of population size do not yet exist, and current models of phenotypic evolution do not allow for decoupling the selection coefficient (e.g. α; sensu Butler & King 2004) across character states.
Previous studies have suggested correlated rates of evolution between morphology and molecular evolution in plants (Davies & Savolainen 2006; Xiang et al. 2008). The results presented here also demonstrate this pattern, but with climate tolerance, and by additionally examining this rate correlation in the context of growth form evolution. Specifically, the same heterogeneity in the rates of molecular evolution associated with growth form is also found in climate evolution. As it relates to molecular evolution, the generation time hypothesis posits that producing more generations per unit time will lead to a higher observed nucleotide substitution rate per unit time (Wu & Li 1985; Li et al. 1987; Gaut et al. 1992, 1996; Smith & Donoghue 2008). This mechanism could link the observed differences in rates of climate evolution among growth forms. Herbaceous plants that often reproduce in the first or second year of life can exhibit very high rates of nucleotide substitution and niche evolution. Trees and shrubs typically take longer to reach reproductive age (Verdú 2002; Petit & Hampe 2006), and high rates of nucleotide substitution and phenotypic evolution are correspondingly rare.
The consistent detection of two rates of phenotypic evolution, one for each growth form, inherently violates the assumption of a single rate of BM when incorporating phylogeny in a comparative test. If phenotypic rate heterogeneity is consistent with other traits (i.e. plant functional traits), comparative tests consisting of data for both woody and herbaceous species should test for differences in the strength of trait correlations. This would effectively treat branches of differing growth form as separate in a comparative test (Revell & Collar 2009). Growth-form-dependent trait correlations may help to understand additional factors (e.g. diversification rates) also associated with the disparity in phenotypic variation among woody and herbaceous species (e.g. Beaulieu et al. 2008; table 2, this study).
Further study is necessary in order to fully understand not only the extent but also the conditions under which molecular rates and phenotypic rates are correlated. For example, an examination of among-species variation is of particular interest. In this study, we discuss large-scale patterns, and more fine-scaled analyses are needed to determine what differences are apparent in among-species variation between different growth forms. Additionally, the examination of other phenotypic traits (aside from those related to climate tolerance is important), though one limitation is the size of available datasets. For plants, many of these patterns become apparent at very large scales (Davies & Savolainen 2006; Smith & Donoghue 2008) and therefore large-scale morphological datasets are required to fully address these questions.
Acknowledgements
Valuable feedback was received from Brian O'Meara, Jeff Oliver and two anonymous reviewers. S.A.S. has been supported through the National Evolutionary Synthesis Center (NESCent; NSF no. EF-0423641). J.M.B. has been supported through an NSF Assembling the Tree of Life (AToL) award.
References
- Akaike H.1974A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723 (doi:10.1109/TAC.1974.1100705) [Google Scholar]
- Andreasen K., Baldwin B. G.2001Unequal evolutionary rates between annual and perennial lineages of checker mallows (Sidalcea, Malvaceae): evidence from 18S-26S rDNA internal and external transcribed spacers. Mol. Biol. Evol. 18, 936–944 [DOI] [PubMed] [Google Scholar]
- Beaulieu J. M., Leitch I. J., Patel S., Pendharkar A., Knight C. A.2008Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986 (doi:10.1111/j.1469-8137.2008.02528.x) [DOI] [PubMed] [Google Scholar]
- Blomberg S. P., Garland T., Jr, Ives A. R.2003Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 [DOI] [PubMed] [Google Scholar]
- Bousquet J., Strauss S. H., Doerksen A. H., Price R. A.1992Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc. Natl Acad. Sci. USA 89, 7844–7848 (doi:10.1073/pnas.89.16.7844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J.1986Rates of DNA sequence evolution differ between taxonomic groups. Science 231, 1393–1398 (doi:10.1126/science.3082006) [DOI] [PubMed] [Google Scholar]
- Bromham L., Rambaut A., Harvey P.1996Determinants of rate variation in mammalian DNA sequence evolution. J. Mol. Evol. 43, 610–621 (doi:10.1007/BF02202109) [DOI] [PubMed] [Google Scholar]
- Bromham L., Woolfit M., Lee M. S. Y., Rambaut A.2002Testing the relationship between morphological and molecular rates of change along phylogenies. Evolution 56, 1921–1930 [DOI] [PubMed] [Google Scholar]
- Butler M. A., King A. A.2004Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am. Nat. 164, 683–695 [DOI] [PubMed] [Google Scholar]
- Cantino P. D., Doyle J. A., Graham S. W., Judd W. S., Olmstead R. G., Soltis D. E., Soltis P. S., Donoghue M. J.2007Toward a phylogenetic nomenclature of Traecheophyta. Taxon 56, 822–846 [Google Scholar]
- Cronquist A.1988The evolution and classification of flowering plants New York, NY: New York Botanical Garden [Google Scholar]
- Davies T. J., Savolainen V.2006Neutral theory, phylogenies, and the relationship between phenotypic change and evolutionary rates. Evolution 60, 476–483 [PubMed] [Google Scholar]
- Donoghue M. J.2008A phylogenetic perspective on the distribution of plant diversity. Proc. Natl Acad. Sci. USA 105, 11 549–11 555 (doi:10.1073/pnas.0801962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M. J., Doyle J. A.1989Phylogenetic analysis of angiosperms and the relationships of Hamamelidae. In Evolution, systematics, and fossil history of the Hamamelidae, vol. 1 (eds Crane P. R., Blackmore S.), pp. 14–45 Oxford, UK: Clarendon [Google Scholar]
- Field T. S., Arens N. C., Doyle J. A., Dawson T. E., Donoghue M. J.2004Dark and disturbed: a new image of early angiosperm ecology. Paleobiology 30, 82–107 [Google Scholar]
- Friis E. M., Doyle J. A., Endress P. K., Leng Q.2003Archaefructus: angiosperm precursor or specialized early angiosperm? Trends Plant Sci. 8, 369–373 (doi:10.1016/S1360-1385(03)00161-4) [DOI] [PubMed] [Google Scholar]
- Garland T., Jr, Ives A. R.2000Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 155, 346–364 [DOI] [PubMed] [Google Scholar]
- Garland T., Jr, Dickerman A. W., Janis C. M., Jones J. A.1993Phylogenetic analysis of covariance by computer simulation. Syst. Biol. 42, 265–292 [Google Scholar]
- Gaut B. S., Muse S. V., Clark W. D., Clegg M. T.1992Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J. Mol. Evol. 35, 292–303 (doi:10.1007/BF00161167) [DOI] [PubMed] [Google Scholar]
- Gaut B. S., Morton B. R., McCaig B. C., Clegg M. T.1996Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl Acad. Sci. USA 93, 10 274–10 279 (doi:10.1073/pnas.93.19.10274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Clark L. G., Wendel J. F., Muse S. V.1997Comparisons of the molecular evolutionary process at rbcL and ndhF in the grass family (Poaceae). Mol. Biol. Evol. 14, 769–777 [DOI] [PubMed] [Google Scholar]
- Gillooly J. F., Allen A. P., West G. B., Brown J. H.2005The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc. Natl Acad. Sci. USA 102, 140–145 (doi:10.1073/pnas.0407735101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick J. L., Godt M. J. W.1996Effects of life history traits on genetic diversity in plants species. Phil. Trans. R. Soc. Lond. B 351, 1291–1298 (doi:10.1098/rstb.1996.0112) [Google Scholar]
- Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W.2008GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A.2005Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (doi:10.1002/joc.1276) [Google Scholar]
- Ho S. Y. W.2009An examination of phylogenetic models of substitution rate variation among lineages. Biol. Lett. 5, 421–424 (doi:10.1098/rsbl.2008.0729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay K. M., Whittall J. B., Hodges S. A.2006A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol. Biol. 6, 36 (doi:10.1186/1471-2148-6-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M.1983The neutral theory of molecular evolution Cambridge, UK: Cambridge University Press [Google Scholar]
- Kohne D. E.1970Evolution of higher-organism DNA. Q. Rev. Biophys. 3, 327–375 (doi:10.1017/S0033583500004765) [DOI] [PubMed] [Google Scholar]
- Kozak K. H., Wiens J. J.2006Does niche conservatism promote speciation? A case study in North American salamanders. Evolution 60, 2604–2621 [PubMed] [Google Scholar]
- Laird C. D., McConaughy B. L., McCarthy B. J.1969Rate of fixation of nucleotide substitutions in evolution. Nature 224, 149–154 (doi:10.1038/224149a0) [DOI] [PubMed] [Google Scholar]
- Laroche J., Peng L., Maggia L., Bousquet J.1997Molecular evolution of angiosperm mitochondrial introns and exons. Proc. Natl Acad. Sci. USA 94, 5722–5727 (doi:10.1073/pnas.94.11.5722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Tanimura M., Sharp P.1987An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 25, 330–342 (doi:10.1007/BF02603118) [DOI] [PubMed] [Google Scholar]
- Lynch M.2007The origin of genome architecture Sunderland, MA: Sinauer Associates, Inc. Publishers [Google Scholar]
- Magallón S. A., Sanderson M. J.2001Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780 [DOI] [PubMed] [Google Scholar]
- Martin A. P., Palumbi S. R.1993Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA 90, 4087–4091 (doi:10.1073/pnas.90.9.4087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins E. P., Hansen T. F.1997Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667 [Google Scholar]
- Mooers A., Harvey P.1994Metabolic rate, generation time, and the rate of molecular evolution in birds. Mol. Phylogenet. Evol. 3, 344–350 (doi:10.1006/mpev.1994.1040) [DOI] [PubMed] [Google Scholar]
- Ohta T.1992The nearly neutral theory of molecular evolution. Ann. Rev. Ecol. Syst. 23, 263–286 (doi:10.1146/annurev.es.23.110192.001403) [Google Scholar]
- O'Meara B. C., Ané C., Sanderson M. J., Wainwright P. C.2006Testing for different rates of continuous trait evolution using likelihood. Evolution 60, 922–933 [PubMed] [Google Scholar]
- Petit R. J., Hampe A.2006Some evolutionary consequences of being a tree. Annu. Rev. Ecol. Evol. Syst. 37, 187–214 (doi:10.1146/annurev.ecolsys.37.091305.110215) [Google Scholar]
- R Development Core Team 2009R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org [Google Scholar]
- Revell L. J.In press Size-correction and principal components for interspecific comparative studies. Evolution (doi:10.1111/j.1558-5646.2009.00804.x) [DOI] [PubMed] [Google Scholar]
- Revell L. J., Collar D. C.2009Phylogenetic analysis of the evolutionary correlation using likelihood. Evolution 63, 1090–1100 (doi:10.1111/j.1558-5646.2009.00616.x) [DOI] [PubMed] [Google Scholar]
- Rohlf F. J.2001Comparative methods for the analysis of continuous variables: geometric interpretations. Evolution 55, 2143–2160 [DOI] [PubMed] [Google Scholar]
- Sarich V. M., Wilson A. C.1973Generation time and genomic evolution in primates. Science 179, 1144–1147 (doi:10.1126/science.179.4078.1144) [DOI] [PubMed] [Google Scholar]
- Schluter D., Price T., Mooers A. Ø, Ludwig D.1997Likelihood of ancestral states in adaptive radiation. Evolution 51, 1699–1711 (doi:10.2307/2410994) [DOI] [PubMed] [Google Scholar]
- Sibley C. G., Ahlquist J. E.1984The phylogeny of the hominoid primates, as indicated by DNA-DNA hybridization. J. Mol. Evol. 20, 2–15 (doi:10.1007/BF02101980) [DOI] [PubMed] [Google Scholar]
- Simpson G. G.1944Tempo and mode of evolution New York, NY: Columbia University Press [Google Scholar]
- Sinnott E. W.1916Rapidity of evolution in various plant types. Am. Nat. 50, 466–478 (doi:10.1086/279557) [Google Scholar]
- Smith S. A., Donoghue M. J.2008Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89 (doi:10.1126/science.1163197) [DOI] [PubMed] [Google Scholar]
- Smith S. A., Beaulieu J. M., Donoghue M. J.2009Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol. Biol. 9, 37 (doi:10.1186/1471-2148-9-37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. L.1965The probable growth habit of the earliest flowering plants. Ann. Mo. Bot. Gard. 52, 457–468 (doi:10.2307/2394810) [Google Scholar]
- Sturtevant A. H.1965A history of genetics New York, NY: Harper and Row [Google Scholar]
- Sugiura N.1978Further analysis of the data by Akaike's information criterion and the finite corrections. Commun. Stat. Theory Methods A7, 13–26 [Google Scholar]
- Sun G., Ji Q., Dilcher D. L., Zheng S., Nixon K. C., Weng X.2002Archaefructaceae, a new basal angiosperm family. Science 296, 899–904 (doi:10.1126/science.1069439) [DOI] [PubMed] [Google Scholar]
- Taylor D. W., Hickey L. J.1992Phylogenetic evidence for the herbaceous origin of angiosperms. Plant Syst. Evol. 180, 137–156 (doi:10.1007/BF00941148) [Google Scholar]
- Verdú M.2002Age at maturity and diversification in woody angiosperms. Evolution 56, 1352–1361 [DOI] [PubMed] [Google Scholar]
- Wiens J. J.2004Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution 58, 193–197 [DOI] [PubMed] [Google Scholar]
- Wiens J. J., Donoghue M. J.2004Historical biogeography, ecology, and species richness. Trends Ecol. Evol. 19, 639–644 (doi:10.1016/j.tree.2004.09.011) [DOI] [PubMed] [Google Scholar]
- Wiens J. J., Graham C. H.2005Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Syst. 36, 519–539 [Google Scholar]
- Wu C. I., Li W.1985Evidence for higher rates of nucleotide substitution in rodents than in man. Proc. Natl Acad. Sci. USA 82, 1741–1745 (doi:10.1073/pnas.82.6.1741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q. Y., Thorne J. L., Seo T. K., Zhang W. H., Thomas D. T., Ricklefs R. E.2008Rates of nucleotide substitution in Cornaceae (Cornales)—pattern of variation and underlying causal factors. Mol. Phylogenet. Evol. 49, 327–342 (doi:10.1016/j.ympev.2008.07.010) [DOI] [PubMed] [Google Scholar]