Abstract
The butterfly family Nymphalidae contains some of the most important non-drosophilid insect model systems for evolutionary and ecological studies, yet the evolutionary history of the group has remained shrouded in mystery. We have inferred a robust phylogenetic hypothesis based on sequences of 10 genes and 235 morphological characters for exemplars of 400 of the 540 valid nymphalid genera representing all major lineages of the family. By dating the branching events, we infer that Nymphalidae originated in the Cretaceous at 90 Ma, but that the ancestors of 10–12 lineages survived the end-Cretaceous catastrophe in the Neotropical and Oriental regions. Patterns of diversification suggest extinction of lineages at the Cretaceous/Tertiary boundary (65 Ma) and subsequent elevated speciation rates in the Tertiary.
Keywords: Lepidoptera, Nymphalidae, mass extinction, times of divergence, diversification
1. Introduction
Butterflies in the family Nymphalidae are among the most charismatic insects in many habitats, and their beauty and diversity inspire a lifelong passion for the natural world among scientists and enthusiasts alike. As adults, nymphalids can be visually striking, and their behaviours are amenable to easy observation. These attributes have led scientists to use many nymphalid species as model systems in evolutionary and ecological studies (Boggs et al. 2003). Species of Nymphalidae are among the first taxa that helped us begin to understand the complex relationships between insects and their host plants (Ehrlich & Raven 1964), the effects of habitat fragmentation on the population dynamics of endangered species (Hanski 1999), the genetic mechanisms behind the developmental pathways of morphological features (Beldade & Brakefield 2002), and the coevolutionary interactions between organisms in mimicry rings and aposematic coloration (Brower 1996; Mallet et al. 1998). However, such important studies have been impeded by a lack of knowledge of the time scale involved in evolutionary processes leading to the patterns we see today (Vane-Wright 2004; Wahlberg 2006), be it the evolution of vagility or the evolution of host plant defence resistance.
The age of butterflies (including Nymphalidae) has been the subject of longstanding debate (Forbes 1947; Shields & Dvorak 1979; Scott & Wright 1990; de Jong 2007), with the consensus being that the five families of Papilionoidea originated and diversified in the Tertiary (Vane-Wright 2004). This is mainly based on the occurrence of the few butterfly fossils known from Oligocene and Eocene deposits (Scott & Wright 1990; Emmel et al. 1992). However, several recent studies on subgroups within butterflies, extrapolating rates of DNA divergence from minimum age estimate calibrations of the fossils, suggest that the butterflies may be older, with most subfamilies originating before the KT boundary (Wahlberg 2006; Nazari et al. 2007; Wheat et al. 2007; Peña & Wahlberg 2008). The implications of such an age for the evolutionary history of Nymphalidae have not been investigated yet.
The timeframe over which a group of species has evolved has a strong influence on their implied historical biogeography. The distributions of extant butterflies have variously been attributed to vicariance following the break up of Gondwana (Miller & Miller 1997; Braby et al. 2005) and to dispersal events in more recent times (Kodandaramaiah & Wahlberg 2007, 2009). Obviously, the period of time during which a group of species has diversified will dictate which geological processes have possibly affected their evolution—for example, if a group is much younger than the splitting of the continents on which it is found today, trans-oceanic dispersal is likely to be favoured over vicariance as an explanation of current distributions (de Queiroz 2005).
Here we provide a robust phylogenetic hypothesis for the butterfly family Nymphalidae based on the most comprehensive sampling of genera to date (400 out of 540 valid genera) and the largest dataset currently available (up to 10 genes sequenced and up to 235 morphological characters coded for each exemplar). Based on these data we also infer times of divergence, historical biogeography and the tempo of diversification in this family of approximately 6000 species.
2. Material and methods
Butterfly samples were collected by the authors and contributed by colleagues from around the world (see electronic supplementary material, table S1 for the list of taxa). In most cases, genomic DNA was extracted from two legs of dried specimens; for some specimens the legs were preserved in 95 per cent ethanol in the field prior to extraction. DNA was extracted using the Qiagen DNEasy extraction kit following a slightly modified insect protocol. Polymerase chain reaction and sequencing protocols have been published previously (Wahlberg & Zimmermann 2000; Wahlberg et al. 2003, 2005b; Wahlberg & Wheat 2008), with the majority of new sequences being generated according to Wahlberg & Wheat (2008). Morphological characters were coded according to Freitas & Brown (2004) for those genera that had sufficient available material (electronic supplementary material, table S1).
We compiled a data matrix of 10 gene regions and 235 morphological characters, providing taxonomic representation of all subfamilies, tribes and subtribes of Nymphalidae, as well as 75 per cent of all currently accepted genera. Of the missing genera, almost all are considered to be closely related to sampled genera, and thus their exclusion is not expected to affect inferred relationships substantially. All taxa (400 genera plus 29 outgroups) were sequenced for 3127 base pairs of the gene regions COI, EF-1α and wingless, the standard gene regions employed for molecular systematics of Lepidoptera (Sperling 2003). In addition, seven novel gene regions (Wahlberg & Wheat 2008) were sequenced for multiple exemplars representing the phylogenetic diversity of the family: 86 taxa were sequenced for 596 bp of ArgKin, 131 for 850 bp of CAD, 174 for 691 bp of GAPDH, 104 for 710 bp of IDH, 168 for 733 bp of MDH, 107 for 411 bp of RpS2 and 199 for 615 bp of RpS5. Furthermore, 238 taxa (including outgroups) were coded for up to 235 morphological characters (Freitas & Brown 2004). Molecular data for outgroups were taken from Wahlberg et al. (2005a), with the seven novel gene regions being sequenced for at least one representative of each outgroup family.
The combined morphological and molecular data were analysed using maximum parsimony in the program TNT (Goloboff et al. 2008). The data were treated with equal weights and morphological characters were coded as ordered or unordered according to Freitas & Brown (2004). Molecular data were treated as unordered. We used the New Search Technology algorithms in TNT, using level 10 of search intensity, holding up to 10 000 trees in each cycle, using default values for tree fusing, drifting and sectorial searches, and 50 ratchet cycles. The trees obtained underwent branch-swapping to obtain additional equally parsimonious trees. Stability of the nodes of the most parsimonious trees was assessed using 1000 bootstrap pseudoreplicates.
In addition, the molecular data were subjected to maximum likelihood analyses using the online version of RAxML (Stamatakis et al. 2008). The data were modelled with the default GTR+G model, and analysed unpartitioned, partitioned into mitochondrial and nuclear gene regions, and partitioned by individual gene regions. For the unpartitioned data, 10 separate analyses were done, each including a search for the maximum likelihood topology, as well as 100 bootstrap pseudoreplicates. The bootstrapped trees were combined into a single file to calculate bootstrap values for all nodes.
Times of divergences and topology were estimated simultaneously using the program BEAST v. 1.4.8 (Drummond & Rambaut 2007). Two kinds of analyses were done—one that included outgroup sequences and one that included only nymphalid sequences. Both analyses were repeated twice (each run took two to three months of CPU time on a 64-bit dual core desktop computer). Both analyses had the same calibration points.
The data were partitioned into the mitochondrial gene region (COI) and the combined nuclear genes (ArgKin, CAD, EF1a, GAPDH, IDH, MDH, RpS2, RpS5 and wingless), owing to the high proportion of A-T nucleotides in the mitochondrial gene compared with the approximately equal base ratios of the nuclear genes. The tree prior was set to the Yule process, the independent models for the two partitions were both set to the GTR+G model, while all other priors were left to defaults. The branch lengths were allowed to vary under a relaxed clock model with an uncorrelated lognormal distribution. The analyses were run for 40 million generations with every 4000th generation sampled. Using the Tracer program (part of the BEAST package) we confirmed that the two runs had converged to a stationary distribution after 5000 samples (burn-in), which left 5000 samples in each run describing the posterior distribution.
Maximum times of divergence were constrained by the ages of plant families for six clades of butterflies whose larvae are oligotrophic on those families. The maximum age of the clade Danainae was constrained to be 83 Ma based on the estimated age of Gentianales (Bremer et al. 2004). This plant clade is not used by any other nymphalid butterflies, but is commonly used by the danaines and inferred to be the ancestral host plant family of the group (Janz et al. 2006). The maximum age of the heliconiine+nymphaline clade was constrained to be 110 Ma based on the estimated age of Rosales (Anderson et al. 2005), which is a plant clade very commonly used in these two clades and inferred to be the ancestral host plant family (Janz et al. 2006; Nylin & Wahlberg 2008). The maximum age of Biblidinae was constrained to be 65 Ma based on the estimated age of the split between Ricinus and Dalechampia (Davis et al. 2005), which represents the clade of Euphorbiaceae on which species of Biblidinae are highly specialized (DeVries 1987). The maximum age of the nymphaline clade including Kallimini, Junoniini, Victorinini and Melitaeini was constrained to be 74 Ma based on the age estimate for the family Acanthaceae (Bremer et al. 2004), which is a plant family not used outside this clade (Nylin & Wahlberg 2008). The maximum age of Melitaeini, excluding Euphydryas, was constrained to be 40 Ma based on the estimated age of Asteraceae (Kim et al. 2005), which is a plant family very rarely used outside of this clade (Nylin & Wahlberg 2008). Finally, the maximum age of Satyrinae was constrained to be 65 Ma based on the estimated age of Poaceae (Prasad et al. 2005), which is a plant family very rarely used outside this clade (Peña & Wahlberg 2008).
Minimum age constraints were based on seven fossils, four of which are from the Florissant formation from the Eocene/Oligocene boundary (Emmel et al. 1992), two are from Dominican amber from the Miocene (Peñalver & Grimaldi 2006) and one is from Oligocene deposits in southeastern France (Nel et al. 1993). The minimum age of the split between Libythea and Libytheana was constrained to be 34 Ma based on two fossils, Libytheana vagabunda and L. florissanti, both from the Florissant formation in Colorado and recently found to be sister to extant Libytheana in a cladistic analysis (Kawahara 2009). The minimum age of the split between Vanessa and its sister group was constrained to be 34 Ma based on the fossil Vanessa amerindica (Emmel et al. 1992), which has been placed in the extant genus (Miller & Brown 1989). This fossil was also used as a calibration point by Wahlberg (2006). The minimum age of the split between Hypanartia and its sister group was constrained to be 34 Ma based on the fossil Prodryas persephone (Emmel et al. 1992), which is clearly related to Hypanartia based on morphological features of the wings (K. Willmott 2006, personal communication). This fossil was also used as a calibration point by Wahlberg (2006). The minimum age of the split between Dynamine and its sister group was constrained to be 20 Ma based on the Dominican amber fossil of Dynamine alexae, which clearly belongs to this extant genus (Peñalver & Grimaldi 2006). The minimum age of the split between Smyrna and its sister group was constrained to be 20 Ma based on the Dominican amber fossil larva of Smyrna (Hammond & Poinar 1998). The minimum age of the split between Lethe and its sister group was constrained to be 25 Ma based on the fossil Lethe corbieri found in the Oligocene deposits of southeast France (Nel et al. 1993). This fossil has been used as a calibration point in the study by Peña & Wahlberg (2008).
The BEAST analysis resulted in an ultrametric tree with 400 terminal taxa for the ingroup and 399 internal nodes. To analyse the tempo of speciation during the evolutionary history of Nymphalidae, the most probable age and 95 per cent credibility intervals for each node were transferred into a spreadsheet program. The cumulative number of lineages through time was plotted using a logarithmic scale: a constant rate of speciation in the lineage together with a lower constant rate of extinction should produce an approximately exponentially increasing number of lineages until the final stages of diversification (Harvey et al. 1994), which are not of concern here owing to incomplete lineage sampling within genera. According to Harvey et al. (1994), a mass extinction leads to a severe drop in the lineages through time (LTT) plot, and this would be observed as a slowdown in speciation rate of extant lineages earlier on in the LTT plot.
Dispersal-vicariance optimization of ancestral areas implemented in DIVA v. 1.2 (Ronquist 1997) was used to infer the biogeographic history of the group. The distributions of genera were divided into six areas corresponding to the zoogeographic realms—Neotropical, Nearctic, Palaearctic, Oriental, Australasian and Afrotropical. Nodes subtending genera with the same distributions were collapsed and treated as a single terminal. Wherever published hypotheses were available about the origin of a clade—for instance Junonia (Kodandaramaiah & Wahlberg 2007), Coenonympha (Kodandaramaiah & Wahlberg 2009) and Phyciodina (Wahlberg & Freitas 2007)—the inferred area of origin of the clade was used instead of the current distribution of its extant species. DIVA assigns a cost of one for dispersals and extinctions, while vicariance and within-area speciation events receive no cost. The ancestral optimization with the smallest cost is chosen as the preferred scenario. The optimization was run on the BEAST topology without constraints on the maximum number of inferred ancestral areas.
3. Results and discussion
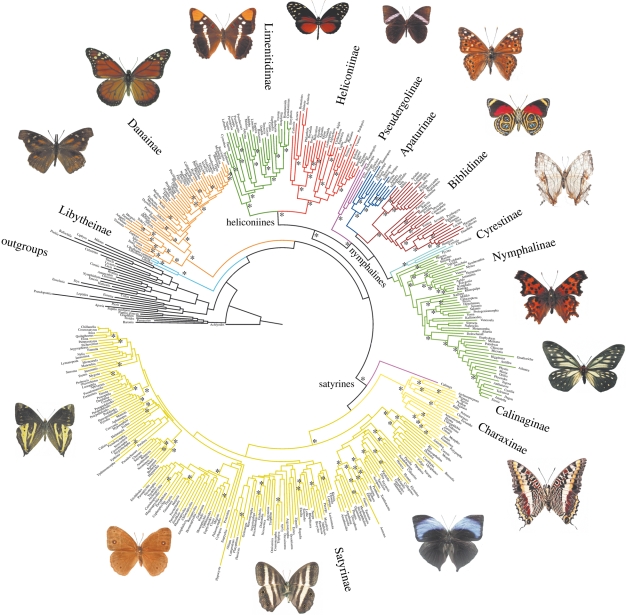
All methods of analysis recovered generally similar topologies (figure 1; nodes with asterisks were very stable in all analyses). The trees were rooted with Hesperiidae, the putative sister taxon to the five families of Papilionoidea (Wahlberg et al. 2005a). We find the monophyly and relationships among 12 nymphalid subfamilies to be strongly supported in all analyses (figure 1; electronic supplementary material, figs S1–S3). Contrary to earlier studies based on molecular data (Brower 2000; Wahlberg et al. 2005a; Wahlberg & Wheat 2008), the position of Danainae is now stable and well-supported as the sister to all Nymphalidae except Libytheinae, a result congruent with a previous morphological study (Freitas & Brown 2004). The subfamily Libytheinae appears to be the sister taxon to the rest of Nymphalidae, a hypothesis suggested 50 years ago (Ehrlich 1958; Kristensen 1976), but never corroborated by molecular work thus far (Brower 2000; Wahlberg et al. 2005a; Wahlberg & Wheat 2008). The subfamily Pseudergolinae is very clearly an independent lineage sister to the other nymphaline subfamilies (figure 1) and not the sister group of Cyrestinae, as found in a previous study (Wahlberg et al. 2003).
Figure 1.
The phylogenetic relationships of 400 genera of Nymphalidae based on a maximum likelihood analysis, along with outgroups. Clades representing subfamilies are coloured. Asterisks (*) indicate nodes that have more than 90 per cent bootstrap in the parsimony and likelihood analyses, as well as more than 0.95 posterior probabilities in the Bayesian analysis.
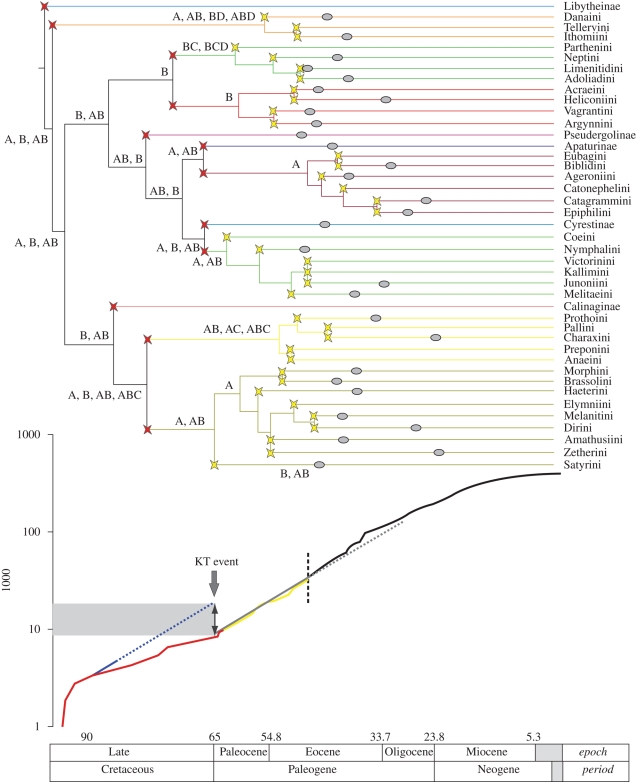
The analysis of times of divergence suggests that Nymphalidae began diversifying in the late Cretaceous around 90 Ma, with the libytheine lineage branching off from the common ancestor of the rest of Nymphalidae (figure 2). Not long after that, the danaine and satyrine lineages diverged at about 89 and 85 Ma, respectively, and finally the heliconiine and nymphaline lineages diverged from each other at about 78 Ma. Given the inferred credibility intervals of the times of divergence, it appears that all the cladogenesis leading to the lineages currently recognized as subfamilies occurred in the late Cretaceous or early Tertiary (figure 2), suggesting that Nymphalidae was already morphologically and ecologically diversified before the end of the Cretaceous period. The estimated origin of the family is coincident with the global rise of the angiosperms at about 100 Ma, which is likely to have been particularly favourable for a radiation of an angiosperm-feeding group such as Nymphalidae.
Figure 2.
Bayesian ultrametric tree showing the phylogenetic relationships of subfamilies (clade colours as in figure 1) and tribes of Nymphalidae, with ages given in millions of years ago (Ma). The red and yellow stars show the nodes of the divergence events that have led to the subfamilies and the tribes, respectively. The grey ellipses indicate the time of the first divergence event within subfamilies/tribes (based on electronic supplementary material, fig. S4). The curve below the tree shows the cumulative number of lineages through time (the analysis includes the complete dataset, up to the genus level). Coloured parts of the curves refer to the inferred divergences leading to the major extant taxonomic subdivisions. The subfamilies (in red) all originated before or immediately after the Cretaceous–Tertiary (KT) event, whereas all the lineages leading to the tribes (in yellow) diversified several million years after the KT boundary. The average rate of net diversification from the Paleocene to the mid-Eocene (grey line) has been traced on top of the lineage through time plot (in yellow). The average rate was then extrapolated as the potential rate of diversification in the late Cretaceous (blue line) where the observed rate of diversification is observed to be twice as slow (probably owing to extinctions), leading to an estimate of up to 60 per cent loss of diversity at 65 Ma. The last part of the curve (in black) represents the time period in which genera have diversified to produce extant species (47 Ma to present). Owing to non-exhaustive species-level sampling, this part of the curve underestimates the net rate of diversification.
It is thought that the KT event that led to the extinction of non-avian dinosaurs also had a large impact on phytophagous insects (such as butterflies; Labandeira et al. 2002). Our inferred slowing down of diversification rate prior to the KT boundary (figure 2) can be explained by an increased number of lineage extinctions (Harvey et al. 1994) occurring at 65 Ma. In the lineage accumulation curve of figure 2, the red region corresponds to the period when all the extant subfamilies originated (94–64 Ma). The yellow region corresponds to the time interval when the subfamily lineages diversified, giving rise to tribes; notice that none of the divergence events leading to the tribes is older than a divergence event leading to a subfamily. The black zone of the curve comprises the period after the beginning of diversification into modern genera. As the sampling is not exhaustive in this time period (one sampled species per genus), the curve underestimates the cumulative number of lineages.
The shape of the curve found in the LTT plot for Nymphalidae (figure 2) is very similar to that found in the simulations of a mass extinction event in Harvey et al. (1994). The rapid increase in diversification rate at the beginning is expected in this kind of analysis; the lineages which survived to the present day are the ones, on average, that got off to a flying start (Nee et al. 1994). This ‘push of the past’ is followed by a period of stability (90–64 Ma) with a low net rate of diversification. This rate changes dramatically at 64 Ma and the time of lineage doubling halves (10 instead of 20 Myr) right after the KT boundary. This dramatic change in the net rate of diversification can reflect a change in the rate of speciation or extinction.
As argued above, a major extinction event in Nymphalidae that took place at about 64 Ma, such as the KT event, would explain the shape of the curve. Extrapolating the average rate of net diversification after the crisis (grey line over yellow part of the curve in figure 2) to the apparent origin of the period preceding the KT boundary (blue dotted line in figure 2), we find that 60 per cent of all the lineages have gone extinct under this hypothesis, suggesting that the KT event may have had a major impact on nymphalid butterflies. Harvey et al. (1994) suggested that only a large mass extinction event would leave such a signal in the LTT plot. It is possible that the ancestral lineages of extant subfamilies of Nymphalidae survived in small populations in geographically restricted areas and became on average more genetically diversified from one another than before the event.
Further support for extinction of nymphalid butterflies around the time of the KT event comes from the historical biogeography of the family. A dispersal-vicariance analysis with no restriction on maximum ancestral areas shows the surprising result that the implied distributions of the ancestral taxa living between 94 and 33 Ma, a span of nearly 60 million years, were almost all in the Neotropical and/or Oriental regions (figure 2; electronic supplementary material, figs S3 and S4). Given that the Neotropics and the Oriental region were never in geological contact during this period, the implied absence of butterflies in the other major biomes is puzzling. We hypothesize that the ancestors of the various lineages in Nymphalidae were widespread prior to the KT boundary, but only 10–12 lineages in the Neotropical and Oriental regions survived the great extinction wave. These lineages then diversified in the Neotropical and Oriental regions before dispersing to the rest of the world 57–40 Myr (electronic supplementary material, fig. S5).
Subsequent diversification of subfamilies occurred in the Tertiary, with almost all lineages leading to currently recognized tribes diverging before the Oligocene (about 34 Ma; figure 2) and almost all sampled genera diverging from sister genera before the Late Miocene (about 11 Ma; electronic supplementary material, fig. S4). Of note here are the divergence times of the genus Heliconius from its sister genus Eueides at about 18 Ma (95% credibility interval 12–23 Ma) and of the genus Bicyclus from its sister genus Henotesia at about 24 Ma (95% credibility interval 18–28 Ma). Heliconius and Bicyclus are the subjects of intense research and are candidates for genomic sequencing (Papa et al. 2008). Also noteworthy is that the estimated times of divergence for genera in the subfamily Nymphalinae are somewhat younger here (between 1 and 13 Ma depending on the clade) than in a previous study (Wahlberg 2006), and the estimated times of divergence for the subfamily Satyrinae are somewhat older (about 10 Myr older) than in a previous study (Peña & Wahlberg 2008), both of which used only butterfly fossils as calibration points. These 10 to 15 per cent fluctuations imply that inferred dates may shift further with the addition of data to the matrix or more external calibrations.
The optimal biogeographical hypothesis in DIVA indicates a widespread tropical ancestor at the base of Nymphalidae. The sister clade to Libytheinae may have arisen either in the Oriental or in the Neotropical region or both. As mentioned above, these two regions were inferred to be the ancestral areas from the base of the tree up to the nodes defining subfamilies as well as many of the tribes (electronic supplementary material, fig. S5). During the time span 57–34 Ma (the Eocene/Oligocene boundary), the Afrotropical region was colonized independently at least three times from the Oriental region: once by the common ancestor of Limenitidinae at around 57 Ma, once or twice by the common ancestor of the Vagrantini+Argynnini tribes in Heliconiinae (between 40 and 50 Ma) and once by the common ancestor of Charaxini+Pallini in Charaxinae (also between 40 and 50 Ma). In addition, it appears that the Afrotropical region was colonized from the Neotropical region between 40 and 30 Ma by the common ancestors of Biblidini and Catonephelini (both Biblidinae). The Palaearctic region appears to have been colonized before the Eocene/Oligocene boundary by the ancestor of Nymphalini around 48 Ma and by the ancestor of Apaturinae around 40 Ma, both from the Oriental region. Also, the Nearctic region appears to have been colonized from the Oriental region by the ancestor of Melitaeini around 40 Ma. Extant distributions of genera have largely formed after the Eocene/Oligocene boundary (around 34 Ma), which is known as a period of cooling and drying on Earth, when large swaths of moist forests were replaced with dry grasslands (Willis & McElwain 2002), mainly because of the separation of remnant Gondwana into South America, Australia and Antarctica, allowing cold ocean currents to cool down the southern polar region. Species groups associated with open landscapes and grasslands appear to have diversified at this period, such as Satyrini (Peña & Wahlberg 2008) and Melitaeini (Leneveu et al. 2009).
Examining the temporal and spatial scale of macroevolutionary processes in phytophagous insects has only recently become possible with comprehensively sampled phylogenetic hypotheses and robust estimates of patterns and times of divergence (McKenna et al. 2009). We have been able to infer novel aspects of the deep evolutionary history of a species-rich group of phytophagous insects, suggesting that the event(s) causing mass extinction of vertebrates and other organisms with a well-preserved fossil record around the KT boundary also affected the taxa with a scant fossil record. These extinctions were putatively caused by a massive bolide impact, which could have cooled down Earth substantially for a long period of time. This clearly would have affected ephemeral, ectothermic insects dependent on the warmth of the sun for daily activities such as feeding and reproduction, and here we show for the first time that phylogenetic evidence supports this hypothesis. Extinctions could have been further compounded as a result of widespread extinctions of angiosperm host plants in the same period, disrupting obligate butterfly–plant interactions (Wilf et al. 2006). The robust, comprehensive phylogenetic hypothesis, and historical, biogeographical and divergence time estimates for this model group, will provide a long-awaited framework for comparative studies and open up new avenues of research that can capitalize on the extensive knowledge about the ecology and natural history of Nymphalidae.
Acknowledgements
We are grateful to the numerous people who have contributed specimens to this study, too numerous to name individually, although we would like to point out the importance of contributions by the African Butterfly Research Institute and Area de Conservación Guanacaste, Costa Rica. We are grateful to Folmer Bokma and an anonymous reviewer for useful comments on a previous version of the manuscript. This work has been supported by grants from the Academy of Finland to N.W. (no. 118369), the Swedish Research Council to S.N. and N.W., and NSF (no. DEB0640301) to A.V.Z.B. A.V.L.F. acknowledges FAPESP (no. 98/05101-8 and no. 04/05269-9), Brazilian CNPq (fellowship no. 300315/2005-8) and NSF (DEB grant no. 0527441).
References
- Anderson C. L., Bremer K., Friis E. M.2005Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am. J. Bot. 92, 1737–1748 (doi:10.3732/ajb.92.10.1737) [DOI] [PubMed] [Google Scholar]
- Beldade P., Brakefield P. M.2002The genetics and evo-devo of butterfly wing patterns. Nat. Rev. 3, 442–452 [DOI] [PubMed] [Google Scholar]
- Boggs C. L., Watt W. B., Ehrlich P. R.2003Butterflies: evolution and ecology taking flight Chicago, IL: University of Chicago Press [Google Scholar]
- Braby M. F., Trueman J. W. H., Eastwood R.2005When and where did troidine butterflies (Lepidoptera: Papilionidae) evolve? Phylogenetic and biogeographic evidence suggests an origin in remnant Gondwana in the Late Cretaceous. Invertebr. Syst. 19, 113–143 (doi:10.1071/IS04020) [Google Scholar]
- Bremer K., Friis E. M., Bremer B.2004Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Syst. Biol. 53, 496–505 (doi:10.1080/10635150490445913) [DOI] [PubMed] [Google Scholar]
- Brower A. V. Z.1996Parallel race formation and the evolution of mimicry in Heliconius butterflies: a phylogenetic hypothesis from mitochondrial DNA sequences. Evolution 50, 195–221 (doi:10.2307/2410794) [DOI] [PubMed] [Google Scholar]
- Brower A. V. Z.2000Phylogenetic relationships among the Nymphalidae (Lepidoptera), inferred from partial sequences of the wingless gene. Proc. R. Soc. Lond. B 267, 1201–1211 (doi:10.1098/rspb.2000.1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. C., Webb C. O., Wurdack K. J., Jaramillo C. A., Donoghue M. J.2005Explosive radiation of Malphigiales supports a mid-Cretaceous origin of modern tropical rain forests. Am. Nat. 165, E36–E65 (doi:10.1086/428296) [DOI] [PubMed] [Google Scholar]
- de Jong R.2007Estimating time and space in the evolution of the Lepidoptera. Tijdschr. Entomol. 150, 319–346 [Google Scholar]
- de Queiroz A.2005The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 20, 68–73 [DOI] [PubMed] [Google Scholar]
- DeVries P. J.1987The butterflies of Costa Rica and their natural history. Volume I: Papilionidae, Pieridae, Nymphalidae Princeton, NJ: Princeton University Press [Google Scholar]
- Drummond A. J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich P. R.1958The comparative morphology, phylogeny and higher classification of the butterflies (Lepidoptera: Papilionoidea). Univ. Kansas Sci. Bull. 39, 305–370 [Google Scholar]
- Ehrlich P. R., Raven P. H.1964Butterflies and plants: a study in coevolution. Evolution 18, 586–608 (doi:10.2307/2406212) [Google Scholar]
- Emmel T. C., Minno M. C., Drummond B. A.1992Florissant butterflies: a guide to the fossil and present-day species of central Colorado Stanford, CA: Stanford University Press [Google Scholar]
- Forbes W. T. M.1947Buckeyes and Wegener. Entomologist 80, 56–58 [Google Scholar]
- Freitas A. V. L., Brown K. S. J.2004Phylogeny of the Nymphalidae (Lepidoptera: Papilionoidea). Syst. Biol. 53, 363–383 (doi:10.1080/10635150490445670) [DOI] [PubMed] [Google Scholar]
- Goloboff P. A., Farris J. S., Nixon K. C.2008TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786 (doi:10.1111/j.1096-0031.2008.00217.x) [Google Scholar]
- Hammond P. C., Poinar G., Jr1998A larval brush-footed butterfly (Lepidoptera: Nymphalidae) in Dominican amber, with a summary of fossil Nymphalidae. Entomol. Scand. 29, 275–279 [Google Scholar]
- Hanski I.1999Metapopulation ecology Oxford Series in Ecology and Evolution Oxford, UK: Oxford University Press [Google Scholar]
- Harvey P. H., May R. M., Nee S.1994Phylogenies without fossils. Evolution 48, 523–529 (doi:10.2307/2410466) [DOI] [PubMed] [Google Scholar]
- Janz N., Nylin S., Wahlberg N.2006Diversity begets diversity: host expansions and the diversification of plant-feeding insects. BMC Evol. Biol. 6, 4 (doi:10.1186/1471-2148-6-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara A. Y.2009Phylogeny of snout butterflies (Lepidoptera: Nymphalidae: Libytheinae): combining evidence from morphology of extant, fossil, and recently extinct taxa. Cladistics 25, 263–278 (doi:10.1111/j.1096-0031.2009.00251.x) [DOI] [PubMed] [Google Scholar]
- Kim K. J., Choi K. S., Jansen R. K.2005Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792 (doi:10.1093/molbev/msi174) [DOI] [PubMed] [Google Scholar]
- Kodandaramaiah U., Wahlberg N.2007Out-of-Africa origin and dispersal mediated diversification of the butterfly genus Junonia (Nymphalidae: Nymphalinae). J. Evol. Biol. 20, 2181–2191 (doi:10.1111/j.1420-9101.2007.01425.x) [DOI] [PubMed] [Google Scholar]
- Kodandaramaiah U., Wahlberg N.2009Phylogeny and biogeography of Coenonympha butterflies (Nymphalidae: Satyrinae)—patterns of colonization in the Holarctic. Syst. Entomol. 34, 315–323 (doi:10.1111/j.1365-3113.2008.00453.x) [Google Scholar]
- Kristensen N. P.1976Remarks on the family-level phylogeny of butterflies (Insecta, Lepidoptera, Rhopalocera). Zeitschrift für Zoologische Systematik und Evolutionsforschung 14, 25–33 [Google Scholar]
- Labandeira C. C., Johnson K. R., Wilf P.2002Impact of the terminal Cretaceous event on plant–insect associations. Proc. Natl Acad. Sci. USA 99, 2061–2066 (doi:10.1073/pnas.042492999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leneveu J., Chichvarkhin A., Wahlberg N.2009Varying rates of diversification in the genus Melitaea (Lepidoptera: Nymphalidae) during the past 20 million years. Biol. J. Linn. Soc. 97, 346–361 (doi:10.1111/j.1095–8312.2009.01208.x) [Google Scholar]
- Mallet J., McMillan W. O., Jiggins C. D.1998Mimicry and warning color at the boundary between races and species. In Endless forms: species and speciation (eds Howard D. J., Berlocher S. H.), pp. 390–403 Oxford, UK: Oxford University Press [Google Scholar]
- McKenna D. D., Sequeira A. S., Marvaldi A. E., Farrell B.2009Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl Acad. Sci. USA 106, 7083–7088 (doi:10.1073/pnas.0810618106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Y., Brown F. M.1989Description of a new Oligocene fossil butterfly, Vanessa amerindica (Lepidoptera: Nymphalidae) from the Florissant formation, Colorado. Bull. Allyn Museum 26, 1–9 [Google Scholar]
- Miller L. D., Miller J. Y.1997Gondwanan butterflies: the Africa–South America connection. Metamorphosis 3(Suppl.), 42–51 [Google Scholar]
- Nazari V., Zakharov E. V., Sperling F. A. H.2007Phylogeny, historical biogeography, and taxonomic ranking of Parnassiinae (Lepidoptera, Papilionidae) based on morphology and seven genes. Mol. Phylogenet. Evol. 42, 131–156 (doi:10.1016/j.ympev.2006.06.022) [DOI] [PubMed] [Google Scholar]
- Nee S., Holmes E. C., May R. M., Harvey P. H.1994Extinction rates can be estimated from molecular phylogenies. Phil. Trans. R. Soc. Lond. B 344, 77–82 (doi:10.1098/rstb.1994.0054) [DOI] [PubMed] [Google Scholar]
- Nel A., Nel J., Balme C.1993Un nouveau Lépidoptère Satyrinae fossile de l'Oligocène du sud-est de la France (Insecta, Lepidoptera, Nymphalidae). Linn. Belg. 14, 20–36 [Google Scholar]
- Nylin S., Wahlberg N.2008Does plasticity drive speciation? Host-plant shifts and diversification in nymphaline butterflies (Lepidoptera: Nymphalidae) during the Tertiary. Biol. J. Linn. Soc. 94, 115–130 (doi:10.1111/j.1095-8312.2008.00964.x) [Google Scholar]
- Papa R., et al. 2008Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genom. 9, 345 (doi:10.1186/1471-2164-9-345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C., Wahlberg N.2008Prehistorical climate change increased diversification of a group of butterflies. Biol. Lett. 4, 274–278 (doi:10.1098/rsbl.2008.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalver E., Grimaldi D.2006New data on Miocene butterflies in Dominican amber (Lepidoptera: Riodinidae and Nymphalidae) with the description of a new nymphalid. Am. Mus. Novit. 3519, 1–17 (doi:10.1206/0003-0082(2006)3519[1:NDOMBI]2.0.CO;2) [Google Scholar]
- Prasad V., Strömberg C. A. E., Alimohammadian H., Sahni A.2005Dinosaur coprolites and the early evolution of grasses and grazers. Science 310, 1177–1180 (doi:10.1126/science.1118806) [DOI] [PubMed] [Google Scholar]
- Ronquist F.1997Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 46, 195–203 [Google Scholar]
- Scott J. A., Wright D. M.1990Butterfly phylogeny and fossils. In Butterflies of Europe, vol. 2 (ed. Kudrna O.), pp. 152–208 Wiesbaden, Germany: Aula-Verlag [Google Scholar]
- Shields O., Dvorak S. K.1979Butterfly distribution and continental drift between the Americas, the Caribbean and Africa. J. Nat. Hist. 13, 221–250 (doi:10.1080/00222937900770161) [Google Scholar]
- Sperling F. A. H.2003Butterfly species and molecular phylogenies. In Butterflies: evolution and ecology taking flight (eds Boggs C. L., Watt W. B., Ehrlich P. R.), pp. 431–458 Chicago, IL: University of Chicago Press [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J.2008A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75, 758–771 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- Vane-Wright R.2004Butterflies at that awkward age. Nature 428, 477–480 (doi:10.1038/428477a) [DOI] [PubMed] [Google Scholar]
- Wahlberg N.2006That awkward age for butterflies: insights from the age of the butterfly subfamily Nymphalinae. Syst. Biol. 55, 703–714 (doi:10.1080/10635150600913235) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., Freitas A. V. L.2007Colonization of and radiation in South America by butterflies in the subtribe Phyciodina (Lepidoptera: Nymphalidae). Mol. Phylogenet. Evol. 44, 1257–1272 (doi:10.1016/j.ympev.2007.04.012) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., Wheat C. W.2008Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 57, 231–242 (doi:10.1080/10635150802033006) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., Zimmermann M.2000Pattern of phylogenetic relationships among members of the tribe Melitaeini (Lepidoptera: Nymphalidae) inferred from mtDNA sequences. Cladistics 16, 347–363 (doi:10.1111/j.1096-0031.2000.tb00355.x) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., Weingartner E., Nylin S.2003Towards a better understanding of the higher systematics of Nymphalidae (Lepidoptera: Papilionoidea). Mol. Phylogenet. Evol. 28, 473–484 (doi:10.1016/S1055-7903(03)00052-6) [DOI] [PubMed] [Google Scholar]
- Wahlberg N., et al. 2005aSynergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc. R. Soc. B 272, 1577–1586 (doi:10.1098/rspb.2005.3124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N., Brower A. V. Z., Nylin S.2005bPhylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. 86, 227–251 (doi:10.1111/j.1095-8312.2005.00531.x) [Google Scholar]
- Wheat C. W., Vogel H., Wittstock U., Braby M. F., Underwood D., Mitchell-Olds T.2007The genetic basis of a plant–insect coevolutionary key innovation. Proc. Natl Acad. Sci. USA 104, 20 427–20 431 (doi:10.1073/pnas.0706229104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilf P., Labandeira C. C., Johnson K. R., Ellis B.2006Decoupled plant and insect diversity after the end-Cretaceous extinction. Science 313, 1112–1115 (doi:10.1126/science.1129569) [DOI] [PubMed] [Google Scholar]
- Willis K. J., McElwain J. C.2002The evolution of plants Oxford, UK: Oxford University Press [Google Scholar]