Abstract
Both the monophyly and inter-relationships of the major annelid groups have remained uncertain, despite intensive research on both morphology and molecular sequences. Morphological cladistic analyses indicate that Annelida is monophyletic and consists of two monophyletic groups, the clitellates and polychaetes, whereas molecular phylogenetic analyses suggest that polychaetes are paraphyletic and that sipunculans are crown-group annelids. Both the monophyly of polychaetes and the placement of sipunculans within annelids are in conflict with the annelid fossil record—the former because Cambrian stem taxa are similar to modern polychaetes in possessing biramous parapodia, suggesting that clitellates are derived from polychaetes; the latter because although fossil sipunculans are known from the Early Cambrian, crown-group annelids do not appear until the latest Cambrian. Here we apply a different data source, the presence versus absence of specific microRNAs—genes that encode approximately 22 nucleotide non-coding regulatory RNAs—to the problem of annelid phylogenetics. We show that annelids are monophyletic with respect to sipunculans, and polychaetes are paraphyletic with respect to the clitellate Lumbricus, conclusions that are consistent with the fossil record. Further, sipunculans resolve as the sister group of the annelids, rooting the annelid tree, and revealing the polarity of the morphological change within this diverse lineage of animals.
Keywords: molecular palaeobiology, phylogeny, non-coding RNA
1. Introduction
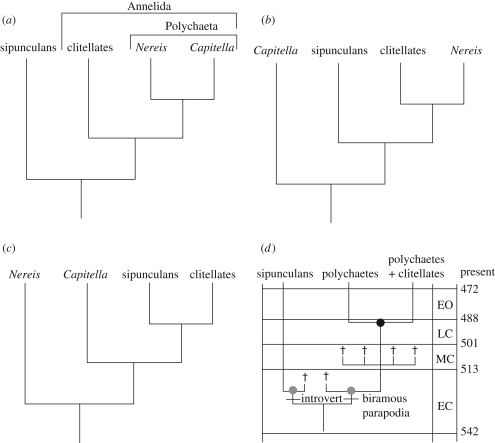
Annelids are a spectacularly diverse and widespread group of animals, inhabiting both marine and terrestrial habitats, and exhibiting a variety of lifestyles. The lack of a robust phylogenetic tree, however, has hindered our understanding of the evolution of this group, especially for higher level taxa. Morphological cladistic analyses recovered annelids as monophyletic, and identified the clitellates and polychaetes as reciprocally monophyletic lineages (Rouse & Fauchald 1997) (figure 1a). However, when this hypothesis was tested with molecular phylogenetics, the results suggested that the clitellates are nested within the polychaetes, making the latter paraphyletic. Curiously, however, non-annelid taxa like phoronids, nemerteans and/or various molluscan taxa (e.g. aplacophorans and gastropods) are also nested within the polychaetes (Bleidorn et al. 2003; Hall et al. 2004; Colgan et al. 2006; Rousset et al. 2007; Helmkampf et al. 2008), rendering the position of the annelid root, and hence the polarity of morphological changes, uncertain (Rousset et al. 2007).
Figure 1.
Four hypotheses for the inter-relationships of four taxa, the sipunculans and the three annelids: the clitellate Lumbricus and the two polychaetes Nereis and Capitella. (a) Morphological cladistic analysis (Rouse & Fauchald 1997) suggests that Annelida is monophyletic, as is Polychaeta, and that the last common ancestor of Nereis and Capitella is the last common ancestor of all living polychaetes. (b,c) Molecular studies suggest that annelids are paraphyletic with respect to sipunculans and that polychaetes are paraphyletic with respect to clitellates, although the exact position of key taxa varies: (b) some studies suggest a basal position for Capitella (Rousset et al. 2007); while others (c) suggest a basal position for Nereis (Colgan et al. 2006; Struck et al. 2007). (d) The fossil record suggests that annelids are monophyletic with respect to sipunculans, but that polychaetes are paraphyletic with respect to clitellates. Note that no indication is given whether Lumbricus is more closely related to Nereis or to Capitella, just that the presence of biramous parapodia is primitive for Annelida. Black circle indicates the annelid crown group; grey circles the sipunculan and annelid total groups. Crosses indicate the age of stem-group members of each of the two phyla (Conway Morris 1979; Huang et al. 2004; Conway Morris & Peel 2008); note that the Burgess Shale (Middle Cambrian) polychaetes are shown as a simple polytomy for illustrative purposes only. Abbreviations are as follows: EC, Early Cambrian; MC, Middle Cambrian; LC, Late Cambrian; EO, Early Ordovician.
While the likelihood that molluscs and phoronids lie within the Annelida appears small, a consensus has emerged that at least some of the unsegmented protostome phyla lie near or within the modern diversity of annelids (Halanych et al. 2002; Rouse & Pleijel 2007). In particular, virtually every recent molecular phylogenetic study, including studies using data as diverse as ribosomal DNA, complete mitochondrial genomes and expressed sequence tags, finds Sipuncula nested within what are traditionally considered annelids (Colgan et al. 2006; Hausdorf et al. 2007; Rousset et al. 2007; Struck et al. 2007; Dunn et al. 2008; Xin et al. 2009)—only Mwinyi et al. (2009) found sipunculans outside of what are traditionally considered annelids. But interpreting these results is problematic as no study shows a statistically robust signal—indeed as lamented by Rousset et al. (2007), ‘ … resolution remains discouraging: rarely so many taxa have been sequenced for so many nucleotides with such sparing results’—and there is an almost total lack of congruence between one study and the next. Indeed, when only four taxa are considered (sipunculans, clitellates and the two polychaete taxa Nereis and Capitella), two different and completely non-overlapping hypotheses were generated by the two most recent and large-scale analyses: Rousset et al. (2006) found that Capitella was basal with clitellates the sister group to Nereis (figure 1b), whereas Struck et al. (2007) found that Nereis was basal with clitellates the sister group to sipunculans (figure 1c).
A previously unremarked feature of these results is that both the monophyly of polychaetes with respect to clitellates (figure 1a) and the paraphyly of annelids with respect to sipunculans (figure 1b,c) are in direct conflict with the fossil record of both annelids and sipunculans (figure 1d). Annelids first appear in the fossil record in the Early Cambrian Sirius Passet fauna of North Greenland: Phragmochaeta bears biramous parapodia with notochaetae and neurochaetae (Conway Morris & Peel 2008), a feature characteristic of living polychaetes, but not clitellates. Six additional genera are known from the Middle Cambrian Burgess Shale, all with parapodia, and all apparently representing stem-group annelids (Eibye-Jacobsen 2004). The annelid crown group does not appear until the latest Cambrian with the appearance of scolecodonts, the jaws of polychaete worms (Hints & Eriksson 2007). These jaw elements are present among a subset of living polychaete groups, but not present in any Early or Middle Cambrian polychaete (Conway Morris 1979; Budd & Jensen 2000; Eibye-Jacobsen 2004). Thus, based on our current knowledge of the fossil record, the polychaete, rather than the clitellate, body plan is primitive for Annelida, as opposed to suggestions from the cladistic morphological perspective that supports the reciprocal monophyly of Polychaeta and Clitellata (Rouse & Fauchald 1997) (figure 1a) and the primitiveness of the clitellate body plan for Annelida (Bartolomaeus et al. 2005).
The paraphyly of annelids with respect to sipunculans is also problematic when the fossil record is taken into account because sipunculans first appear in the Early Cambrian Chengjiang fauna of China (Huang et al. 2004) (figure 1d). If sipunculans were crown-group annelids (figure 1b,c), this would indicate diversification of the annelid crown group before the Early Cambrian (approx. 520 Ma), even though it is not represented in the fossil record until the latest Cambrian (approx. 490 Ma) (figure 1d). This striking discordance suggests that either the fossil record of annelids or that most of the molecular hypotheses of their relationships are unreliable.
In view of this conflict, and the fact that adding more taxa and more sequences to molecular phylogenetic analyses has not resolved these problems, we approached the problem of deep annelid systematics by using an independent molecular dataset, the presence or absence of specific microRNAs (miRNAs). miRNAs, which are an emerging new dataset for metazoan phylogenetics (Sperling & Peterson 2009), show four properties that make them excellent phylogenetic markers: (i) miRNAs experience very few substitutions to the mature sequence over time, (ii) new miRNA families are continually incorporated into metazoan genomes through time; (iii) miRNAs are almost impossible to evolve convergently, and (iv) miRNAs show only rare instances of secondary loss (Sempere et al. 2006; Wheeler et al. 2009). Because of these four properties, miRNAs can be applied to virtually any area of the metazoan tree, from the inter-relationships of Drosophila species to metazoan superphyla (Sperling & Peterson 2009). Here we demonstrate that the presence/absence pattern of miRNAs strongly supports the monophyly of annelids with respect to the sipunculans, at least for the taxa tested, and the paraphyly of the polychaetes with respect to clitellates, results that are consistent with the known fossil record.
2. Material and methods
(a). Taxon sampling
Using 454 sequencing of small RNA libraries, coupled with genomic searches, Wheeler et al. (2009) demonstrated that the two polychaete taxa Capitella sp. and Nereis diversicolor share seven miRNA families that are not present in any other metazoan analysed to date, including the two gastropod molluscs Haliotis rufescens and Lottia gigantea and the nemertean Cerebratulus lacteus. These two polychaetes were chosen (Wheeler et al. 2009) because the genome of Capitella sp. has been sequenced, and the morphological cladistic analysis of Rouse & Fauchald (1997) resolved the last common ancestor of Capitella and Nereis as the last common ancestor of all living polychaetes. Furthermore, although virtually all molecular analyses suggest that polychaetes are paraphyletic, all show that clitellates are more closely related either to Capitella or to Nereis among the polychaetes considered (Bleidorn et al. 2003; Hall et al. 2004; Colgan et al. 2006; Rousset et al. 2006; Struck et al. 2007; Dunn et al. 2008). Thus, for an initial investigation into miRNA evolution in annelids, analysing the descendants of the last common ancestor of Capitella and Nereis captures much of modern polychaete, if not modern annelid, diversity.
To determine whether miRNAs could resolve the inter- and intra-relationships of annelids, we built and sequenced small RNA libraries from the clitellate Lumbricus sp. (collected in Hanover, NH, USA) and the sipunculan Phascolosoma agassizii (collected in Friday Harbor, WA, USA, and kindly donated by R. Elahi) and compared these data with previously published data from the polychaetes N. diversicolor and Capitella sp. (Wheeler et al. 2009). To test the monophyly of Annelida with respect to other lophotrochozoan phyla, we built and sequenced small RNA libraries from the aplacophoran mollusc Chaetoderma nitidulum (collected at Kristineberg, Sweden, and kindly donated by M. Obst) and the phoronid Phoronis architecta (purchased from Gulf Specimens Marine Supply, Panacea, FL, USA), and compared these data with those from the annelids and with previously published data from the gastropod molluscs Haliotis and Lottia and the nemertean Cerebratulus (Wheeler et al. 2009). Pooled, bar-coded small RNA libraries were constructed as described by Wheeler et al. (2009) and were sequenced at the Yale Center for Genomics and Proteomics using 454 sequencing technology (Margulies et al. 2005). The numbers of parsed and non-redundant reads for each taxon are listed in electronic supplementary material, file 1.
(b). miRMiner
An updated version of the program ‘miRMiner’ (Wheeler et al. 2009) was used to identify known miRNAs and to generate a list of potential novel miRNAs. Shared sequences between two or more taxa were BLASTed against the Capitella sp. genomic trace archive and any resulting hit was folded using mfold (Zuker et al. 1999) as described in Wheeler et al. (2009). To identify candidate miRNA genes specific to the Capitella output, a semi-automated method written in Python (available from the authors upon request) was developed that annotates non-conserved transcripts from the 454 small RNA library. The input file containing the sequence of small RNAs obtained by 454 sequencing was parsed to retain sequences between 19 and 25 bp long, as Wheeler et al. (2009) found no miRNAs outside this size range. These retained sequences were blasted against the Capitella sp. whole genome sequence (release v. 1.0, 23 August 2007, http://genome.jgi-psf.org/Capca1/Capca1.home.html), and sequences matching the genome more than 10 times were considered repeats and discarded. A 140 nucleotide (nt) sequence fragment (called a ‘putative pre-miRNA’) around each putative mature sequence in the remaining dataset was extracted from the whole genome sequence extending 60 nt upstream of the putative mature sequence and 140 nt long in total. Two minimum energy secondary structures for each of these putative pre-miRNAs were predicted (one for positions 1–100 and the other for positions 40–140) using the Vienna RNA Package (RNAfold v. 1.7, http://www.tbi.univie.ac.at/RNA) (Gruber et al. 2008). Folds with a minimum energy lower than −18.5 kcal mol−1 were retained if they showed a single ‘stem’ in the predicted fold and if the putative mature sequence matched the other arm for at least 16 of the first 22 nt (Ambros et al. 2003).
(c). Northern analyses and genome walking
Northern analyses were performed as described by Wheeler et al. (2009), using 10 µg of total RNA per organism. Thelepus crispus and Abarenicola sp. were collected at Friday Harbor. Amphitrite sp. and Pectinaria sp. were purchased from Marine Biological Laboratories, Woods Hole, MA, USA. Diopatra cuprea and Chaetopterus variopedatus were purchased from Gulf Specimens Marine Supply. Scoloplos armiger was collected at Roskilde Fjord, Denmark; Mytilus californianus was collected at the SIO pier, La Jolla, CA, USA. Genome-walker libraries were constructed for Phascolosoma, Lumbricus, Nereis and Chaetopterus using the Clontech Genomewalker Universal Kit. PCR conditions, cloning and sequencing of genome-walker products were as described by Wheeler et al. (2009).
(d). Phylogenetic analyses
Seventy-three miRNA families were coded as presence/absence for 11 taxa with data generated during this study, and taken from miRBase v. 13 using MacClade v. 4.08 (Maddison & Maddison 2005). Phylogenetic analyses used PAUP* v. 4.0b10 (Swofford 2002). Bremer support indexes (Bremer 1994) were calculated using TreeRot. v. 3 (Sorenson & Franzosa 2007).
3. Results and discussion
(a). The monophyly of Annelida and Polychaeta
First, we tested the monophyletic status of Annelida with respect to Sipuncula by determining whether any of the complement of miRNAs specific to Capitella were found in both Nereis and Lumbricus with respect to Phascolosoma. Although morphological analyses indicate that this should be the case (e.g. figure 1a), virtually all molecular analyses (figure 1b,c) resolve sipunculans as annelid worms, nested within the current diversity of annelids (Bleidorn et al. 2003, 2006; Hall et al. 2004; Colgan et al. 2006; Rousset et al. 2007; Struck et al. 2007; Dunn et al. 2008; Xin et al. 2009), and therefore as crown-group (Jefferies 1979; Budd 2001) annelids. Indeed, recent investigations into neural patterning suggest that, as in echiurans (Hessling 2002), most signs of segmentation may have been secondarily lost in sipunculans (Kristof et al. 2008).
Second, we determined whether Polychaeta (Nereis + Capitella) is monophyletic or paraphyletic with respect to Lumbricus—the former hypothesis predicts that Capitella shares a subset of miRNAs with Nereis, which are not found in Lumbricus; the latter predicts that either Capitella or Nereis shares miRNAs with Lumbricus, but not with the other polychaete. In order to test the monophyly of both Annelida and Polychaeta, we identified all known and novel miRNAs found in our Capitella small RNA library. In addition to the 50 known families that annelids share with other metazoans (electronic supplementary material, file 1), and the seven miRNA families restricted to annelids identified by Wheeler et al. (2009), we identified another 37 novel families of miRNAs in Capitella sp. (electronic supplementary material, file 2). Each of the miRNA genes constituting these 37 families was expressed in our small RNA library at least once, and the surrounding genomic region folds into a diagnostic hairpin structure (Ambros et al. 2003). Further, eight of these genes express both arms of the hairpin and, as described below, nine of these miRNA families are phylogenetically conserved in other taxa. This brings the total known miRNA diversity of Capitella to 123 genes grouped into 94 miRNA families (electronic supplementary material, table S1).
miRMiner uses cross-species conservation to help identify novel miRNAs (Wheeler et al. 2009). miRMiner found five sequences that are conserved in the annelid taxa under consideration, but absent in the sipunculan and in all other taxa explored thus far for their respective miRNA complements. Three of these five sequences were the previously identified ‘annelid-specific’ miRNAs miR-1987, -1998 and -1999 (Wheeler et al. 2009). The other two genes are two new genes identified herein, miR-2688 and miR-2692 (electronic supplementary material, file 1). These data support the monophyly of Annelida with respect to Sipuncula. Our data also support the paraphyly of Polychaeta because the clitellate Lumbricus shares four novel miRNA families with the polychaete Capitella that are not found in Nereis or Phascolosoma (or any other metazoan taxon): miR-2686, -2687, -2690, and -2693 (electronic supplementary material, file 1). One of these families, miR-2686, consists of multiple genes that are expressed copiously in both Capitella and Lumbricus (electronic supplementary material, file 1); all are on the same genomic trace in Capitella, suggesting that they are transcribed as a polycistron (figure 2a). Further, Capitella and Lumbricus both express the antisense strand of a paralogue of miR-10, miR-10c, a transcript not detected in any other taxon analysed (figure 2b).
Figure 2.
miRNAs suggest that annelids are monophyletic with respect to the sipunculan P. agassizii, but that polychaetes are paraphyletic with respect to the clitellate Lumbricus sp. (a) miRNA family 2686, transcripts of which were found only in Capitella and in Lumbricus. In both taxa, multiple mature reads were found (see alignment), and in Capitella all three genes are embedded in the same genomic trace (shown in bold capital letters) and are likely to be processed from a single polycistronic transcript. The star sequence of miR-2686 was also found in the Capitella small RNA library (shown in bold small letters both on the polycistron and in the structure of miR-2686a). (b) Predicted secondary structure (top, miR-1995) and Northern analyses (bottom) for miRNAs restricted to the sipunculan + annelid clade. As expected from the 454 library reads (electronic supplementary material, file 1), transcripts of miR-1997 were detected in the sipunculan Phascolosoma (P) and the two annelids Nereis (N) and Lumbricus (L), but not in the mollusc Mytilus (M). In addition, transcripts of miR-2688 were only detected in the two annelids, and transcripts of miR-10c-antisense were detected only in Lumbricus; transcripts of miR-1990 were only detected in the bivalve, but not in the sipunculan nor in any of the annelids, consistent with the 454 read data (electronic supplementary material, file 1). (c) Maximum parsimony analysis of 71 miRNA families in 13 metazoan taxa using the demosponge Amphimedon as the outgroup (electronic supplementary material, file 3). The maximum parsimony analysis with all characters unordered and unweighted finds 11 equally shortest trees at 107 steps (CI = 0.66; RI = 0.75). Bremer support values are shown at each node. Black circles indicate that the miRNA family is known to occur in that taxon; empty circles indicate that the miRNA family was not cloned in the library (e.g. Chaetoderma) or not found in the genomic traces (e.g. Lottia) in that specific taxon. miRNAs not detected in our Capitella small RNA library, but found in the genome, are shown in grey. Note that lowly expressed miRNAs are often absent from miRNA libraries (e.g. miR-124), but present in genomic sequences when available (electronic supplementary material, file 1).
Because these comparisons are necessarily made with respect to Capitella, the only annelid with a sequenced genome, it is possible that Phascolosoma, Nereis and/or Lumbricus share miRNAs not found in Capitella, which would affect our phylogenetic inferences. However, examination of all the shared small RNA sequences (i.e. potential miRNAs) identified by miRMiner indicates that this is unlikely. One hundred and thirty small RNA sequences between 20 and 24 nt in length are shared between at least two of the four taxa: the 10 novel miRNAs discussed above; 68 edits and/or seedshifts (Wheeler et al. 2009) of known (miRBase v. 13) or novel miRNAs; and 46 degraded tRNAs, rRNA, snRNAs and mRNAs, as ascertained by BLAST. Only three unidentified RNAs are shared between Phascolosoma and Nereis, and only two between Phascolosoma and Lumbricus. Even if all five of these were miRNAs, which is unlikely given the ratio between degraded non-miRNA gene products and bona fide miRNAs in our libraries (Wheeler et al. 2009), this would not refute our main conclusion, that the hypotheses presented in figure 1a–c are inconsistent with our miRNA data.
(b). Exploration of molluscan microRNAs
Because there is a long standing hypothesis that molluscs and sipunculans are closely related (Scheltema 1993), and because aplacophoran and/or gastropod molluscs sometimes appear within Annelida in molecular phylogenetic analyses (e.g. Rousset et al. 2007), we built and analysed a small RNA library from the aplacophoran C. nitidulum, and analysed these data in conjunction with those from the gastropods Haliotis and Lottia (Wheeler et al. 2009). Chaetoderma shares with the two gastropods a novel miRNA family, miR-2722, a miRNA not found in the small RNA libraries of either the sipunculan or any of the annelids (electronic supplementary material, file 1). However, the sipunculan shares with the annelids seven families of miRNAs: miR-1995, -1996, -1997, -2000, -2685, -2689 and -2692—none of these miRNAs were found in any of the mollusc RNA libraries (electronic supplementary material, file 1) or in the genomic traces from Lottia. These data support the hypothesis that sipunculans are more closely related to annelids than either are to molluscs.
(c). Experimental validation of microRNA results
We used Northern analysis to confirm that some of these miRNAs are indeed expressed as approximately 22mers in total RNA preparations. As expected, we were able to show the expression of miR-1997 in the two annelids (Nereis and Lumbricus) as well as the sipunculan at the correct size, but transcripts were not detected in the bivalve mollusc Mytilus (figure 2b). In addition, we detected transcripts of miR-2688 in Lumbricus and Nereis, but not in Phascolosoma or Mytilus, and transcripts of miR-10c-antisense in Lumbricus, but in none of the other taxa queried (figure 2b). We also confirmed that the mature reads of several of these miRNAs are processed from a genomic DNA region that folds into a stable hairpin structure (Ambros et al. 2003) in taxa from which the genome has not yet been sequenced (figure 2b, and electronic supplementary material, file 2).
(d). The root of the annelid tree
We next investigated whether sipunculans are the sister taxon of annelids. In addition to echiurans, sipunculans and, on occasion, various molluscan taxa (see above), molecular phylogenetic studies have found other non-annelid taxa, such as phoronids and nemerteans, nested within the annelids as well (Bleidorn et al. 2003; Hall et al. 2004; Colgan et al. 2006; Rousset et al. 2007; Helmkampf et al. 2008). Because identifying close relatives that are not actually annelids has proved so problematic, the position of the root of the annelid tree is effectively unknown (Rousset et al. 2007). Hence, we constructed and sequenced one additional 454 library, from the phoronid P. architecta, and analysed these data in conjunction with those from the nemertean Cerebratulus (Wheeler et al. 2009). As in the case with molluscs, none of the annelid+sipunculan-specific miRNAs, nor any of the annelid-specific miRNAs, were found in either the phoronid or the nemertean (electronic supplementary material, file 1), strongly suggesting that annelids are monophyletic, and that sipunculans are the sister group of annelids.
To test this hypothesis, we coded 13 taxa for the presence/absence of 71 miRNA families and analysed the resulting data matrix (electronic supplementary material, figure S3) with PAUP* (Swofford 2002) using maximum parsimony and decay analysis (Bremer 1994). The resulting strict-consensus tree (figure 2c) confirms that annelids are monophyletic and that sipunculans are indeed the annelid sister taxon. Further, both the nemertean and phoronid are outside of the clade Sipuncula + Annelida, and with Mollusca cluster into an unresolved polytomy. Finally, although annelids are monophyletic, polychaetes are not—Capitella is more closely related to Lumbricus than to Nereis (figure 2a).
(e). Exploring the annelid crown group for conserved microRNAs
One difficulty in addressing the monophyly of annelids with respect to sipunculans is identifying the annelid crown group. Although we chose Capitella and Nereis as the two taxa whose last common ancestor is most likely the last common ancestor of all living annelids (see above, and figure 3a), it remains possible, even likely, that other annelid taxa are more basal. A recent large-scale EST analysis, for example, suggested that the polychaete worm Chaetopterus is either basal to or falls within a clade that consisted of all other annelids plus the sipunculan (Dunn et al. 2008). Other studies support a relationship between sipunculans and a specific polychaete group—Struck et al. (2007) suggested a relationship between sipunculans and terebellid polychaetes, whereas Rousset et al. (2007) suggested a relationship between sipunculans and orbiniid polychaetes (see also Hall et al. 2004; Bleidorn et al. 2006). Because the mature sequence of most metazoan miRNAs is so conserved (Wheeler et al. 2009), we explored the annelid phylum with Northern analysis using miR-2688 and miR-10c-antisense as probes (two of the highest expressed miRNAs, electronic supplementary material, file 1). We examined the total RNA of three terebellid polychaetes, Thelepus, Amphitrite and Pectinaria, as well as Chaetopterus, the orbiniid Scoloplos, the onuphid Diopatra and the arenicolid Abarenicola (figure 3a). Northern analysis detected transcripts of the correct size (approx. 22 nt) hybridizing to the miR-2688 probe in all of these polychaete species (figure 3b). Interestingly, when miR-10c-antisense was used as a probe, only Abarenicola showed a hybridization signal (figure 3b), consistent with the hypothesized close relationship between arenicolids and capitellids (Rouse & Fauchald 1997).
Figure 3.
Exploration of the annelid crown group for annelid-specific miRNAs. (a) Phylogenetic perspective of the annelid taxa explored by Northern analysis and (b,c) genome walking in relation to Nereis and Capitella, according to Rouse & Fauchald (1997). (b) Northern analysis using probes derived from the sequences of miR-2688 and miR-10c-antisense against the total RNA derived from the indicated taxon (see electronic supplementary material, file 2). Note that all annelids queried express miR-2688 as an approximately 22 nt RNA, but only Abarenicola appears to express miR-10c-antisense. (c) Predicted secondary structure of miR-2688 from the polychaete annelid Chaetopterus sp. derived from genome walking (§2).
To determine whether these transcripts arise from miRNA loci, we constructed a genome-walker library from the polychaete Chaetopterus. Using the inferred mature sequence as a primer (§2), we amplified not only miR-2688 (figure 3c), as expected from the Northern result (figure 3b), but two other loci as well: miR-1997 and miR-1988 (electronic supplementary material, file 2). These data demonstrate that the sipunculan remains phylogenetically outside Annelida even when a broad spectrum of annelids is analysed using both Northern analysis and genome walking.
4. Conclusions
These results suggest that the hypotheses shown in figure 1a–c are incorrect. The miRNA data, like the evidence derived from the fossil record (figure 1d) and a recent mitochondrial gene study (Mwinyi et al. 2009), support: (i) the paraphyly of polychaetes with respect to clitellates and the primitiveness of the polychaete body plan and (ii) the monophyly of annelids and a sister taxon relationship between annelids and sipunculans. The concordance of the miRNA phylogeny and the fossil evidence suggests that the earliest annelids were epibenthic, vagile, segmented organisms (Westheide 1997; Bartolomaeus et al. 2005) and not burrowing worms as sometimes assumed (Clark 1964; Fauchald 1974), and that the absence of segmentation in sipunculans may be primitive. Finally, this study demonstrates the potential of miRNAs to reveal the broad pattern not only of the annelid evolutionary tree, but also that of other metazoan groups (Sperling & Peterson 2009).
Acknowledgements
K.J.P. is supported by the National Science Foundation. E.A.S. is funded by student grants from the Systematics Association and the Yale Enders Fund. J.V. is funded by the Carlsberg Foundation. We thank J. Grassle, R. Elahi, C. Tanner and M. Obst for material; D. Eibye-Jacobsen, F. Oyarzun, R. Strathmann and B. Swalla for help in collecting; A. Heimberg for technical assistance; P. Donoghue for his usual perspicacity; and E. Champion for help with figure 1.
References
- Ambros V., et al. 2003A uniform system for microRNA annotation. RNA 9, 277–279 (doi:10.1261/rna.2183803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomaeus T., Purschke G., Hausen H.2005Polychaete phylogeny based on morphological data—a comparison of current attempts. Hydrobiologia 535, 341–356 (doi:10.1007/s10750-004-1847-5) [Google Scholar]
- Bleidorn C., Vogt L., Bartolomaeus T.2003New insights into polychaete phylogeny (Annelida) inferred from 18S rDNA sequences. Mol. Phylogenet. Evol. 29, 279–288 (doi:10.1016/S1055-7903(03)00107-6) [DOI] [PubMed] [Google Scholar]
- Bleidorn C., Podsiadlowski L., Bartolomaeus T.2006The complete mitochondrial genome of the orbiniid polychaete Orbinia latreillii (Annelida, Orbiniidae)—a novel gene order for Annelida and implications for annelid phylogeny. Gene 370, 96–103 (doi:10.1016/j.gene.2005.11.018) [DOI] [PubMed] [Google Scholar]
- Bremer K.1994Branch support and tree stability. Cladistics 10, 295–304 (doi:10.1111/j.1096-0031.1994.tb00179.x) [Google Scholar]
- Budd G.2001Climbing life's tree. Nature 412, 487 (doi:10.1038/35087679) [DOI] [PubMed] [Google Scholar]
- Budd G. E., Jensen S.2000A critical reappraisal of the fossil record of the bilaterian phyla. Biol. Rev. Camb. Phil. Soc. 75, 253–295 (doi:10.1017/S000632310000548X) [DOI] [PubMed] [Google Scholar]
- Clark R. B.1964Dynamics in metazoan evolution: the origin of the coelom and segments Oxford, UK: Clarendon Press [Google Scholar]
- Colgan D. J., Hutchings P. A., Braune M.2006A multigene framework for polychaete phylogenetic studies. Organ. Divers. Evol. 6, 220–235 (doi:10.1016/j.ode.2005.11.002) [Google Scholar]
- Conway Morris S.1979Middle Cambrian polychaetes from the Burgess Shale of British-Columbia. Phil. Trans. R. Soc. Lond. B 285, 227–274 (doi:10.1098/rstb.1979.0006) [Google Scholar]
- Conway Morris S., Peel J. S.2008The earliest annelids: Lower Cambrian polychaetes from the Sirius Passet Lagerstätte, Peary Land, North Greenland. Acta Palaeontol. Pol. 53, 137–148 [Google Scholar]
- Dunn C. W., et al. 2008Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749 (doi:10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- Eibye-Jacobsen D.2004A reevaluation of Wiwaxia and the polychaetes of the Burgess Shale. Lethaia 37, 317–335 (doi:10.1080/00241160410002027) [Google Scholar]
- Fauchald K.1974Polychaete phylogeny: a problem in protostome evolution. Syst. Zool. 23, 493–506 (doi:10.2307/2412467) [Google Scholar]
- Gruber A. R., Lorenz R., Bernhart S. H., Neuboock R., Hofacker I. L.2008The Vienna RNA Websuite. Nucleic Acids Research 36, W70–W74 (doi:10.1093/nar/gkn188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halanych K. M., Dahlgren T. G., McHugh D.2002Unsegmented annelids? Possible origins of four lophotrochozoan worm taxa. Integr. Comp. Biol. 42, 678–684 (doi:10.1093/icb/42.3.678) [DOI] [PubMed] [Google Scholar]
- Hall K. A., Hutchings P. A., Colgan D. J.2004Further phylogenetic studies of the Polychaeta using 18S rDNA sequence data. J. Mar. Biol. Assoc. UK 84, 949–960 (doi:10.1017/S0025315404010240h) [Google Scholar]
- Hausdorf B., Helmkampf M., Meyer A., Witek A., Herlyn H., Bruchhaus I., Hankeln T., Struck T., Lieb B.2007Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol. Biol. Evol. 24, 2723–2729 (doi:10.1093/molbev/msm214) [DOI] [PubMed] [Google Scholar]
- Helmkampf M., Bruchhaus I., Hausdorf B.2008Multigene analysis of lophophorate and chaetognath phylogenetic relationships. Mol. Phylogenet. Evol. 46, 206–214 (doi:10.1016/j.ympev.2007.09.004) [DOI] [PubMed] [Google Scholar]
- Hessling R.2002Metameric organisation of the nervous system in developmental stages of Urechis caupo (Echiura) and its phylogenetic implications. Zoomorphology 121, 221–234 (doi:10.1007/s00435-002-0059-7) [Google Scholar]
- Hints O., Eriksson M. E.2007Diversification and biogeography of scolecodont-bearing polychaetes in the Ordovician. Palaeogeogr. Palaeoclimatol. Palaeoecol. 245, 95–114 (doi:10.1016/j.palaeo.2006.02.029) [Google Scholar]
- Huang D.-Y., Chen J.-Y., Vannier J., Salinas J. I. S.2004Early Cambrian sipunculan worms from southwest China. Proc. R. Soc. Lond. B 271, 1671–1676 (doi:10.1098/rspb.2004.2774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies R. P. S.1979The origin of chordates—a methodological essay. In The origin of major invertebrate groups (ed. House M. R.), pp. 443–477 London, UK: Academic Press [Google Scholar]
- Kristof A., Wollesen T., Wanninger A.2008Segmental mode of neural patterning in Sipuncula. Curr. Biol. 18, 1129–1132 (doi:10.1016/j.cub.2008.06.066) [DOI] [PubMed] [Google Scholar]
- Maddison D. R., Maddison W. P.2005MacClade 4 Sunderland, UK: Sinauer Associates [Google Scholar]
- Margulies M., et al. 2005Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (doi:10.1038/nature03959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwinyi A., Meyer A., Bleidorn C., Lieb B., Bartolomaeus T., Podsiadlowski L.2009Mitochondrial genome sequence and gene order of Sipunculus nudus give additional support for an inclusion of Sipuncula into Annelida. BMC Genom. 10, 27 (doi:10.1186/1471-2164-10-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse G. W., Fauchald K.1997Cladistics and polychaetes. Zoologica Scripta 26, 139–204 (doi:10.1111/j.1463-6409.1997.tb00412.x) [Google Scholar]
- Rouse G. W., Pleijel F.2007Annelida. Zootaxa 1668, 245–264 [Google Scholar]
- Rousset V., Pleijel F., Rouse G. W., Erséus C., Siddall M. E.2006A molecular phylogeny of annelids. Cladistics 23, 41–63 (doi:10.1111/j.1096-0031.2006.00128.x) [DOI] [PubMed] [Google Scholar]
- Rousset V., Pleijel F., Rouse G. W., Erséus C., Siddall M. E.2007A molecular phylogeny of annelids. Cladistics 23, 41–63 (doi:10.1111/j.1096-0031.2006.00128.x) [DOI] [PubMed] [Google Scholar]
- Scheltema A. H.1993Aplacophora as progenetic aculiferans and the coelomate origin of mollusks as the sister taxon of Sipuncula. Biol. Bull. 184, 57–78 (doi:10.2307/1542380) [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Cole C. N., McPeek M. A., Peterson K. J.2006The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. (Mol. Dev. Evol.) 306B, 575–588 (doi:10.1002/jez.b.21118) [DOI] [PubMed] [Google Scholar]
- Sorenson M. D., Franzosa E. A.2007TreeRot. Boston, MA: Boston University [Google Scholar]
- Sperling E. A., Peterson K. J.2009microRNAs and metazoan phylogeny: big trees from little genes. In Animal evolution—genomes, trees and fossils (eds Telford M. J., Littlewood D. T. J.), pp. 157–170 Oxford, UK: Oxford University Press [Google Scholar]
- Struck T. H., Schult N., Kusen T., Hickman E., Bleidorn C., McHugh D., Halanych K. M.2007Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 7, 57 (doi:10.1186/1471-2148-7-57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L.2002PAUP* phylogenetic analysis using parsimony (* and other methods) v.4.0b10 for Macintosh Sunderland, UK: Sinauer Associates [Google Scholar]
- Westheide W.1997The direction of evolution within the Polychaeta. J. Nat. Hist. 31, 1–15 (doi:10.1080/00222939700770011) [Google Scholar]
- Wheeler B. M., Heimberg A. M., Moy V. N., Sperling E. A., Holstein T. W., Heber S., Peterson K. J.2009The deep evolution of metazoan microRNAs. Evol. Dev. 11, 50–68 (doi:10.1111/j.1525-142X.2008.00302.x) [DOI] [PubMed] [Google Scholar]
- Xin S., Ma X., Ren J., Zhao F.2009A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genom. 10, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Mathews D. H., Turner D. H.1999Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In RNA Biochemistry and biotechnology (eds Barciszewski J., Clark B. F. C.), pp. 11–43 Dordrecht, The Netherland: Kluwer Academic Publishers [Google Scholar]