Abstract
Recent genetic studies have challenged the traditional view that the ancestors of British Celtic people spread from central Europe during the Iron Age and have suggested a much earlier origin for them as part of the human recolonization of Britain at the end of the last glaciation. Here we propose that small mammals provide an analogue to help resolve this controversy. Previous studies have shown that common shrews (Sorex araneus) with particular chromosomal characteristics and water voles (Arvicola terrestris) of a specific mitochondrial (mt) DNA lineage have peripheral western/northern distributions with striking similarities to that of Celtic people. We show that mtDNA lineages of three other small mammal species (bank vole Myodes glareolus, field vole Microtus agrestis and pygmy shrew Sorex minutus) also form a ‘Celtic fringe’. We argue that these small mammals most reasonably colonized Britain in a two-phase process following the last glacial maximum (LGM), with climatically driven partial replacement of the first colonists by the second colonists, leaving a peripheral geographical distribution for the first colonists. We suggest that these natural Celtic fringes provide insight into the same phenomenon in humans and support its origin in processes following the end of the LGM.
Keywords: colonization history, Microtus agrestis, mitochondrial DNA, Myodes glareolus, replacement, Sorex minutus
1. Introduction
The settlement of Britain by early modern humans commenced before the last glacial maximum (LGM, 19–23 000 BP; Mix et al. 2001), but the ice advance during it led to the British Isles being abandoned by temperate species, including humans (Barton et al. 2003). Although a variety of scenarios have been proposed based on archaeological and genetic data, there is still debate over the nature and timing of the reoccupation of the British Isles by humans after the LGM (Oppenheimer 2006; Conneller 2007; Jacobi & Higham 2009). Likewise, linked in with these scenarios, there is much discussion on the regional impact (both biologically and culturally) of subsequent human migrations into the British Isles.
In terms of current regional distributions in the British Isles, the long-standing human occupants of the western and northern periphery are distinct culturally and genetically from those in central and eastern Britain (Davies 1999; Forster et al. 2004; Oppenheimer 2006). The cultural distinctiveness is most evident at present through the separation of the peripheral people and areas into the nations of Scotland, Ireland (Northern Ireland and the Republic of Ireland) and Wales, the dependency of the Isle of Man and the most westerly English county of Cornwall. The central and eastern areas and the people within them constitute England. The traditional languages in the peripheral areas (Scottish Gaelic, Irish Gaelic, Welsh, Manx and Cornish) are closely related to each other linguistically (Forster & Toth 2003) and distinctive from English. Genetically, there are differences between people in the peripheral areas and those in England; for example, in peripheral areas there are high frequencies of individuals with the mtDNA marker J/16192 and belonging to the Y-chromosomal gene cluster R1b-14, while J/16231 and I1a occur at high frequencies in England (Forster et al. 2004; Oppenheimer 2006).
While the pattern is clear, the processes that led to that pattern remain contentious. The vernacular name for the peripheral regions and people within them is the Celtic fringe, which also includes Brittany in north-western France. This reflects a traditional scenario that Celts spread into and occupied the whole of the British Isles by 2600 BP, replacing earlier inhabitants. It is generally assumed that the Celts remained numerically dominant and geographically widespread until they were themselves partially replaced by Anglo-Saxons at approximately 1600–1200 BP, resulting in the Anglo-Saxon territory of ‘England’ and a peripheral fringe of Celtic people, which has persisted to the present day (Davies 1999). However, this scenario is controversial among historians and human geneticists and, while supported by some, alternative scenarios involve, for example, greater persistence of pre-Celtic people, and cultural spread rather than population replacement by the Anglo-Saxons (Davies 1999; Burmeister 2000; Weale et al. 2002; Capelli et al. 2003; Oppenheimer 2006). Therefore, interpretation of the human pattern may need to take into account the aspects of human migration leading all the way back to the recolonization of the British Isles after the LGM.
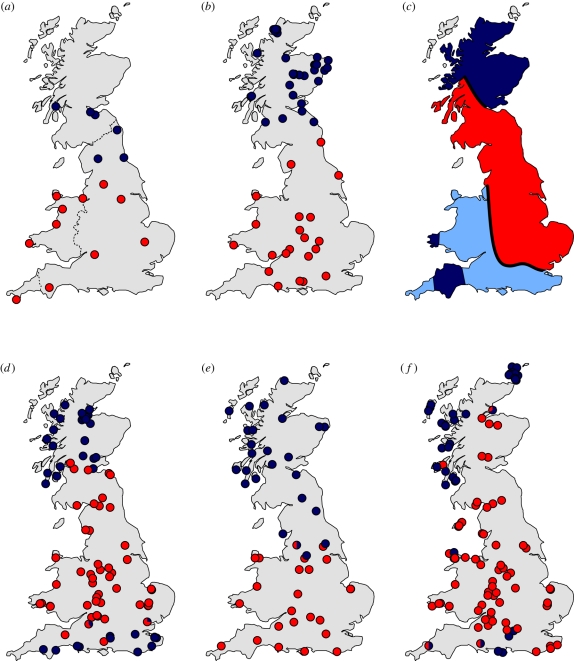
Given this controversy with regard to the origin and timing of the human Celtic fringe, it is of interest to understand similar geographical distributions in other organisms, as they may shed light on the human phenomenon (Matisoo-Smith & Robins 2004; Larson et al. 2007; Searle et al. 2009). It has already been noted that in Britain common shrews (Sorex araneus) with particular chromosomal characteristics (Searle & Wilkinson 1987) and water voles of a specific mtDNA lineage (Piertney et al. 2005) have peripheral distributions with striking similarities to that of Celtic people (figure 1). Here we have carried out detailed phylogeographic analyses of three other small mammal species, the bank vole (Myodes glareolus), field vole (Microtus agrestis) and pygmy shrew (Sorex minutus). These species already occupied Britain before the LGM, but, like humans, they were displaced by glacial advances and subsequently recolonized Britain following glacial retreat (Yalden 1999). We show that mtDNA lineages of these three small mammal species also form a ‘Celtic fringe’. We have combined our new data with earlier results on other species (including those mentioned above); overall, there is now a rich and varied dataset involving seven species of small mammals in Britain. Although our results build on earlier work, only with our new findings is the dataset adequate to provide a compelling explanation for Celtic fringes in small mammals and to allow comparisons with humans. In this paper, our first consideration is Celtic fringes on the island of Britain that were attained through natural colonization. We later address Celtic fringes involving either Ireland and/or human introduction.
Figure 1.
Celtic fringes in British small mammals. The distribution of the peripheral (blue) and central (red) phylogroup is shown for each species and genetic marker. The figures are based on data on (a) haemoglobin electrophoresis polymorphism for bank voles (M. glareolus), (c) chromosome characteristics (darker blue, the ko, np and qr chromosomal race within the peripheral ko chromosomal form; see text) of common shrews (S. araneus), and mitochondrial DNA sequences for (b) water voles (A. terrestris), (d) bank voles, (e) field voles (M. agrestis) and (f) pygmy shrews (S. minutus). Thin dashed lines in (a) show the borders of Scotland, Wales and Cornwall within Britain. See text for references to previously published data.
2. Material and methods
(a). Molecular biological techniques
Genomic DNA was extracted from 157 bank voles (M. glareolus), 41 field voles (M. agrestis) and 162 pygmy shrews (S. minutus) using tissue samples of liver, spleen, tail or toe clips stored in 95 per cent ethanol and using the Qiagen (Valencia, CA, USA) DNeasy Tissue Kit. For the bank vole, a 1074 bp fragment of the mtDNA gene cytochrome b (cytb) was PCR amplified as described previously (Kotlík et al. 2006) with primers designed for this species within the cytb and Thr tRNA genes, respectively. For the field vole the entire cytb (1143 bp) was amplified with primers located within the Glu tRNA and Thr tRNA genes (Jaarola & Searle 2002). For the pygmy shrew, 1110 bp of cytb were amplified in two overlapping segments using a combination of published (Irwin et al. 1991) forward and reverse primers located within the Glu tRNA and Thr tRNA genes, respectively, and newly designed forward (5′-GAGGACAAATGTCATTCTGAGGC-3′) and reverse (5′-GTAGTAGGGGTGGAAAGGAATT-3′) primers located within the cytb gene. For all three species, the resulting PCR products were purified using the Qiagen QIAquick PCR Purification Kit and were directly cycle sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, MA, USA). The extension products were run on ABI 3730 automated sequencers. Sequences were aligned manually and ambiguities were resolved by sequencing the complementary strand, and identical sequences were collapsed into haplotypes using MacClade 4.08 (Maddison & Maddison 2003). Nucleotide sequences of each unique haplotype have been deposited in the GenBank database under the accession numbers FJ619746–FJ619786, FJ623774–FJ623893, FJ640865–FJ640869 and FJ640871–FJ640953.
(b). Phylogeographic analysis
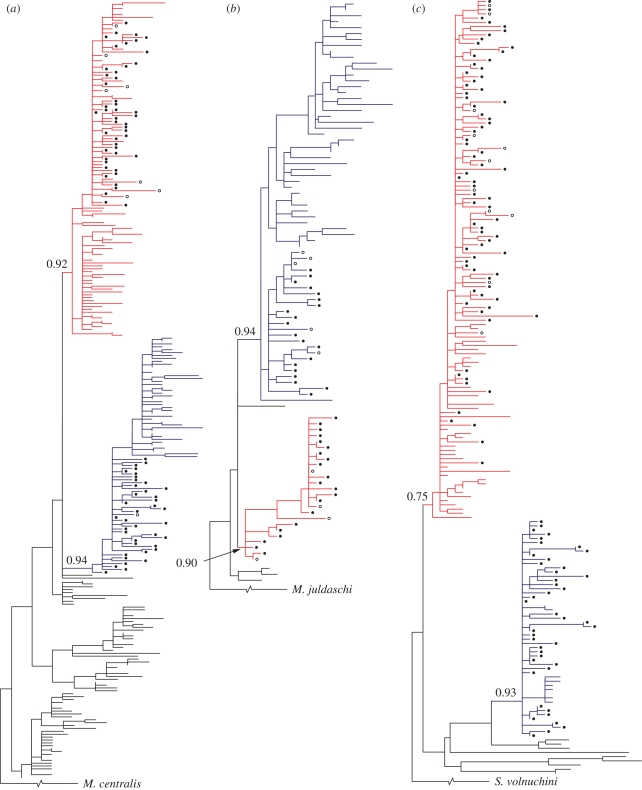
The phylogenetic relationships among the sequences were reconstructed using the maximum-likelihood optimality criterion. In addition to the newly obtained sequences (appendices 1–3 in the electronic supplementary material), the tree for each species also included published sequences from continental Europe (Jaarola & Searle 2002; Mascheretti et al. 2003; Deffontaine et al. 2005; Kotlík et al. 2006) in order to determine the relatedness and position of the British sequences within the phylogeography of the entire species. The small number of published British sequences were also included (12 bank voles from five localities, Deffontaine et al. 2005; nine field voles from nine localities, Jaarola & Searle 2002; 11 pygmy shrews from eight localities, Mascheretti et al. 2003). The respective trees were rooted by using Myodes centralis, Microtus juldaschi and Sorex volnuchini as the outgroup (Mascheretti et al. 2003; Jaarola et al. 2004; Kotlík et al. 2006). The program jModelTest 0.1.1 (Posada 2008) was used to determine the best-fit model of sequence evolution for cytb for each species using the Akaike information criterion method of measure of fit of 88 different nested models to the data. The maximum-likelihood phylogenetic analyses were performed by the BEST approach implemented in PhyML 3.0.1, which combines the nearest neighbour interchanges with subtree pruning and regrafting algorithms to maximize tree likelihood (Guindon & Gascuel 2003), and using the TIM + I + G evolutionary model (where TIM stands for Transitional Model, I is for the proportion of invariable sites, and G is for the gamma-distributed rates across sites; Posada 2003) for the bank and field voles and the GTR + G model (where GTR stands for general time-reversible; Tavaré 1986) for the pygmy shrew. Branch support for the phylogenetic partitioning of the British sequences was quantified by the approximate likelihood ratio test (Anisimova & Gascuel 2006) using the non-parametric Shimodaira–Hasegawa-like (SH-like) procedure implemented in PhyML (Guindon & Gascuel 2003). Model-corrected pairwise genetic distances between haplotypes were estimated with PAUP* 4.0 (Swofford 2003) and the resulting distance matrices were imported into MEGA 4.0.2 (Tamura et al. 2007) to estimate the average sequence divergence between distinct phylogenetic groups (phylogroups) identified for each species. Distributions of the phylogroups in Britain were plotted using a Geographical Information Systems (GIS) map produced using the ArcMap application in ArcGIS 9.3 (Environmental Systems Research Institute Inc., Redlands, CA, USA). Previously published genetic data for other small mammal species and genetic systems were mapped in the same way: haemoglobin variants detected by protein electrophoresis for the bank vole (Hall 1979), chromosomal forms of the common shrew S. araneus (Searle & Wilkinson 1987; Searle 1988) and mtDNA phylogroups of the water vole Arvicola terrestris (Piertney et al. 2005). For these maps, the total numbers of individuals/sampling localities were as follows: haemoglobin, bank voles: 322/17; mtDNA, water vole: 57/53; chromosomes, common shrew: 1412/151; mtDNA, bank vole: 169/86; mtDNA, field vole: 50/48; mtDNA, pygmy shrew: 173/109.
3. Results and discussion
Our phylogenetic analyses for the bank vole, field vole and pygmy shrew divided British sequences among two distinct phylogenetic groups (phylogroups) with high statistical support in each species (figure 2). Remarkably, in all of these species, we demonstrate peripheral distributions for one phylogroup relative to a second one, with striking similarities to that of Celtic people (figure 1). In all cases, the phylogroups are too genetically distinctive (1.2–2.2% average sequence divergence) to have evolved from one another in situ in Britain over the maximum time period available since the LGM (approx. 19 000 years; Avise et al. 1998; Yalden 1999), and, in all species, both phylogroups have been found in continental Europe as well as in Britain (figure 2; from the figure this is not evident for the central lineage of the field vole, but becomes clear on further analysis with more samples: J. S. Herman, unpublished data).
Figure 2.
Maximum-likelihood phylogenies of British small mammals. (a) Bank vole (M. glareolus), (b) field vole (M. agrestis), (c) pygmy shrew (S. minutus). The sequences are from throughout the species range with new and previously reported British sequences labelled. Filled circle, new from this study; open circle, published. The phylogroups found in Britain have been labelled ‘central’ (red) or ‘peripheral’ (blue) according to their distribution there. Numbers indicate the SH-like aLRT supports for the phylogroups.
These findings build on the earlier data for the common shrew and water vole. For the common shrew, a chromosomal race characterized by the chromosomes ko, np and qr (where each letter defines a chromosome arm) was found extensively in Scotland, and also in southwest Wales and Cornwall (Searle & Wilkinson 1987; Searle 1988), matching closely the human Celtic fringe (figure 1). This is one race among several which together constitute a chromosomal form (a group of chromosomal races) characterized by chromosome ko with a peripheral distribution to the north, west and south of Britain, as opposed to the central and eastern kq form. For the water vole (Piertney et al. 2005), the two mtDNA phylogroups meet along a line extraordinarily close to the current English–Scottish border (figure 1). Interestingly, a much earlier report showed a pattern of distribution of haemoglobin variants in the bank vole similar to that of the water vole, but with a boundary between the two types running through northern England (Hall 1979; figure 1).
For the field vole, the distribution of mtDNA phylogroups that we found is almost identical to that of the bank vole haemoglobin variants (figure 1). The bank vole mtDNA phylogroups have similarities to the common shrew chromosomal forms, in that there is a peripheral distribution in the north and south, and the northern boundary is at the same location in the two systems (figure 1). For the pygmy shrew mtDNA phylogroups, again there is a peripheral distribution to the north, west and south, but the peripheral mtDNA phylogroup is much more scattered than in the bank vole (figure 1). The peripheral phylogroup of the pygmy shrew is particularly evident on islands surrounding Britain (Isle of Wight, Anglesey, ten islands of the Inner Hebrides, three islands of the Outer Hebrides and three Orkney islands). The current study greatly extends an earlier mtDNA analysis of pygmy shrews by Mascheretti et al. (2003), which, through insufficient sampling, failed to detect the peripheral phylogroup in Britain, although did demonstrate its presence in Ireland.
The explanation provided by Searle & Wilkinson (1987) and Piertney et al. (2005) for the Celtic fringe distributions of the common shrew and water vole was a two-phase colonization that involved one genetic type colonizing the whole of Britain, before being partially replaced by a second invading genetic type from everywhere except the periphery. We believe that this explanation can apply to all cases presented in figure 1. To accommodate all the distributions seen, it would appear that the second, replacement colonization would have had to have come from the east, in this way generating peripheral distributions in the north, west and south. The occurrence of pygmy shrews of the peripheral type on the islands immediately surrounding Britain suggests natural or human-mediated colonization of those islands from the nearby mainland before the peripheral type was almost completely replaced on the mainland.
For another small mammal, the stoat Mustela erminea, the Celtic fringe has not been observed within Britain itself but is represented by the presence of a distinctive mtDNA phylogroup in Ireland (Martínková et al. 2007). The authors also surmised two-phase colonization for this case. They inferred that the first-colonizing mtDNA phylogroup occupied both Britain and Ireland during the LGM, at the time that they were part of a common landmass with continental Europe. A water barrier then formed between Britain and Ireland and it was deduced that subsequently the first-colonizing phylogroup was completely replaced in Britain by another phylogroup invading overland from continental Europe (Martínková et al. 2007). The stoat is the only species of those considered here believed to have been sufficiently cold-hardy to have colonized Ireland naturally (Searle 2008), although the pygmy shrew (above), the bank vole and house mouse (below) also occur there, apparently through human introduction (Searle 2008).
Considering events in Britain from the LGM onwards, apparently only the cold-hardy stoat was already present in its vicinity at the LGM (see above). The other, less cold-tolerant species for which two-phase colonization has been inferred were almost certainly not present in the vicinity of Britain then (but see Stewart & Lister 2001). Instead, for these species, the initial occupation of Britain would have been during the climatic improvement after the LGM (i.e. the temperate period of the Late Glacial; within the interval 12 900–19 000 BP; Mix et al. 2001; Steffensen et al. 2008). During this time, there was a landbridge to Britain, the climate and habitat would have been suitable and there is subfossil evidence for the presence of bank, field and water voles and common and pygmy shrews (Yalden 1982, 1999; Price 2003). On this basis, a likely key factor in the replacement events that followed the initial colonization would have been the cold period at the very end of the last glaciation, the Younger Dryas (11 700–12 900 BP: Steffensen et al. 2008). This followed the temperate period of the Late Glacial (see above) and preceded the temperate period of the Holocene (11 700 BP until the present).
For all the species for which two-phase colonization has been inferred, if, as we suggest, the first-colonizing genetic type was already present in Britain during the Younger Dryas, it probably would have had a small and dispersed population there, due to the unfavourable conditions (see Price 2003). This would have made that genetic type susceptible to replacement by another form invading from continental Europe after the Younger Dryas, before the separation of Britain from continental Europe. Conditions changed very rapidly from extreme cold to warm temperate conditions in Britain at this time (Coope 1979), which would have promoted rapid population growth in all the species of small mammals. The variation between species in the extent of replacement may therefore reflect the distribution and population size of the first colonizing genetic type at the time of colonization of the second type and the relative performance of the two forms under different temperature conditions. Also the topography of Britain (with uplands in the periphery to the north, west and south and several major rivers) is likely to have been of importance. Genetic forms within species tend to stabilize their contacts at geographical boundaries (such as large rivers) and transitions (such as between lowland and upland), responding to barriers of migration and adaptation to different environmental conditions, respectively (Barton & Hewitt 1985; Swenson & Howard 2005).
The extent to which the proposed replacement events involved invasion by whole new genomes, or more limited selective sweeps of particular loci and markers closely linked to them, is poorly known. For the bank vole, the nuclear and mitochondrial markers show some differences in the pattern of replacement (figure 1), and for the stoat, the British and Irish populations are morphologically distinct subspecies as well as differing in mtDNA (Martínková et al. 2007).
The scenario that the replacements came from the east fits well with the location of the landbridge connecting Britain and continental Europe at the end of the Younger Dryas. The replacing form thus would have colonized over ‘Doggerland’, a landmass now submerged beneath the North Sea (Spinney 2008). That submersion and the formation of the English Channel occurred at approximately 8000 BP (Weninger et al. 2008), which resulted in Britain being severed from continental Europe and halting any further land colonizations from there by small mammals.
Replacement is likely to have an impact on genetic patterns in other organisms in the British Isles as well, including the larger mammals, as noted recently for otters Lutra lutra (Stanton et al. 2009). But what about humans? What is the relationship between the Celtic fringes in voles and shrews and that in humans? There are striking similarities in terms of patterns and process. For the chromosomal forms of the common shrew, there is a boundary close to the English–Welsh border, and for the water vole, the mtDNA phylogroups meet at the current English–Scottish border (figure 1). In other cases, the boundaries between the two genetic types found in Britain are not particularly close to the current national borders. However, in five of the six panels in figure 1, the peripheral type shows a widespread northern distribution that broadly matches the northern part of the human Celtic fringe. In considering the determinants of the human Celtic fringe, it seems reasonable to suggest that cultural and/or genetic adaptations relating to environmental conditions on either side of the natural frontiers between what is currently England (generally lowland) and what is currently Scotland or Wales (generally upland) may have been involved in its formation.
Was it possible that the Celtic fringe in humans—that is, the occurrence of a distinctive population in peripheral parts of Britain—was initially the result of a replacement event at the end of the Younger Dryas, and that the replacing population invaded over Doggerland? Of course this timing (approx. 11 700 BP) would not fit with the traditional scenario that the Celtic fringe was formed by Celts and Anglo-Saxons. However, there are genetic data consistent with a colonization of Britain from Iberia before the Younger Dryas, with subsequent genetically distinctive migrations over Doggerland (Pereira et al. 2005; Oppenheimer 2006). Therefore, what we name the Celtic fringe may actually have been formed long before the Celtic spread through Europe, perhaps at the same time as in small mammals. Under those circumstances, of course, the Celts and Anglo-Saxons would have ‘reinforced’ a pre-existing distribution of peripheral and central populations in Britain. There is evidence from historic times that such a geographical distribution has been reinforced by culturally different people at various intervals. Indeed, it is already clear that the Anglo-Saxons were not the first group to occupy ‘England’; they reinforced an area that had been occupied by the Romans, and the Normans later took control of the same territory (Davies 1999).
One of the reinforcing invasions, that of the Norwegian Vikings around 1200–1000 BP, needs special mention. These seafarers occupied parts of the Celtic fringe including Ireland, the Isle of Man and northern and western Scotland (Logan 2005). Apparently, they also accidentally brought along with them a particular mtDNA phylogroup of house mice (Mus musculus) which now occurs over the same distribution (Searle et al. 2009). The presence of house mice in western Europe reflects their transport as stowaways on human boats and vehicles, primarily in agricultural materials (Searle et al. 2009). Therefore, it can be suggested that the same processes that led to the formation of Celtic fringes in naturally colonizing small mammals also influenced the formation of the Celtic fringe in humans, and that the Celtic fringe in humans generated a Celtic fringe in a commensal small mammal that was accidentally introduced by them. In this way, there can be a unified theory to explain the Celtic fringes described in seven species of small mammal that occupy the British Isles (water, bank and field voles, common and pygmy shrew, stoat and house mouse).
The results of our study have broader implications beyond understanding Celtic fringes in the British Isles. For five species of small mammal (water, bank and field vole, pygmy shrew and stoat), the distribution of two mtDNA phylogroups in the British Isles is best explained by replacement of one phylogroup by another. There is every reason to believe that the cause of the replacement (e.g. a response to climatic change at the end of the Younger Dryas, as suggested here) is something that could occur in a continental setting as well. However, such replacements may be considerably more difficult to detect than in Britain. That Britain was a peninsula and then later an island, with a high-altitude periphery, probably assists in the retention of the signal and detection of partial replacement events. If replacement or partial replacement of mtDNA phylogroups is common in a continental setting, then that makes suspect phylogeographic interpretations based solely on current genetic patterns (for a review, see Hewitt 2000). Techniques that can be applied to samples of varying antiquity are particularly valuable to test for replacement in both humans (Fregel et al. 2009) and wild mammals, and, for the latter, ancient DNA studies and studies using museum specimens have demonstrated replacement events unequivocally in a continental context (Barnes et al. 2002; Pergams et al. 2003; Hofreiter et al. 2007). If, as we suggest, climate is driving the replacement events, then not only does this influence the interpretation of phylogeographic patterns created in the past, it also has implications for the persistence and genetic architecture of species during current and future global warming.
Acknowledgements
We gratefully acknowledge the many colleagues who provided specimens (listed in appendices 1–3 in the electronic supplementary material). We also thank Rodrigo Vega and Fríđa Jóhannesdóttir for help with data formatting and Angela Douglas, John Stewart and an anonymous referee for comments on the manuscript. The work was supported by grants from the Royal Society of London, Grant Agency of the Academy of Sciences of the Czech Republic (grants IAA600450701, IAA600450901 and IRP IAPG AV0Z50450515), the Skye Foundation, South African National Research Foundation and the Irish Research Council for Science, Engineering and Technology and Enterprise Ireland.
REFERENCES
- Anisimova M., Gascuel O.2006Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552 (doi:10.1080/10635150600755453) [DOI] [PubMed] [Google Scholar]
- Avise J. C., Walker D., Johns G. C.1998Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc. R. Soc. Lond. B 265, 1707–1712 (doi:10.1098/rspb.1998.0492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes I., Matheus P., Shapiro B., Jensen D., Cooper A.2002Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295, 2267–2270 (doi:10.1126/science.1067814) [DOI] [PubMed] [Google Scholar]
- Barton N. H., Hewitt G. M.1985Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148 (doi:10.1146/annurev.es.16.110185.000553) [Google Scholar]
- Barton R. N. E., Jacobi R. M., Stapert D., Street M. J.2003The Late-glacial reoccupation of the British Isles and the Creswellian. J. Q. Sci. 18, 631–643 (doi:10.1002/jqs.772) [Google Scholar]
- Burmeister S.2000Archaeology and migration—approaches to an archaeological proof of migration. Curr. Anthropol. 41, 539–567 (doi:10.1086/317383) [Google Scholar]
- Capelli C., et al. 2003A Y chromosome census of the British Isles. Curr. Biol. 13, 979–984 (doi:10.1016/S0960-9822(03)00373-7) [DOI] [PubMed] [Google Scholar]
- Conneller C.2007Inhabiting new landscapes: settlement and mobility in Britain after the last glacial maximum. Oxford J. Archaeol. 26, 215–237 (doi:10.1111/j.1468-0092.2007.00282.x) [Google Scholar]
- Coope G. R.1979Cenozoic fossil Coleoptera: evolution, biogeography, and ecology. Annu. Rev. Ecol. Syst. 10, 247–267 (doi:10.1146/annurev.es.10.110179.001335) [Google Scholar]
- Davies N.1999The isles: a history London, UK: Macmillan [Google Scholar]
- Deffontaine V., Libois R., Kotlík P., Sommer R., Nieberding C., Paradis E., Searle J. B., Michaux J. R.2005Beyond the Mediterranean peninsulas: evidence of central European glacial refugia for a temperate forest mammal species, the bank vole (Clethrionomys glareolus). Mol. Ecol. 14, 1727–1739 (doi:10.1111/j.1365-294X.2005.02506.x) [DOI] [PubMed] [Google Scholar]
- Forster P., Toth A.2003Toward a phylogenetic chronology of ancient Gaulish, Celtic, and Indo-European. Proc. Natl Acad. Sci. USA 100, 9079–9084 (doi:10.1073/pnas.1331158100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Romano V., Cali F., Röhl A., Hurles M.2004MtDNA markers for Celtic and Germanic language areas in the British Isles. In Traces of ancestry: studies in honour of Colin Renfrew (ed. Jones M.), pp. 99–111 Cambridge, UK: McDonald Institute for Archaeological Research [Google Scholar]
- Fregel R., Gomes V., Gusmão L., González A. M., Cabrera V. M., Amorim A., Larruga J. M.2009Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC Evol. Biol. 9, 181 (doi:10.1186/1471-2148-9-181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O.2003A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- Hall S. J. G.1979Haemoglobin polymorphism in the Bank vole, Clethrionomys glareolus, in Britain. J. Zool. 187, 153–160 (doi:10.1111/j.1469-7998.1979.tb03939.x) [Google Scholar]
- Hewitt G.2000The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (doi:10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- Hofreiter M., Münzel S., Conard N. J., Pollack J., Slatkin M., Weiss G., Pääbo S.2007Sudden replacement of cave bear mitochondrial DNA in the late Pleistocene. Curr. Biol. 17, R122–R123 (doi:10.1016/j.cub.2007.01.026) [DOI] [PubMed] [Google Scholar]
- Irwin D. M., Kocher T. D., Wilson A. C.1991Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32, 128–144 (doi:10.1007/BF02515385) [DOI] [PubMed] [Google Scholar]
- Jaarola M., Searle J. B.2002Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol. Ecol. 11, 2613–2621 (doi:10.1046/j.1365-294X.2002.01639.x) [DOI] [PubMed] [Google Scholar]
- Jaarola M., et al. 2004Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 33, 647–663 (doi:10.1016/j.ympev.2004.07.015) [DOI] [PubMed] [Google Scholar]
- Jacobi R. M., Higham T. F. G.2009The early Lateglacial re-colonization of Britain: new radiocarbon evidence from Gough's Cave, southwest England. Q. Sci. Rev. 28, 1895–1913 (doi:10.1016/j.quascirev.2009.03.006) [Google Scholar]
- Kotlík P., Deffontaine V., Mascheretti S., Zima J., Michaux J. R., Searle J. B.2006A northern glacial refugium for bank voles (Clethrionomys glareolus). Proc. Natl Acad. Sci. USA 103, 14 860–14 864 (doi:10.1073/pnas.0603237103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G., et al. 2007Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc. Natl Acad. Sci. USA 104, 4834–4839 (doi:10.1073/pnas.0607753104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan F. D.2005The Vikings in history, 3rd edn London, UK: Routledge [Google Scholar]
- Maddison D. R., Maddison W. P.2003MacClade: analysis of phylogeny and character evolution, v. 4.08 Sunderland, MA: Sinauer; [DOI] [PubMed] [Google Scholar]
- Martínková N., McDonald R. A., Searle J. B.2007Stoats (Mustela erminea) provide evidence of natural overland colonization of Ireland. Proc. R. Soc. B 274, 1387–1393 (doi:10.1098/rspb.2007.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti S., Rogatcheva M. B., Gündüz İ., Fredga K., Searle J. B.2003How did pygmy shrews colonize Ireland? Clues from a phylogenetic analysis of mitochondrial cytochrome b sequences. Proc. R. Soc. Lond. B 270, 1593–1599 (doi:10.1098/rspb.2003.2406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matisoo-Smith E., Robins J. H.2004Origins and dispersals of Pacific peoples: evidence from mtDNA phylogenies of the Pacific rat. Proc. Natl Acad. Sci. USA 101, 9167–9172 (doi:10.1073/pnas.0403120101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mix A. C., Bard E., Schneider R.2001Environmental processes of the ice age: land, oceans, glaciers (EPILOG). Q. Sci. Rev. 20, 627–657 (doi:10.1016/S0277-3791(00)00145-1) [Google Scholar]
- Oppenheimer S.2006The origins of the British: a genetic detective story London, UK: Constable and Robinson [Google Scholar]
- Pereira L., et al. 2005High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 15, 19–24 (doi:10.1101/gr.3182305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergams O. R. W., Barnes W. M., Nyberg D.2003Rapid change in mouse mitochondrial DNA. Nature 423, 397 (doi:10.1038/423397a) [DOI] [PubMed] [Google Scholar]
- Piertney S. B., Stewart W. A., Lambin X., Telfer S., Aars J., Dallas J. F.2005Phylogeographic structure and postglacial evolutionary history of water voles (Arvicola terrestris) in the United Kingdom. Mol. Ecol. 14, 1435–1444 (doi:10.1111/j.1365-294X.2005.02496.x) [DOI] [PubMed] [Google Scholar]
- Posada D.2003Using Modeltest and PAUP* to select a model of nucleotide substitution. In Current protocols in bioinformatics (eds Baxevanis A. D., Davison D. B., Page R. D. M., Petsko G. A., Stein L. D., Stormo G. D.), pp. 6.5.1–6.5.14 New York, NY: Wiley; [DOI] [PubMed] [Google Scholar]
- Posada D.2008jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- Price C. R.2003Late Pleistocene and Early Holocene small mammals in south west Britain: environmental and taphonomic implications and their role in archaeological research British Archaeological Reports 347 Oxford, UK: Archaeopress [Google Scholar]
- Searle J. B.1988Karyotypic variation and evolution in the common shrew, Sorex araneus. In Kew chromosome Conference III (ed. Brandham P. E.), pp. 97–107 London, UK: HMSO [Google Scholar]
- Searle J. B.2008The colonization of Ireland by mammals. In Mind the gap: Postglacial colonisation of Ireland Special supplement to The Irish Naturalists' Journal (eds Davenport J. L., Sleeman D. P., Woodman P. C.), pp. 109–115 [Google Scholar]
- Searle J. B., Wilkinson P. J.1987Karyotypic variation in the common shrew (Sorex araneus) in Britain—a ‘Celtic fringe’. Heredity 59, 345–351 (doi:10.1038/hdy.1987.141) [DOI] [PubMed] [Google Scholar]
- Searle J. B., et al. 2009Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proc. R. Soc. B 276, 201–207 (doi:10.1098/rspb.2008.0958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinney L.2008The lost world. Nature 454, 151–153 (doi:10.1038/454151a) [DOI] [PubMed] [Google Scholar]
- Stanton D. W. G., Hobbs G. I., Chadwick E. A., Slater F. M., Bruford M. W.2009Mitochondrial genetic diversity and structure of the European otter (Lutra lutra) in Britain. Conserv. Genet. 10, 733–737 (doi:10.1007/s10592-008-9633-y) [Google Scholar]
- Steffensen J. P., et al. 2008High-resolution Greenland ice core data show abrupt climate change happens in few years. Science 321, 680–684 (doi:10.1126/science.1157707) [DOI] [PubMed] [Google Scholar]
- Stewart J. R., Lister A. M.2001Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 16, 608–613 (doi:10.1016/S0169-5347(01)02338-2) [Google Scholar]
- Swenson N. G., Howard D. J.2005Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am. Nat. 166, 581–591 (doi:10.1086/491688) [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2003PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0 Sunderland, MA: Sinauer [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Tavaré S.1986Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17, 57–86 [Google Scholar]
- Weale M. E., Weiss D. A., Jager R. F., Bradman N., Thomas M. G.2002Y chromosome evidence for Anglo-Saxon mass migration. Mol. Biol. Evol. 19, 1008–1021 [DOI] [PubMed] [Google Scholar]
- Weninger B., et al. 2008The catastrophic final flooding of Doggerland by the Storegga Slide tsunami. Doc. Praehist. 35, 1–24 [Google Scholar]
- Yalden D. W.1982When did the mammal fauna of the British Isles arrive? Mamm. Rev. 12, 1–57 (doi:10.1111/j.1365-2907.1982.tb00007.x) [Google Scholar]
- Yalden D. W.1999The history of British mammals London, UK: Poyser [Google Scholar]