Abstract
The major histocompatibility complex (MHC) contains the most variable genes in vertebrates, but despite extensive research, the mechanisms maintaining this polymorphism are still unresolved. One hypothesis is that MHC polymorphism is a result of balancing selection operating by overdominance, but convincing evidence for overdominant selection in natural populations has been lacking. We present strong evidence consistent with MHC-specific overdominance in a free-living population of Arctic charr (Salvelinus alpinus) in northernmost Europe. In this population, where just two MHC alleles were observed, MHC heterozygous fish had a lower parasite load, were in better condition (as estimated by a fatness indicator) and had higher survival under stress than either of the homozygotes. Conversely, there was no consistent association between these fitness measures and assumedly neutral microsatellite variability, indicating an MHC-specific effect. Our results provide convincing empirical evidence consistent with the notion that overdominance can be an important evolutionary mechanism contributing to MHC polymorphism in wild animal populations. They also support a recent simulation study indicating that the number of alleles expected to be maintained at an MHC loci can be low, even under strong heterozygote advantage.
Keywords: major histocompatibility complex, overdominance, heterozygote advantage, parasite, condition, survival
1. Introduction
The extreme polymorphism of the major histocompatibility complex (MHC) genes has fascinated researchers for decades, but the mechanisms of how this extensive genetic variation is maintained are hotly debated (Klein 1986; Hughes & Nei 1988; Potts & Slev 1995; Apanius et al. 1997; Edwards & Hedrick 1998; Paterson et al. 1998; Wegner et al. 2003a). The primary role of the MHC is to encode molecules that recognize foreign proteins, present them to T-cells and initiate an immune response (Langefors et al. 2001; Piertney & Oliver 2006). The crucial function of MHC molecules in the adaptive immune response suggests that pathogens and parasites are the most important agents of selection for MHC polymorphism (Landry & Bernatchez 2001; Wegner et al. 2004; Froeschke & Sommer 2005; Harf & Sommer 2005; de Eyto et al. 2007; Turner et al. 2007).
Hypotheses proposed to maintain MHC polymorphism include the overdominance (heterozygote superiority) hypothesis which predicts that heterozygous individuals are fitter than individuals homozygous for an MHC allele because two different alleles will identify a broader range of peptides (Doherty & Zinkernagel 1975) compared with one. Although overdominance has been convincingly argued to contribute to the evolution of MHC polymorphism (Doherty & Zinkernagel 1975; Hughes & Nei 1989, 1992; Takahata & Nei 1990), more recent studies suggest that the ability of overdominance to create and maintain MHC polymorphism may be more limited (Hedrick 1999, 2002; Borghans et al. 2004; De Boer et al. 2004; Stoffels & Spencer 2008). Considerable empirical support has been presented for the importance of MHC variability in pathogen and parasite resistance from studies on humans, or on other vertebrates under experimental conditions (Jeffery & Bangham 2000; Mitton 2002; Bernatchez & Landry 2003; Wegner et al. 2003b, 2004; Sommer 2005). However, only a few studies have found experimental evidence for overdominance (Penn 2002; McClelland et al. 2003), and unequivocal support for the overdominance hypothesis has been completely lacking in wild animal populations.
In the present study, we investigated the relationship between MHC diversity and fitness in a natural salmonid population. Several studies have reported fitness differences between MHC heterozygotes and homozygotes when comparing all different heterozygous or homozygous genotypes in two groups (population heterozygote advantage (Sommer 2005). However, such a group comparison approach does not enable testing of the original allele-specific hypothesis, whereby heterozygotes are expected to be superior to each homozygote class (Doherty & Zinkernagel 1975) and, therefore, cannot be considered as support for overdominance. In fact, population heterozygote advantage can sometimes occur even when the fitness of heterozygotes is lower than that of either corresponding homozygote (Lipsitch et al. 2003). In addition, only a handful of studies where a population heterozygote advantage has been observed have combined estimates of MHC diversity with estimates of genome-wide diversity by also assessing assumedly neutral markers (Wegner et al. 2003a; Turner et al. 2007). Therefore, possible genome-wide effects often cannot be ruled out.
We investigated genetic variability of the MHC class IIβ peptide-binding region (PBR) in a wild Arctic charr (Salvelinus alpinus) population (Lake Peltojärvi, northernmost Finland). Furthermore, we investigated 11 assumedly neutral microsatellite loci and compared the parasite load, condition (fatness) and survival during fishing stress between different MHC and microsatellite genotypes. Our results provide strong empirical evidence consistent with the notion that overdominance can be an important evolutionary mechanism contributing to MHC polymorphism in wild animal populations.
2. Material and methods
(a). Data collection, measurements and parasitological examination
Arctic charr (S. alpinus) individuals were gillnetted in April and September 2006 from Lake Peltojärvi in northern Finland (69°7′ N, 26°34′ E), which contains a presumably indigenous Arctic charr population (Finnish Game and Fisheries Research Institute, broodstock registry). Gillnets were examined once a day (approximately at the same time every day) and fish were frozen and transported to the laboratory for later analyses. During the gillnetting periods, the total numbers of dead and alive fish were counted. In the laboratory, fish were measured, weighed, sexed, their stomach contents analysed and the adipose fins placed in 95 per cent ethanol for genetic analyses. In addition, Diphyllobothrium cysts from the body cavity and muscles were counted and thickness of the belly flap was measured for charr over 300 mm using a micrometer (Mitutoyo Corporation) to 0.01 mm accuracy from four standard locations (two measurements per fillet). Fish less than 300 mm in length (n = 10) were excluded from belly flap measurements, as the size of the fillets was too small for the instrument used for thickness to be measured accurately. Fitness measurements and genetic analyses (see below) were conducted blind, i.e. by separate researchers, without one knowing the results of the other.
(b). DNA extraction, major histocompatibility complex primer design and polymerase chain reaction
DNA was extracted from tissue samples as described in Vähä et al. (2007). Arctic charr MHC II β sequences were initially characterized using Atlantic salmon designed primers MG7 and ÅL 1002 (Langefors et al. 2000). Because cross-species amplification of Arctic charr MHC with salmon primers proved to be difficult in some cases, Arctic charr sequence-specific primers MHC-int-F and MHC-int-R were designed (electronic supplementary material, table S1). They amplified a 212 bp long fragment of the MHC IIβ PBR. The primers were designed with the program Primer3 (Rozen & Skaletsky 2000), using full-length second exon sequences (see below). The MHC-int-F-primer had a 40 bp ‘CG clamp’ added to the 3′-end to prevent total denaturation of samples (Sheffield et al. 1989) in denaturing gradient gel electrophoresis (DGGE) (Myers et al. 1987). The 25 µl PCR mixture contained approximately 100 ng of DNA, 0.75 U of BioTaq polymerase (Bioline), 12.5 pmol of each primer, 1× PCR reaction buffer, 60 pmol of MgCl2 and 6.25 pmol of dNTPs. The PCR program consisted of an initial 2 min denaturation at 95°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 56°C and extension for 1 min at 72°C. The final extension was for 10 min at 72°C.
(c). Denaturing gradient gel electrophoresis
DGGE was performed using 9.5 µl of the PCR product in 9 per cent 19 : 1 acrylamide : bisacrylamide gels containing 1× TAE buffer and a gradient of urea and formamide (Hayes et al. 1999). The gels were cast with a gravitational gradient maker and the DGGE was performed in a DGGE system buffer tank (C.B.S. Scientific Inc.) filled with 0.5× TAE and heated to 60°C. Separation of different alleles was found to be optimal with a 25–55% denaturant gradient and with an electrophoresis running time of approximately 900 Vh. The gels were stained with ethidium bromide for 10 min after the electrophoresis and stained DNA was visualized with a BioRad ChemiDoc XRS imaging system, after which the individuals were genotyped by eye.
(d). Cloning and sequencing
The genotypes of 14 individuals from the Peltojärvi population were confirmed by sequencing directly from both DGGE PCR products and full-length PBR fragments cloned to pGEM-T plasmids (Promega Corporation) according to manufacturers' instructions. The MHC-int-F primer without the CG clamp was used for direct sequencing. The full-length PBRs were amplified with the same PCR program as described above, using salmon-specific primers MG7 and ÅL 1002 (Langefors et al. 2000). Up to five clones were sequenced from each individual with an Applied Biosystems 3130xl Genetic Analyzer following the recommendations of the manufacturers, using vector-specific primers M13F and M13R. The sequences were aligned and analysed using Vector NTI Advance 10 (Invitrogen). In all 62 cases, the sequence of the allele(s) matched the genotypes obtained by DGGE. Sequences were deposited in GenBank (accession numbers GQ165716 and GQ165717).
(e). Microsatellite genotyping and data analyses
The level of neutral microsatellite variation in the studied fish was ascertained using 11 microsatellite markers (electronic supplementary material, table S1). The PCR was carried out in two 10 µl multiplexes using QIAGEN Multiplex PCR Kit according to the manufacturer's instructions with the annealing temperature of the recommended PCR program set to 56°C. The forward primer for each locus was end labelled with a fluorescent dye and a GTTT tail was added to each reverse primer to ensure addition of an A-overhang to each PCR fragment by Taq polymerase (Brownstein et al. 1996). Microsatellites were genotyped using the same methods as described in Vähä et al. (2007). The observed and expected heterozygosities were calculated using Microsatellite Toolkit (Park 2001), allelic richness and the inbreeding coefficient, FIS, were calculated using FSTAT (Goudet 1995) and the possible deviation from Hardy–Weinberg equilibrium was estimated using GenePop (Raymond & Rousset 1995). Severe reductions in the effective size of the populations (i.e. bottlenecks) were tested using the program Bottleneck (Piry et al. 1999) Statistical significance was tested with three tests: the sign test, the one-way Wilcoxon sign-rank test and the ‘mode-shift’ test. For two-phase mutation model tests, 30 per cent non-stepwise mutations were allowed. A measure of individual inbreeding level, internal relatedness (IR), was calculated according to Amos et al. (2001).
(f). Statistical analyses
The effect of MHC genotype and season (fixed factors) on Diphyllobothrium parasite numbers, belly flap thickness, number of sticklebacks in stomachs, mean observed microsatellite heterozygosity (HOBS) and IR were studied with ANOVA or ANCOVA, with fish standard length as a covariate (table 1; electronic supplementary material, table S2). The effect of microsatellite genotype (homozygous versus heterozygous) on parasite numbers and belly flap thickness was tested by ANCOVA (electronic supplementary material, table S3). ANCOVA was used also to study the effect of survival status (fixed factor) on belly flap thickness, Diphyllobothrium numbers (table 1) and HOBS. As the seasonal differences in belly flap thickness and Diphyllobothrium numbers were not significant, the spring and autumn samples were combined in ANCOVAs studying the effect of survival status. It was verified that none of the interactions between the covariate and any of the factors was significant (equal slopes assumption). Normality was assessed using Shapiro-Wilk statistics and when needed, natural-logarithm transformation was applied to the dependent variable to satisfy the requirement of normal distribution. Tukey's least significant difference post hoc tests were used to study between-group differences in ANCOVAs. Associations between Diphyllobothrium numbers and belly flap thickness and between HOBS and Diphyllobothrium abundance or belly flap thickness were studied with partial correlations, fish standard length being a controlling variable. All presented p-values are from two-tailed tests with α = 0.05.
Table 1.
ANCOVA and ANOVA statistics assessing the effect of MHC genotype and season (spring versus autumn) on two fitness measures (1 and 2) and number of sticklebacks in stomach (3), as well as an assessment of associations between survival (dead versus alive) and the two fitness measures (4 and 5).
| parameter | analysis | details of the test | n | F | p |
|---|---|---|---|---|---|
| 1. number of Diphyllobothrium versus genotype | ANCOVA | genotypea | 50 | 4.522 | 0.017 |
| seasona | 0.506 | 0.481 | |||
| genotype×season | 0.662 | 0.521 | |||
| standard lengthb | 40.576 | <0.001 | |||
| 2. belly flap thickness versus genotype | ANCOVA | genotypea | 37 | 5.176 | 0.012 |
| seasona | 2.238 | 0.145 | |||
| genotype×season | 0.719 | 0.495 | |||
| standard lengthb | 4.381 | 0.045 | |||
| 3. number of sticklebacks in stomach | ANOVA | genotypea | 50 | 0.067 | 0.935 |
| seasona | 28.339 | <0.001 | |||
| genotype×season | 0.509 | 0.605 | |||
| 4. belly flap thickness versus survival | ANCOVA | survivala | 37 | 6.410 | 0.016 |
| standard lengthb | 8.413 | 0.006 | |||
| 5. number of Diphyllobothrium versus survival | ANCOVA | survivala | 50 | 0.825 | 0.368 |
| standard lengthb | 31.951 | <0.001 |
aFixed factor.
bCovariate.
To test if the proportion of survived fish among MHC heterozygotes differed from both MHC homozygotes, logistic regression with simple contrast was used with the forward stepwise method based on the log-likelihood ratio so that the spring and autumn samples were combined, and the genotype and fish standard length were used as explanatory variables (electronic supplementary material, table S4). Possible survival differences between microsatellite heterozygotes and homozygotes in seven microsatellite loci were investigated in a similar manner (each of the seven polymorphic loci analysed separately), but without contrasting (electronic supplementary material, table S5).
3. Results
(a). Genetic variation of Arctic charr in study lakes
The level of genetic variability in Lake Peltojärvi was low: only two MHC IIβ alleles (Saal-DAB*001 and Saal-DAB*002) were identified in the 50 individuals assessed and the mean number of alleles in 11 microsatellite loci known to be polymorphic in some north European Arctic charr populations was 2.6 (table 2; electronic supplementary material, table S6). The sequence divergence between the two MHC PBR alleles was 29 nucleotides (9%) and 16 amino acids (13%) with a dN/dS ratio of 3.17 (electronic supplementary material, figure S1). To assess the uniqueness of the low MHC variability found in Lake Peltojärvi, we also genotyped Arctic charr from three Norwegian lakes (26–35 individuals per population) and also observed relatively low levels of variation; 3–9 MHC alleles and an average of 2.5–6.2 microsatellite alleles per locus (table 2; electronic supplementary material, table S6). There was no evidence for recent population bottlenecks in Lake Peltojärvi (all tests p > 0.24). In addition, there were no indications of inbreeding (as estimated by FIS, electronic supplementary material, table S6) in any population. Also neither the mean observed microsatellite heterozygosity (HOBS) nor IR differed between the MHC genotypes in Lake Peltojärvi charr (ANCOVA, F1,49 = 0.070, p = 0.932 and F1,49 = 0.031, p = 0.969, respectively, electronic supplementary material, table S2).
Table 2.
Diversity indices for the MHC locus and microsatellites (averaged over 11 loci) in Arctic charr populations of Lake Peltojärvi and three presumably unstocked small Norwegian lakes from similar latitudes (Primmer et al. 1999). See electronic supplementary material, table S6 for more detailed information. no. A, number of observed alleles; Ar, allelic richness; Ho, observed heterozygosity.
| Peltojärvi 69°07′ N, 26°34′ E |
Haukejavri 69°57′ N, 29°14′ E |
Lisma 70°08′ N, 28°04′ E |
Bluevatnet 70°37′ N, 30°05′ E |
mean |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| no. A | Ar | Ho | no. A | Ar | Ho | no. A | Ar | Ho | no. A | Ar | Ho | total no. A | A | Ar | Ho | |
| MHC IIβ | 2 | 2.0 | 0.38 | 3 | 3.0 | 0.38 | 6 | 5.9 | 0.56 | 9 | 9.0 | 0.52 | 16 | 5 | 5.0 | 0.46 |
| microsatellites | 2.6 | 2.5 | 0.25 | 2.5 | 2.8 | 0.23 | 3.5 | 3.4 | 0.36 | 6.2 | 6.4 | 0.53 | 8.6 | 3.92 | 6.0 | 0.34 |
(b). Diphyllobothrium abundance
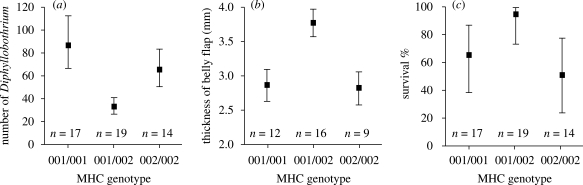
There were up to 1500 Diphyllobothrium (Cestoda) plerocercoid cysts, mainly 4–8 mm in diameter each, encapsulated by the host in the body cavity and muscles of the charr. We opened and examined approximately five Diphyllobothrium cysts from most of the fish finding plerocercoids resembling Diphyllobothrium dendriticum, but we cannot exclude the possibility that Diphyllobothrium ditremum also occurred in the charr. Diphyllobothrium plerocercoids were clearly the most abundant parasite in the charr population. Three other parasite taxa were also detected—Eubothrium salvelini (Cestoda) in the intestine, Diplostomum sp. (Trematoda) in the eyes and Acanthobdella peledina (Hirudinae) on the skin—but the materials and numbers of infected individuals were insufficient for statistical analyses for those parasites. We found a significant difference in the mean number of Diphyllobothrium cysts between the three MHC genotypes (ANCOVA, F1,49 = 4.522, p = 0.017, table 1). Post hoc tests indicated that parasite load was lower in MHC heterozygotes than in either of the homozygotes (p = 0.007 compared with Saal-DAB*001 and p = 0.043 for Saal-DAB*002), but there was no difference between the homozygotes (p = 0.439) (figure 1a). The effect of sampling period (April versus September) was not significant, but parasite load increased with the covariate, fish standard length (table 1). However, there was no difference in the mean length of individuals between different MHC genotypes (electronic supplementary material, table S2 and supplementary data).
Figure 1.
(a) Size-adjusted mean (±s.e.) number of Diphyllobothrium plerocercoid cysts, (b) mean (±s.e.) belly flap thickness and (c) survival during gillnetting (proportion of survived individuals ±95 per cent CI) in Arctic charr with different MHC genotypes.
The number of digested three-spined sticklebacks (Gasterosteus aculeatus) in the stomachs of Arctic charr did not differ between MHC genotypes (ANOVA, F1,49 = 0.067, p = 0.935) (table 1). Sticklebacks are the most abundant prey and presumably the main vector of Diphyllobothrium in those large size classes included in the present study (210–595 mm) (electronic supplementary material, supplementary data). These observations suggest that the lower Diphyllobothrium abundance in MHC heterozygotes is due to higher parasite resistance rather than lower exposure among the heterozygous fish.
(c). Thickness of the belly flap and survival
Thickness of the belly flap differed between the MHC genotypes (ANCOVA, F1,36 = 5.176, p = 0.012, table 1), with heterozygotes having a thicker belly flap than both Saal-DAB*001 (p = 0.011) and Saal-DAB*002 homozygotes (p = 0.012), but the belly flap thickness of homozygotes did not differ between each other (p = 0.940) (figure 1b). In the data combined over spring and autumn samples, to test if survival during gillnetting was higher in MHC heterozygotes than in either of the homozygotes the final logistic regression model included the variable ‘genotype’ but not ‘fish standard length’ (forward stepwise method based on likelihood ratio, −2 log likelihood = 49.318, Nagelkerke R2 = 0.260). Thus, the result of logistic regression analysis indicated that survival of fish during gillnetting stress was significantly higher among MHC heterozygotes (95%, n = 19) than in either of the homozygotes (65%, n = 17, and 50%, n = 14, for Saal-DAB*001 and Saal-DAB*002, respectively) (figure 1c; electronic supplementary material, table S4). Overall, these findings indicate higher fitness of MHC heterozygote individuals (see also Penn et al. 2002).
(d). Associations between fitness measures
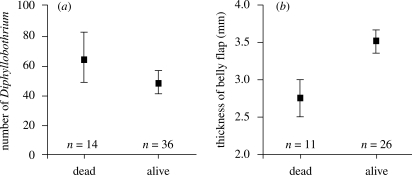
There was a statistically significant negative correlation between the number of Diphyllobothrium and the thickness of the belly flap (partial correlation, with the effect of fish standard length partialled out, r = −0.430, n = 37, p = 0.009). In addition, the mean (±s.e.) thickness of the belly flap was lower among charr that died during the gillnetting (2.82 ± 0.15 mm) than in those that survived (3.50 ± 0.20 mm) (ANCOVA, F1,36 = 6.410, p = 0.016, table 1, figure 2). Although the dead and alive fish did not differ significantly in their numbers of Diphyllobothrium cysts (ANCOVA, F1,49 = 0.825, p = 0.368), the trend was in the expected direction (mean ± s.e.; 201 ± 103 and 90 ± 24, respectively).
Figure 2.
(a) Size-adjusted mean (±s.e.) number of Diphyllobothrium plerocercoid cysts and (b) mean (±s.e.) belly flap thickness in dead and survived Arctic charr.
(e). Genome-wide effects
There was no dependence between locus-specific heterozygosity at any of the seven polymorphic microsatellites and belly flap thickness (ANCOVAs, p > 0.050, in all cases, electronic supplementary material, table S3) or survival during gillnetting (logistic regression analyses, electronic supplementary material, table S5). Furthermore, there was no dependence between HOBS and Diphyllobothrium abundance or belly flap thickness (partial correlations, fish standard length partialled out, r = −0.063, −0.080, n = 49 and 37 and p = 0.671 and 0.641, respectively) or between HOBS and survival (ANCOVA, F1,49 = 1.406, p = 0.242). Abundance of Diphyllobothrium cysts was significantly associated with just one microsatellite locus (ANCOVA, F1,48 = 5.687, p = 0.007, electronic supplementary material, table S3). However, post hoc tests revealed that the parasite number of heterozygotes was only lower than the average of the homozygotes, but not lower than both (p = 0.002 and 0.092), i.e. overdominance was not detected. In addition, this rate of significance at assumedly neutral microsatellite loci (one significant association out of 21 tests conducted) is in line with the rate expected owing to the type I error, a conclusion which is also supported by the fact that this locus was associated with only one of the three fitness measures. Thus, our results indicate an MHC-specific effect, rather than a genome-wide heterozygote advantage.
4. Discussion
Our results indicate clear signals of MHC-specific overdominance with respect to three different parameters: MHC heterozygous charr had significantly lower Diphyllobothrium plerocercoid numbers, thicker belly flaps and higher survival during gillnetting than either of the homozygotes, which together indicate higher fitness of MHC heterozygote individuals. According to the overdominance theory, heterozygotes are expected to have a selective advantage compared with homozygotes, because two different alleles will identify a broader range of pathogens and parasites than one (Doherty & Zinkernagel 1975).
Energy reserves often play an important role in determining fitness. Lipid stores are the primary energy reserves of fish preparing to reproduce (Rowe & Thorpe 1990) and the size of these stores may determine the survival probability during unfavourable feeding conditions, which are not uncommon in Arctic conditions. In salmonids, the major and most readily mobilized fat reserve is in the belly flap (Nanton et al. 2007). Thus, the size of this lipid store can be regarded as an indicator of the overall condition of the fish. The negative associations between the measured fitness parameters (§3) suggest that the impact of Diphyllobothrium parasites on individual fitness may be high. This is not surprising given (i) the high observed numbers of these relatively large parasites in the charr population; (ii) the fact that they are located in the muscles and body cavity, and migrate to their final site of infection through the tissues of the host; and (iii) they have been shown to cause severe pathological tissue damage and mortality in salmonid fishes (Rahkonen et al. 1996).
We did not find any evidence that our results could be somehow related to inbreeding or population bottlenecks in the Lake Peltojärvi charr population, although the genetic variability in both MHC and microsatellite loci was low. Low levels of variation are not unusual for northern Arctic charr populations (table 2; electronic supplementary material, table S6) as has also been demonstrated for MHC in a North American Arctic charr population (Conejeros et al. 2008). Arctic charr juveniles are able to discriminate siblings and non-siblings with respect to both MHC II-linked odours and odours of some other genes when fish cannot rely on information from MHC class II alleles (Olsén et al. 2002; see also Amos & Balmford 2001; Hoffman et al. 2007). This suggests that Arctic charr are capable of avoiding mating with close relatives, which could reduce the probability of inbreeding also in Lake Peltojärvi. Although heterozygote deficiencies were observed at the MHC locus in some of the other Nordic charr populations, this is likely to be due to technical difficulties in visualizing all heterozygotes using the DGGE method (Conejeros et al. 2008), as the DGGE conditions were optimized primarily for the Lake Peltojärvi population and estimates for other lakes are provided primarily to indicate levels of genetic diversity.
A closer examination of genotype frequencies in the spring and autumn samples provides preliminary evidence that the selective regime in Lake Peltojärvi may be more complex than simple parasite-driven overdominance, as has been suggested in other studies (Froeschke & Sommer 2005). More specifically, there was in fact a strong trend towards a decrease in heterozygotes and an increase in Saal-DAB*001 homozygotes in the autumn sample, even though the mortality of homozygotes was substantial (figure 1c). If the mortality differences between homozygotes and heterozygotes are used to estimate a selection coefficient to predict expected genotype frequencies in the autumn (second sampling), this trend is in fact significantly different from the expected genotype frequencies (data not shown).
Although a longer time series is required before any firm conclusions can be drawn, possible explanations include: seasonal variation in selection that would result in selective mortality during one time period being a poor predictor of selection coefficients during other time periods. Second, other non-detected parasites against which Saal-DAB*001 homozygous fish were most resistant could explain an increase in Saal-DAB*001 homozygotes between spring and autumn. Third, the observed mortality rates during gillnetting may not accurately describe the actual selective advantage of MHC heterozygotes. In this case, the higher mortality of homozygotes may simply reflect their lower survival probability (e.g. higher oxygen demand of highly parasitized fish, see e.g. Booth et al. 1993) when caught in gillnets, but not necessarily their higher mortality in more natural conditions. Fourth, if MHC heterozygotes, i.e. fish with lower parasite loads, were better able to avoid capture in gillnets or have better ability to escape from the nets, then the gillnet sample would underestimate the true frequency of heterozygotes. Finally, it is also feasible that the genotype deviations may be simply due to the pooling of samples from different time periods and/or locations, i.e. a possible Wahlund effect. This is further supported by the observation that allele frequencies and also heterozygosity levels at a number of microsatellite loci differed by a similar amount or even more between seasons than did MHC frequencies (data available from CRP on request). Continued monitoring of the population over a longer period to create an extended time series may be of use to distinguish between these possible explanations in the future.
Overdominance is usually assumed to emerge only after simultaneous coinfections when many allele-specific susceptibilities to pathogens will be masked by the resistant allele in heterozygotes (McClelland et al. 2003). Thus, in dual infections different alleles should have opposite susceptibility profiles (the dual-infection heterozygote superiority, DIHS hypothesis). The present results indicate overdominance with respect to a single parasite taxon, Diphyllobothrium, which could be due to the considerable genetic variation observed within local populations of Diphyllobothrium (deVos et al. 1990). In helminths, sexual reproduction creates and maintains high antigenic diversity and may rapidly generate strain diversity, which permits evasion of host immunity (Galvani 2003; Galvani et al. 2001, 2003). Thus, different parasite genotypes may have different surface antigens, giving possibly a selective advantage to MHC heterozygotes in recognizing a broader range of intra-specific parasite genotypes. Therefore, even if there was only one Diphyllobothrium species in Lake Peltojärvi charr, our finding does not necessarily contradict the DIHS hypothesis because it is possible that different MHC alleles may have different levels of recognition degeneracy (i.e. overlap) for a particular pathogen, or pathogen strains within the same species, and hence, overdominance is feasible based on the effects of a single species (Neff et al. 2008; Stoffels & Spencer 2008). The higher variation in homozygotes, compared with heterozygotes, for parasite load, belly flap thickness and survival under stress (figure 1) is a further indication that there may be allele-specific variation in fitness (i.e. susceptibility profiles) against different parasite genotypes, although the overall fitness of homozygote genotypes would remain the same (Stoffels & Spencer 2008).
We have demonstrated signals clearly consistent with MHC-specific overdominance in a free-living animal population. Interestingly, Oliver et al. (2009) recently demonstrated that in a water vole (Arvicola terrestris) population with a low level of MHC variation, MHC heterozygote individuals were infected by fewer parasite types than homozygotes. Therefore, this is the second study within a short period of time presenting empirical evidence supporting the overdominance hypothesis in a population with low MHC polymorphism. But why should such a signal be observed in this population when numerous other studies have failed to detect such an association? A recent simulation study (Stoffels & Spencer 2008) provides a possible answer to this question. Namely, by characterizing the function of MHC molecules by the sets of parasites they recognize (recognition sets) as well as considering the effects of effective population size and genetic drift, Stoffels & Spencer were able to demonstrate that for populations with Ne = 1000 (the smallest Ne considered), the number of alleles expected to be maintained at an MHC locus can be less than five, even with a relatively strong heterozygote advantage.
The lower level of MHC diversity observed in this northernmost Arctic charr population, which appears to be a feature of salmonid populations in northern regions (Dionne et al. 2007), falls within the range of parameters suggested by Stoffels & Spencer (2008) where a strong effect of overdominance can be expected. Similarly, Lewontin et al. (1978) demonstrated that the proportion of stable heterosis equilibria is highest when the number of alleles in a population is just two. In addition, the lower levels of variability may simplify analyses and contribute to the ability to observe such an allele-specific association with moderate sample sizes (Apanius et al. 1997; Oliver et al. 2009). Thus, our results indicate that overdominance can be an important evolutionary mechanism contributing to MHC diversity in wild populations. However, as a two-allele system only enables the investigation of one class of heterozygotes, future analyses, for example, including the investigation of additional populations in the region would be useful to determine the generality of our findings and for further investigating how MHC–pathogen interactions may affect the occurrence of overdominance.
Acknowledgements
We thank Erica Leder, Frode Skarstein and J-P Vähä for advice regarding laboratory procedures, Kirsti Kyyrönen for graphical assistance and Dustin Penn, Petteri Ilmonen and two anonymous reviewers for comments on earlier versions of the manuscript. This study was financially supported by Kone Foundation, The Finnish Academy (grant 7121694) and Department of Physics and Mathematics (University of Joensuu) (to J.K.) and Center of Excellence in Evolutionary Genetics and Physiology, The Population Genetics Graduate School and Nordic Working Group on Fisheries Research (to C.R.P. and J.A.V.).
References
- Amos W., Balmford A.2001When does conservation genetics matter? Heredity 87, 257–265 (doi:10.1046/j.1365-2540.2001.00940.x) [DOI] [PubMed] [Google Scholar]
- Amos W., Worthington-Wilmer J., Fullard K., Burg T. M., Croxall J. P., Bloch D., Coulson T.2001The influence of parental relatedness on reproductive success. Proc. R. Soc. Lond. B 268, 2021–2027 (doi:10.1098/rspb.2001.1751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanius V., Penn D., Slev P. R., Ruff L. R., Potts W. K.1997The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 17, 79–224 [DOI] [PubMed] [Google Scholar]
- Bernatchez L., Landry C.2003MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 (doi:10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- Booth D. T., Clayton D. H., Block B. A.1993Experimental demonstration of the energetic cost of parasitism in free-ranging hosts. Proc. R. Soc. Lond. B 253, 125–129 (doi:10.1098/rspb.1993.0091) [Google Scholar]
- Borghans J. A. M., Beltman J. B., DeBoer R. J.2004MHC polymorphism under host–pathogen coevolution. Immunogenetics 55, 732–739 (doi:10.1007/s00251-003-0630-5) [DOI] [PubMed] [Google Scholar]
- Brownstein M. J., Carpten J. D., Smith J. R.1996Modulation of non-templated nucleotide addition by taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20, 1004–1010 [DOI] [PubMed] [Google Scholar]
- Conejeros P., Phan A., Power M., Alekseyev S., O'Connell M., Dempson B., Dixon D.2008MH class IIα polymorphism in local and global adaptation of Arctic charr (Salvelinus alpinus L.). Immunogenetics 60, 325–337 (doi:10.1007/s00251-008-0290-6) [DOI] [PubMed] [Google Scholar]
- De Boer R. J., Borghans J. A. M., van Boven M., Keşmir C., Weissing F. J.2004Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics 55, 725–731 (doi:10.1007/s00251-003-0629-y) [DOI] [PubMed] [Google Scholar]
- de Eyto E., et al. 2007Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc. R. Soc. B 274, 861–869 (doi:10.1098/rspb.2006.0053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- deVos T., Szalai A. J., Dick T. A.1990Genetic and morphological variability in a population of Diphyllobothrium dendriticum. Syst. Parasitol. 16, 99–105 (doi:10.1007/BF00009609) [Google Scholar]
- Dionne M., Miller K. M., Dodson J. J., Caron F., Bernatchez L.2007Clinal variation in MHC diversity with temperature: evidence for the role of host–pathogen interaction on local adaptation in Atlantic salmon. Evolution 61, 2154–2164 (doi:10.1111/j.1558-5646.2007.00178.x) [DOI] [PubMed] [Google Scholar]
- Doherty P. C., Zinkernagel R. M.1975Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256, 50–52 (doi:10.1038/256050a0) [DOI] [PubMed] [Google Scholar]
- Edwards S. W., Hedrick P. W.1998Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Evol. Ecol. 13, 305–311 (doi:10.1016/S0169-5347(98)01416-5) [DOI] [PubMed] [Google Scholar]
- Froeschke G., Sommer S.2005MHC Class II DRB variability and parasite load in the striped mouse, Rhabdomys pumilio, in the Southern Kalahari. Mol. Biol. Evol. 22, 1254–1259 (doi:10.1093/molbev/msi112) [DOI] [PubMed] [Google Scholar]
- Galvani A. P.2003Epidemiology meets evolutionary ecology. Trends Evol. Ecol. 18, 132–139 (doi:10.1016/S0169-5347(02)00050-2) [Google Scholar]
- Galvani A. P., Coleman R. M., Ferguson N. M.2001Antigenic diversity and the selective value of sex in parasites. Ann. Zool. Fennici 38, 305–314 [Google Scholar]
- Galvani A. P., Coleman R. M., Ferguson N. M.2003The maintenance of sex in parasites. Proc. R. Soc. Lond. B 270, 19–28 (doi:10.1098/rspb.2002.2182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J.1995FSTAT (Version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486 [Google Scholar]
- Harf R., Sommer S.2005Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the Southern Kalahari. Mol. Ecol. 14, 85–91 (doi:0.1111/j.1365-294X.2004.02402.x) [DOI] [PubMed] [Google Scholar]
- Hayes V. M., Wu Y., Osinga J., Mulder I. M., van der Vlies P., Elfferich P., Buys C. H., Hofstra R. M.1999Improvements in gel composition and electrophoretic conditions for broad-range mutation analysis by denaturing gradient gel electrophoresis. Nucleic Acids Res. 27, e29 (doi:10.1093/nar/27.20.e29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W.1999Balancing selection and MHC. Genetica 104, 207–214 (doi:10.1023/A:1026494212540) [DOI] [PubMed] [Google Scholar]
- Hedrick P. W.2002Pathogen resistance and genetic variation at MHC loci. Evolution 56, 1902–1908 (doi:10.1111/j.0014-3820.2002.tb00116.x) [DOI] [PubMed] [Google Scholar]
- Hoffman J. I., Forcada J., Tratham P. N., Amos W.2007Female fur seals show active choice for males that are heterozygous and unrelated. Nature 445, 912–913 (doi:10.1038/nature05558) [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M.1988Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170 (doi:10.1038/335167a0) [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M.1989Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl Acad. Sci. USA 86, 958–962 (doi:10.1073/pnas.86.3.958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M.1992Models of host–parasite interaction and MHC polymorphism. Genetics 132, 863–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery K. J., Bangham C. R.2000Do infectious diseases drive MHC diversity? Microb. Infect. 2, 1335–1341 (doi:10.1016/S1286-4579(00)01287-9) [DOI] [PubMed] [Google Scholar]
- Klein J.1986. In Natural history of the major histocompatibility complex New York, NY: Wiley [Google Scholar]
- Landry C., Bernatchez L.2001Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar). Mol. Ecol. 10, 2525–2539 (doi:10.0000/096132198369887) [DOI] [PubMed] [Google Scholar]
- Langefors A., Lohm J., Von Schantz T., Grahn M.2000Screening of MHC variation in Atlantic salmon (Salmo salar): a comparison of restriction fragment length polymorphism (RFLP), denaturing gradient gel electrophoresis (DGGE) and sequencing. Mol. Ecol. 9, 215–219 (doi:10.1046/j.1365-294x.2000.00838.x) [DOI] [PubMed] [Google Scholar]
- Langefors Å., Lohm J., Grahn M., Andersen Ø., von Schantz T.2001Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. Lond. B 268, 479–485 (doi:10.1098/rspb.2000.1378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Ginzburg L. R., Tuljapurkar S. D.1978Heterosis as an explanation for large amounts of genic polymorphism. Genetics 88, 149–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Bergstrom C. T., Antia R.2003Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med. Genet. 4, 2 (doi:10.1186/1471-2350-4-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland E. E., Penn D. J., Potts W. K.2003Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 71, 2079–2086 (doi:10.1128/IAI.71.4.2079-2086.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton J. B.2002Heterozygous advantage. In Encyclopedia of life sciences Chichester, UK: John Wiley & Sons; See http://www.els.net/(doi:10.1038/npg.els.0001760) [Google Scholar]
- Myers R. M., Maniatis T., Lerman L. S.1987Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 155, 501–527 (doi:10.1016/0076-6879(87)55033-9) [DOI] [PubMed] [Google Scholar]
- Nanton D. A., Vegusdal A., Bencze Rørå A. M., Ruyter B., Baeverfjord G., Torstensen B. E.2007Muscle lipid storage pattern, composition, and adipocyte distribution in different parts of Atlantic salmon (Salmo salar) fed fish oil and vegetable oil. Aquaculture 265, 230–243 (doi:10.1016/j.aquaculture.2006.03.053) [Google Scholar]
- Neff B. D., Garner S. R., Heath J. W., Heath D. D.2008The MHC and non-random mating in a captive population of Chinook salmon. Heredity 101, 175–185 (doi:10.1038/hdy.2008.43) [DOI] [PubMed] [Google Scholar]
- Oliver M. K., Telfer S., Piertney S. B.2009Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. R. Soc. B 276, 1119–1128 (doi:10.1098/rspb.2008.1525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsén K. H., Grahn M., Lohm J.2002Influence of MHC on sibling discrimination in Arctic char, Salvelinus alpinus (L.). J. Chem. Ecol. 28, 783–795 (doi:10.1023/A:1015240810676) [DOI] [PubMed] [Google Scholar]
- Park S. D. E.2001Trypanotolerance in West African cattle and the population genetic effects of selection. PhD thesis, University of Dublin, Ireland [Google Scholar]
- Paterson S., Wilson K., Pemberton J. M.1998Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large undamaged ungulate population (Ovis aries L.). Proc. Natl Acad. Sci. USA 95, 3714–3719 (doi:10.1073/pnas.95.7.3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D. J.2002The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology 108, 1–21 (doi:10.1046/j.1439-0310.2002.00768.x) [Google Scholar]
- Penn D. J., Damjanovich K., Potts W. K.2002MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11260–11264 (doi:10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney S. B., Oliver M. K.2006The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 (doi:10.1038/sj.hdy.6800724) [DOI] [PubMed] [Google Scholar]
- Piry S., Luikart G., Cornuet J. M.1999Bottleneck: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 90, 502–503 (doi:10.1093/jhered/90.4.502) [Google Scholar]
- Potts W. K., Slev P. R.1995Pathogen based models favoring MHC genetic diversity. Immunol. Rev. 143, 181–197 (doi:10.1111/j.1600-065X.1995.tb00675.x) [DOI] [PubMed] [Google Scholar]
- Primmer C. R., Aho T., Piironen J., Estoup A., Cornuet J.-M., Ranta E.1999Microsatellite analysis of hatchery stocks and natural populations of arctic charr, Salvelinus alpinus, from the nordic region: implications for conservation. Hereditas 130, 277–289 (doi:10.1111/j.1601-5223.1999.00277.x) [Google Scholar]
- Rahkonen R., Aalto J., Koski P., Särkkä J., Juntunen K.1996Cestode larvae Diphyllobothrium dendriticum as a cause of heart disease leading to mortality in hatchery-reared sea trout and brown trout. Dis. Aquat. Org. 25, 15–22 (doi:10.3354/dao025015) [Google Scholar]
- Raymond M., Rousset F.1995Genepop (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- Rowe D. K., Thorpe J. E.1990Differences in growth between maturing and non-maturing male Atlantic salmon, Salmo salar L. parr. J. Fish Biol. 36, 643–658 (doi:10.1111/j.1095-8649.1990.tb04319.x) [Google Scholar]
- Rozen S., Skaletsky H. J.2000Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: methods in molecular biology (eds Krawetz S., Misener S.), pp. 365–386 Towota, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M.1989Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl Acad. Sci. USA 86, 232–236 (doi:10.1073/pnas.86.1.232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S.2005The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Frontiers Zool. 2, 16 (doi:10.1186/1742-9994-2-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffels R. J., Spencer H. G.2008An asymmetric model of heterozygote advantage at major histocompatibility complex genes: degenerate pathogen recognition and intersection advantage. Genetics 178, 1473–1489 (doi:10.1534/genetics.107.082131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M.1990Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. M., Faisal M., DeWoody J. A.2007Zygosity at the major histocompatibility class IIB locus predicts susceptibility to Renibacterium salmoninarum in Atlantic salmon (Salmo salar L.). Anim. Genet. 38, 517–519 (doi:10.1111/j.1365-2052.2007.01631.x) [DOI] [PubMed] [Google Scholar]
- Vähä J., Erkinaro J., Niemelä E., Primmer C. R.2007Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol. Ecol. 16, 2638–2654 (doi:10.1111/j.1365-294X.2007.03329.x) [DOI] [PubMed] [Google Scholar]
- Wegner K. M., Reusch T. B. H., Kalbe M.2003aMultiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 16, 224–232 (doi:10.1046/j.1420-9101.2003.00519.x) [DOI] [PubMed] [Google Scholar]
- Wegner K. M., Kalbe M., Kurtz J., Reusch T. B. H., Milinski M.2003bParasite selection for immunogenetic optimality. Science 301, 1343 (doi:10.1126/science.1088293) [DOI] [PubMed] [Google Scholar]
- Wegner K. M., Kalbe M., Schaschl H., Reusch T. B. H.2004Parasites and individual major histocompatibility complex diversity—an optimal choice? Microb. Infect. 6, 1110–1116 (doi:10.1016/j.micinf.2004.05.025) [DOI] [PubMed] [Google Scholar]