Abstract
Faster rates of microevolution have been recorded for plants and marine foraminifera occupying warmer low latitude environments relative to those occurring at higher latitudes. By contrast, because this rate heterogeneity has been attributed to a relationship between thermal habit and mutagenesis via a body temperature linkage, it has been assumed that microevolution in mammals should not also vary systematically with environmental temperature. However, this assumption has not previously been empirically examined. In this study, we tested for a thermally mediated influence on the tempo of microevolution among mammals using a comprehensive global dataset that included 260 mammal species, from 10 orders and 29 families. In contrast to theoretical predictions, we found that substitution rates in the cytochrome b gene have been substantially faster for species living in warmer latitudes and elevations relative to sister species living in cooler habitats. These results could not be attributed to factors otherwise thought to influence rates of microevolution, such as body mass differentials or genetic drift. Instead, the results indicate that the tempo of microevolution among mammals is either responding directly to the thermal environment or indirectly via an ecological mechanism such as the ‘Red Queen’ effect.
Keywords: metabolic rate, species richness, ectotherms, endotherms
1. Introduction
Faster rates of microevolution among ectothermic organisms in warmer environments were predicted by Rohde (1992). Subsequent empirical confirmations of that pattern for plants and foraminifera have been attributed to a relationship between thermal habit and mutagenesis via a body temperature/metabolic linkage (Allen et al. 2006; Wright et al. 2006). The mechanistic explanations for the relationship have been that metabolic rate influences mutagenesis either via the rate of cell division and replication error in the germline or via the total quantum of oxygen free-radical damage to DNA (Martin & Palumbi 1993). In this context, however, it has been assumed that, because euthermic body temperatures and metabolic rates in endotherms do not vary systematically across latitudes, microevolution for endotherms should also not vary systematically with latitude (Mittelbach et al. 2007; Weir & Schluter 2007). Two studies support this view (Bromham & Cardillo 2003; Weir & Schluter 2008), whereas a third by Bleiweiss (1998) found that rates of microevolution in a phylogenetically restricted group of hummingbirds were faster in species living at low elevations compared with those in high elevations. However, there have been no previous attempts to falsify the assumption that the tempo of microevolution among mammals is independent of thermal habit. Here we test for faster rates of microevolution among mammal species occupying warmer habitats (at lower latitudes or elevations) relative to those occurring in cooler environments using a comprehensive global dataset of 130 phylogenetically independent sister species pairs.
2. Material and methods
Rates of microevolution were compared in the cytochrome b gene of mtDNA between sister species pairs for 130 phylogenetically independent mammalian contrasts where one species occurs at lower latitude, or elevation, relative to the comparator species in the pair. These contrasts span a wide range of both placental and marsupial mammals from 10 orders and 29 families. The data were assembled using all possible species pairings available on GenBank (http://www.ncbi.nlm.nih.gov/GenBank) for cytochrome b that met the selection criteria at the time of dataset assembly. Cytochrome b is the most widely used gene for mammalian taxonomy and is therefore the only one that is consistently available for each species in the extensive range of pairings we have used for this study.
Species pairs were identified using published phylogenetic studies (see table S3 electronic supplementary material). However, where several studies were available, each with components of total diversity for a given clade, we reanalysed such clades ourselves by including sequences for all relevant species. Each pair selected was sister species according to the relevant cytochrome b phylogeny or, where such pairs did not meet other selection criteria, at most involved a contrast between a given species and its next most closely related neighbour. In some instances, the pairs included species with different generic names, but in such cases, each species was nonetheless embedded within (but not basal to) larger clades comprising one or other of the two genera involved. The use of such closely related species minimized the potentially confounding influences of more distant phylogenetic comparisons (figure 1). The amount of microevolution measured for each species in a given pair is that which has occurred since those species diverged from a common ancestor. It is therefore a measurement of evolution for each species over an equivalent period of time (figure 2).
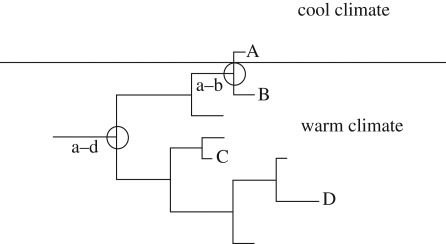
Figure 1.
The potential problem with using distant relatives to compare rates of evolution. Microevolution that has occurred within a species located in a cooler climate is represented above the horizontal line and that for species inhabiting warmer climates is represented below. If distant relatives are compared, such as A with C or D, then the difference in the rate of microevolution since the common ancestor (nodes a–d) is confounded by the large phylogenetic distance between the species. However, by comparing the two most closely related species available (A and B), the potential effect of the different climatic regimes becomes more observable.
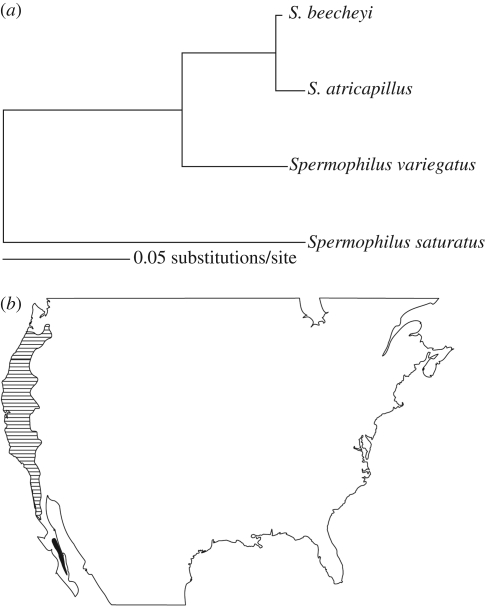
Figure 2.
An example of one of the 130 independent four-species comparisons. (a) The two ingroup species, Spermophilus beecheyi and Spermophilus atricapillus (from higher relative to lower latitudes, respectively), are contrasted against the two most proximate outgroup species. Compared branch lengths are from a common ancestor represented on the tree as the node of ingroup divergence from the two outgroup species. The genetic divergence estimated for each species is therefore measured over an equivalent time period. (b) The distributions of the two ingroup species. Line fill, S. beecheyi; solid fill, S. atricapillus.
Prior to dataset assembly, it was decided that only species weighing 5 kg or less would be selected. This was done because large mammals, which do not constitute a major component of total mammalian diversity, were often migratory prior to the recent expansion of the human population. This means that the historical climates that they experienced are typically likely to have been spatially broad and variable. Therefore, the inclusion of such large species in this study would have made it difficult, based on present day data, to ensure the necessary spatial fidelity for between-species latitudinal, or elevational, separations on which the research is predicated. However, any confounding influences owing to such a lack of spatial fidelity or owing to the recent contractions of range for the species that were in fact included in the study, or owing to gene flow between such included species, would have had a conservative rather than an inflationary effect on the results. Species distributions were obtained from ‘Walker's mammals of the world’ (Nowak 1999), from the International Union for Conservation of Nature and Natural Resources (IUCN) website (http://www.iucnredlist.org) and from a range of other published literature (see table S3 electronic supplementary material). Species pairs were only selected if their latitudinal/elevational overlap was 25 per cent or less of either of the respective ranges.
The minimum sequence length used for comparison was greater than 50 per cent of the total cytochrome b gene, and where there were multiple accessions with greater than 50 per cent of the gene sequence available, the most complete sequence was used. In all instances, where there were two or more species meeting the study criteria, or where multiple GenBank accessions of equivalent proportions of the cytochrome b gene were available for the same species, the accession with the fewest substitutions (assessed on the basis of uncorrected genetic distance) was used. This precaution minimized the risk of artefacts owing to laboratory error, ensured that closest relatives were always compared and provided for consistent treatment across the dataset.
The two nearest phylogenetic neighbours were applied as outgroups to the ingroup pairing. Potential outgroup taxa were initially identified from the DNA-based phylogenetic literature, but where multiple publications existed for a given genus, we optimized outgroup proximity by ourselves generating larger phylogenies that improved tree resolution. This enabled us to apply the closest available pair of outgroup species to each ingroup pairing and thereby to optimize the phylogenetic reconstruction. For each comparison, two outgroup species were applied to provide a more robust basis for the comparison.
Thus, initial four-species alignments were generated for each warmer climate versus cooler climate comparison that comprised the two members of the contrasted ingroup pair, together with the two relevant outgroup species. Then, pair-by-pair comparisons (Sarich & Wilson 1967) were generated for these individual phylogenetic trees using PAUP* v. 4.0b10 (Swofford 2002), whereby branch lengths were derived with maximum likelihood (ML) under the GTR model of evolution with substitution rate matrices, proportions of invariant sites, base frequencies and gamma shape parameters all estimated. The substitution-derived branch length per species was therefore estimated as the length of the branch leading to each member of the ingroup pair being contrasted. The relative rates were summarized as the ratio between them and, because our data are non-normal, ratios were then tested against the null hypothesis of a median ratio of 1 using a Wilcoxon signed-rank test and a sign test.
For verification, we also analysed our results using PAML. Again, branch lengths for each pair of the ingroup species were calculated on the tree topologies, previously obtained from the four-species alignments, under ML with a GTR model of evolution. The local substitution (clock) branch lengths, respectively, leading to each member of a given warmer climate versus cooler climate species pair were estimated using the baseml component of PAML 4.0 (Yang 2007) as the basis for comparison between each of the pairs. These rates were then expressed as pair-by-pair ratios in the manner described earlier.
The ratio of non-synonymous substitution relative to that of synonymous substitution (dN/dS) was similarly estimated using the tree topologies, previously obtained from the four-species alignments, with the codeml component of PAML 4.0 (Yang 2007). A dN/dS ratio was thus simultaneously estimated for the two foreground branches, respectively, leading to the warmer and cooler climate species comprising each pair to provide the dN/dS ratio contrast between the members of those pairings.
For each of these datasets, sequences were aligned at the codon level, so that when computationally translated to amino acids, a coherent protein alignment was obtained. The nucleotide alignments were truncated at the first codon boundary and prior to the first termination codon. Some sequences downloaded from GenBank extended beyond the coding region of the cytochrome b gene, and this overhanging region was discarded. None of the sequences that were compared involved divergences owing to insertions or deletions. All alignments were analysed as nucleotide sequences.
3. Results and discussion
(a). The tempo of microevolution
Substitutions in the cytochrome b gene of mtDNA for mammal species occurring in warmer climates (at lower latitudes or elevations) were on average 1.47 times greater than in those occurring in cooler climates (at higher latitudes or elevations) (Wilcoxon two-tailed signed-rank test, n = 130, p = 0.0003; sign test, p = 0.001). For verification, we also analysed our results using PAML and obtained results that were substantially unchanged (Wilcoxon two-tailed signed-rank test, n = 130, p = 0.0004). Faster rates of microevolution in warmer environments were evident for both elevational (1.59 times faster) and latitudinal (1.43 times faster) comparisons and within each hemisphere and within both the New and Old Worlds (table 1). Substitution ratios for each pairing and GenBank accession numbers for each species are listed in table S4, electronic supplementary material.
Table 1.
Two-tailed Wilcoxon signed-rank sum tests for comparisons of ML branch lengths. Statistical significance at pfamily = 0.05 for multiple comparisons using a Bonferroni correction is shown with an asterisk. Using false discovery rates (Benjamini & Hochberg 1995), all results are significant (α < 0.05) except for that from the Southern Hemisphere.
| comparisons | n | p-values |
|---|---|---|
| latitude | 101 | 0.0077* |
| elevation | 29 | 0.0064* |
| Southern Hemisphere | 46 | 0.0183 |
| Northern Hemisphere | 77 | 0.0033* |
| New World (Americas) | 70 | 0.0127 |
| Old World (Africa/Eurasia) | 46 | 0.0019* |
This study, using 130 phylogenetically independent species pairs for contrasts, is the largest to date that has investigated rates of microevolution for any group of organisms. The compared taxa were either sister species or, where sisters failed to meet other selection criteria, involved at most a phylogenetic distance to the next nearest neighbour. The use of such closely related species minimized the potentially confounding influences of more distant phylogenetic comparisons (figure 1). The application of this criterion resulted in comparisons between species that are not only phylogenetically closely related but also geographically proximate. Thus, elevational and latitudinal effects on rates of microevolution are evident in this instance over relatively small spatial separations. We might therefore expect the pattern that we have identified to be more powerful at continental to global scales.
The method we employed, of selecting for the shortest branch length among multiple accessions, might potentially bias the data towards shorter branches at higher latitudes and elevations if there were generally more accessions for higher latitude/elevation species. We therefore tested for this bias by examining a subset of the data from which all pairs were removed that involved the selection of a sequence for the higher latitude/elevation species from a larger pool of accessions relative to that available for the contrasted lower latitude/elevation species. The result remained statistically significant for this data subset (Wilcoxon two-tailed signed-rank test, n = 92, p = 0.0007), indicating that our results are robust against this potential bias.
Our results are consistent with the faster rates of microevolution found among low elevation hummingbirds by Bleiweiss (1998) and with similar results obtained using nuclear ITS for plants (Wright et al. 2006) and SSU rDNA for foraminifera (Allen et al. 2006). By contrast, two investigations for a relationship between latitude and rates of microevolution with endotherms failed to find statistically significant results (Bromham & Cardillo 2003; Weir & Schluter 2008). However, both the latter studies involved more distantly related species and smaller sample sizes than were employed here.
In this study, faster rates of microevolution for mammals occupying warmer environments were found to be consistently strong across all orders of mammals except for the Australasian marsupials (Peramelemorpha and Dasyuromorphia), where the rate differential, although still favouring lower latitudes, was small (table 2). However, the two New World marsupial comparisons (Didelphimorphia) both showed substantially faster rates of microevolution at lower latitudes. If the faster rates of microevolution we have found for lower latitude/elevation species are related to environmental productivities, rather than to ambient temperature, then the results for Australian marsupials may not be indicative of a difference between placental and non-placental mammals, but may instead reflect the aridity of that continent and the potentially confounding influence of water deficits in the warmer climates there.
Table 2.
Mean ML branch length ratios (for warmer species relative to those for the cooler species) for each order.
| order/family | mean branch length ratio | n |
|---|---|---|
| Carnivora | 1.353 | 6 |
| Primates | 1.806 | 5 |
| Insectivora | 1.303 | 14 |
| Lagomorpha | 1.865 | 7 |
| Rodentia | 1.403 | 74 |
| Scandentia | 6.552 | 1 |
| Chiroptera | 1.653 | 10 |
| Dasyuromorphia | 1.024 | 10 |
| Peramelemorpha | 0.748 | 1 |
| Didelphimorphia | 2.138 | 2 |
(b). Biologically available energy
The slower rates of microevolution shown in these results for mammal species living within habitats with less biologically available energy, whether they live at higher latitudes or elevations, suggest that energy or a linked variable such as productivity may be the limiting factor for such evolution. Although many variables vary with both latitude and elevation, energy availability is the only variable that consistently varies in the same way with both elevation and latitude. Faster rates of microevolution among ectothermic organisms in warmer environments have been previously attributed to a relationship between thermal habit and mutagenesis via a body temperature/metabolic linkage (Gillooly et al. 2005; Allen et al. 2006; Wright et al. 2006). In this context, however, it has been predicted that because euthermic body temperatures and metabolic rates in endotherms vary little across latitudes, that microevolution for mammals should also vary little with latitude (Allen et al. 2006; Mittelbach et al. 2007; Weir & Schluter 2007). Nonetheless, a general mechanism that might explain faster rates of microevolution for mammalian endotherms living at warmer elevations and latitudes could arise from several processes involving adaptive responses to climatic variation.
First, in cooler climates, the average body size of endotherms tends to be greater (Bergmann's rule) (Ashton et al. 2000). Increasing body mass has been related to depressed rates of microevolution (Gillooly et al. 2005) and might therefore result in slower evolution towards the poles. To examine this hypothesis, we obtained comparative body sizes for 95 of the 130 species pairs using the database of Smith et al. (2003) supplemented by other published sources. However, body weights were only greater in the cooler climate species in 53.7 per cent of the pairs, and, for the subset of pairs in which the heavier species occurred at a lower latitude or elevation, rates of microevolution remained 1.55 times faster on average in the warmer climate species (Wilcoxon two-tailed signed-rank test, n = 45, p = 0.007). Furthermore, the general pattern of faster rates for warmer climate species was weaker among the subset of pairs in which the cooler climate species was larger (1.47 times faster, Wilcoxon two-tailed signed-rank test, n = 50, p > 0.05). Body size asymmetries cannot therefore explain the results. The lack of any influence of body weight on our results suggests that it is energy that has the strong mediating effect on rates of molecular evolution.
A second possible climate-related explanation for the results is that despite the relatively constant body temperatures and metabolic rates of mammals, there might be a link between average annual metabolic rates and the tempo of microevolution. Field euthermic metabolic rates in mammals increase with latitude (Anderson & Jetz 2005), and rates of microevolution in mammals have not been found to correlate with basal metabolic rates (Lanfear et al. 2007). However, at higher latitudes, periods of torpor or hibernation that conserve energy in response to diminished energetic circumstances (McKechnie & Lovegrove 2002; Munro et al. 2005) may nonetheless reduce average annual metabolic activity. Torpor also occurs among endotherms at low latitudes where there are energetic constraints (McKechnie & Lovegrove 2002) such as those encountered at high elevations. It is therefore possible that mutagenesis might depend on an annual metabolic average that is reduced at higher elevations and latitudes. However, hibernation alone cannot account for our results because slower rates of microevolution were also recorded for cooler climate species that are known not to hibernate (mean branch length ratio: warmer/cooler = 1.63, Wilcoxon two-tailed signed-rank test, n = 27, p = 0.066). Nonetheless, whether or not hibernation is involved, the possibility of average metabolic rate reductions among mammals living in cooler habitats—owing, for example, to variation in the ratio of active to resting metabolism—might yet be a factor influencing the asymmetry in the tempo of microevolution that we have found and is worthy of further investigation.
(c). The Red Queen hypothesis
A third explanation for our results, indirectly linked to climate, arises from the Red Queen Hypothesis (VanValen 1973), whereby the rate of evolution of a given species in a particular ecosystem is thought to be dependent on the rate of evolution among other species within that community with which it is coevolving. The rate of microevolution among mammals might therefore be dependent on the coevolutionary influence of elevated ectothermic rates within the same community (Brown et al. 2004). Given that microevolution among ectotherms has been found to be more rapid in warmer climates, it is possible that the biotic environment produced by ectotherms may also be more dynamic in warmer climates. Therefore, the probability of a mutation possessing a positive selection coefficient may be greater, if it occurs within a mammal population living within the context of a more rapidly changing ectothermic biotic environment, than if that same mutation were to occur in a population living in a more stable biotic milieu where conditions were cooler. For example, if a mutation in a tropical plant produced a defence against a mammal browser, a subsequent mutation by the browser to overcome this would be rapidly fixed. However, if the same anti-defence mutation were to occur in a temperate browser, where the plant species being predated was evolving its defences more slowly—and the plant species had not produced the relevant protective mutation—it is axiomatic that a corresponding anti-defence mutation would not be selected and fixed in the browser population. Therefore, mammals living within the context of a more rapidly evolving trophic milieu, such as that presented within tropical communities, might also be evolving more rapidly. Thus, the rate of fixation in mammals may, on average, be faster in warmer climates owing to a greater proportion of mutations possessing a selective advantage. Both the non-synonymous and synonymous components of microevolution might be thus affected if synonymous mutations are commonly linked to positively selected mutations (refer to discussion below) or if such mutations are often non-neutral.
With respect to linkage, it is apposite to note that Birky & Walsh (1988) demonstrated that, under the assumptions of neutral and nearly neutral theories (Ohta 1993), the rate of fixation of neutral mutations cannot be affected by linkage. The fixation of a selectively advantageous mutation may sweep linked neutral variants to fixation. However, the same process is expected to simultaneously remove from a population any other neutral variants in that linkage region segregating in genomes without the advantageous mutation. This removal of variants that may otherwise have drifted to fixation is assumed to cancel out the increase in fixation caused by the sweep—thus there is no change predicted in the total rate of fixation for neutral mutations. Therefore, if the substitution rate heterogeneity that we have measured has occurred by drift under neutral or nearly neutral mechanisms, linkage cannot be the cause. On the other hand, if the assumptions of nearly neutral theory do not hold, and our results are not due to nearly neutral effects, the concept of Birky & Walsh (1988) does not apply as their model was entirely based on these assumptions. Linkage can be used to explain positive selection effects, but not nearly neutral effects.
(d). Population size effects
Faster rates of evolution at lower latitudes might occur owing to the influence of smaller average population sizes in those habitats. Genetic drift is predicted to be faster in smaller populations and, where it is warmer, populations might on average be smaller owing to the greater number of species per unit area (Stevens 1989). However, this hypothesis is not able to explain the results with respect to elevation because available land area and, consequently, mammal populations generally decline with increasing elevation (e.g. Patterson et al. 1989).
Further, if the faster rates of evolution at lower elevations and latitudes that we have found were due to nearly neutral effects in small populations, we would expect a higher proportion of substitutions in the smaller populations to be non-synonymous and slightly deleterious than in larger populations at higher latitudes where such mutations should have a higher probability of being purified (Ohta 1993). Therefore, the ratios of non-synonymous to synonymous substitutions (dN/dS) for species in small populations at low latitudes are predicted to be greater than for higher latitude species with larger populations (Woolfit & Bromham 2005). Although a difference in dN/dS ratios between small and large populations cannot distinguish between nearly neutral effects and positive selection effects, a lack of a ratio difference between small and large populations suggests the absence of nearly neutral effects. We therefore tested for a difference in the dN/dS ratio between the cooler climate and warmer climate species. However, the median ratios for the respective cooler and warmer climate species groups were similar (0.023 and 0.025; n = 125, two-tailed Wilcoxon signed-rank test, p = 0.13). Those ratios were also similar for the two species groups when the elevational and latitudinal data subsets were independently contrasted (p = 0.76 and 0.11, respectively). These results suggest that any tendency to smaller population size in warmer habitats does not explain the enhanced rates of substitution at lower latitudes and elevations that we have recorded.
It has also been suggested that small population sizes following speciation events (bottlenecks) might have increased rates of microevolution in the more speciose lower latitudes (Cardillo 1999). However, the ratios of non-synonymous to synonymous substitutions described earlier also suggest that population bottlenecks at the time of speciation is an unlikely explanation for our results. In addition, empirical evidence from plants demonstrates that rates of microevolution are faster in species at low latitudes relative to high regardless of whether there are more or less species in a given taxon at low latitudes (Wright et al. 2006).
(e). Global diversity and origination gradients
The faster rates of evolution at lower latitudes relative to higher latitudes found in this study for mammals, together with similar results for ectotherms (Allen et al. 2006; Wright et al. 2006), are consistent with the ‘Evolutionary Speed’ (Rohde 1992) explanation for typical global species richness gradients that favour low latitude habitats (see also Davies et al. 2004). This explanation invokes faster rates of microevolution in warmer (Rohde 1992), or more productive (Gillman & Wright 2006), environments as the driver of greater rates of speciation and species accumulation there (Allen et al. 2006; Wright et al. 2006). Both latitudinal and elevational patterns of species richness in mammals (including mid-elevational peaks where montane precipitation is higher) have also been found to correlate positively with productivity gradients (McCain 2007).
Fossil evidence also suggests that origination rates have been greater in the tropics relative to temperate latitudes (Allen et al. 2006; Jablonski et al. 2006; Martin et al. 2007). However, in a recent study by Weir & Schluter (2007), using the same cytochrome b gene as was used in this study, it was reported that greater speciation rates were evident at high, not low, latitudes. In that study, genetic distances between pairs of bird species or mammal species from similar latitudes were assumed to represent time since divergence. Then, using the assumption of a stable inter-latitudinal molecular clock, derived divergence times were related to latitude and used to infer a gradient of increasing speciation rates towards the poles. The results presented here demonstrate that the key assumption applied by those researchers that rates of evolution in cytochrome b are unrelated to latitudinal effects is not supported. Thus, in the light of our findings, their data showing more substitutions separating species pairs living in low latitude habitats could be reinterpreted as indicating faster rates of microevolution—not slower rates of diversification—in tropical environments.
4. Conclusions
In this study, the first large-scale examination of spatial effects on microevolutionary tempo between sister species for endotherms, we show that the rate of evolution is not a time constant response but rather one that varies with both latitude and elevation. Body mass differentials (Gillooly et al. 2005) and genetic drift in small populations (Stevens 1989) were not found to contribute to the global pattern in the evolutionary rate differential that we recorded. Instead, because climatic variables change with latitude and elevation in a similar manner, our results suggest that climate has influenced the tempo of genetic change in mammals. It is possible that this effect is either directly due to the ambient thermal context of mammalian habitats or indirectly controlled via a ‘Red Queen’ effect in which endothermic evolution is paced by that of coevolving ectotherms. These findings provide new insights into microevolution that are likely to have important implications for understanding global patterns of biodiversity and diversification.
Acknowledgements
The study was funded by Nga Pae O Te Maramatanga, Maori Center of Research Excellence, under the direction of Michael Walker and Linda Smith and by the Auckland University of Technology. S.D.W. holds the Michael Horton Lectureship in Biogeography at the University of Auckland.
References
- Allen A., Gillooly J., Savage V., Brown J.2006Kinetic effects of temperature on rates of genetic divergence and speciation. Proc. Natl Acad. Sci. USA 103, 9130–9135 (doi:10.1073/pnas.0603587103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. J., Jetz W.2005The broad-scale ecology of energy expenditure of endotherms. Ecol. Lett. 8, 310–318 (doi:10.1111/j.1461-0248.2005.00723.x) [Google Scholar]
- Ashton K. G., Tracy M. C., deQueiroz A.2000Is Bergmann's rule valid for mammals. Am. Nat. 156, 390–415 (doi:10.1086/303400) [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y.1995Controlling the false-discovery rate: a simple and powerful approach to multiple hypothesis testing. J. R. Stat. Soc. Ser. B 57, 289–300 [Google Scholar]
- Birky C. W., Jr, Walsh J. B.1988Effects of linkage on rates of molecular evolution. Proc. Natl Acad. Sci. USA 85, 6414–6418 (doi:10.1073/pnas.85.17.6414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleiweiss R.1998Slow rate of molecular evolution in high-elevation hummingbirds. Proc. Natl Acad. Sci. USA 95, 612–616 (doi:10.1073/pnas.95.2.612) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L., Cardillo M.2003Testing the link between the latitudinal gradient in species richness and rates of molecular evolution. J. Evol. Biol. 16, 200–207 (doi:10.1046/j.1420-9101.2003.00526.x) [DOI] [PubMed] [Google Scholar]
- Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B.2004Toward a metabolic theory of ecology. Ecology 85, 1771–1789 (doi:10.1890/03-9000) [Google Scholar]
- Cardillo M.1999Latitude and rates of diversification in birds and butterflies. Proc. R. Soc. Lond. B 266, 1221–1225 (doi:10.1098/rspb.1999.0766) [Google Scholar]
- Davies T., Savolainen V., Chase M., Moat J., Barraclough T.2004Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. Lond. B 271, 2195–2200 (doi:10.1098/rspb.2004.2849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman L. N., Wright S. D.2006The influence of productivity on the species richness of plants: a critical assessment. Ecology 87, 1234–1243 (doi:10.1890/0012-9658(2006)87[1234:TIOPOT]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Gillooly J. F., Allen A. P., West G. B., Brown J. H.2005The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc. Natl Acad. Sci. USA 102, 140–145 (doi:10.1073/pnas.0407735101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski D., Roy K., Valentine J. W.2006Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102–106 (doi:10.1126/science.1130880) [DOI] [PubMed] [Google Scholar]
- Lanfear R., Thomas J. A., Welch J. J., Brey T., Bromham L.2007Metabolic rate does not calibrate the molecular clock. Proc. Natl Acad. Sci. USA 104, 15 388–15 393 (doi:10.1073/pnas.0703359104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. P., Palumbi S. R.1993Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA 90, 4087–4091 (doi:10.1073/pnas.90.9.4087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P. R., Bonier F., Tewksbury J. J.2007Revisiting Jablonski (1993): cladogenesis and range expansion explain latitudinal variation in taxonomic richness. J. Evol. Biol. 20, 930–936 (doi:10.1111/j.1420-9101.2007.01317.x) [DOI] [PubMed] [Google Scholar]
- McCain C. M.2007Could temperature and water availability drive elevational species richness patterns? A global case study for bats. Global Ecol. Biogeogr. 16, 1–13 (doi:10.1111/j.1466-8238.2006.00263.x) [Google Scholar]
- McKechnie A. E., Lovegrove B. G.2002Avian facultative hypothermic responses: a review. Condor 104, 705–724 (doi:10.1650/0010-5422(2002)104[0705:AFHRAR]2.0.CO;2) [Google Scholar]
- Mittelbach G. G., et al. 2007Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 (doi:10.1111/j.1461-0248.2007.01020.x) [DOI] [PubMed] [Google Scholar]
- Munro D., Thomas D. W., Humphries M. M.2005Torpor patterns of hibernating eastern chipmunks Tamias striatus vary in response to the size and fatty acid composition of food hoards. J. Anim. Ecol. 74, 692–700 (doi:10.1111/j.1365-2656.2005.00968.x) [Google Scholar]
- Nowak R. M.1999Walker's mammals of the world London, UK: The John Hopkins University Press [Google Scholar]
- Ohta T.1993Amino acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc. Natl Acad. Sci. USA 90, 4548–4551 (doi:10.1073/pnas.90.10.4548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B. D., Meserve P. L., Lang B. K.1989Distribution and abundance of small mammals along an elevational transect in temperate rainforests of Chile. J. Mamm. 70, 67–78 (doi:10.2307/1381670) [Google Scholar]
- Rohde K.1992Latitudinal gradients in species diversity: the search for the primary cause. Oikos 65, 514–527 (doi:10.2307/3545569) [Google Scholar]
- Sarich V. M., Wilson A. C.1967Immunological time scale for hominid evolution. Science 158, 1200–1203 (doi:10.1126/science.158.3805.1200) [DOI] [PubMed] [Google Scholar]
- Smith F. A., Lyons S. K., Ernest S. K. M., Jones K. E., Kaufman D. M., Dayan T., Marquet P. A., Brown J. H., Haskell J. P.2003Body mass of Late Quaternary mammals. Ecology 84, 3403 (doi:10.1890/02-9003) [Google Scholar]
- Stevens G. C.1989The latitudinal gradient in geographical range: how so many species coexist in the tropics. Am. Nat. 133, 240–256 (doi:10.1086/284913) [Google Scholar]
- Swofford D.2002PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0 Beta Sunderland, MA: Sinauer Associates: CD-ROM [Google Scholar]
- VanValen L. M.1973A new evolutionary law. Evol. Theory 1, 1–30 [Google Scholar]
- Weir J. T., Schluter D.2007The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science 315, 1574 (doi:10.1126/science.1135590) [DOI] [PubMed] [Google Scholar]
- Weir J. T., Schluter D.2008Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328 (doi:10.1111/j.1365-294X.2008.03742.x) [DOI] [PubMed] [Google Scholar]
- Woolfit M., Bromham L.2005Population size and molecular evolution on islands. Proc. R. Soc. B 272, 2277–2282 (doi:10.1098/rspb.2005.3217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Keeling J., Gillman L.2006The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proc. Natl Acad. Sci. USA 103, 7718–7722 (doi:10.1073/pnas.0510383103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.2007PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]