Abstract
Feathers are known to contain amplifiable DNA at their base (calamus) and have provided an important genetic source from museum specimens. However, feathers in subfossil deposits generally only preserve the upper shaft and feather ‘vane’ which are thought to be unsuitable for DNA analysis. We analyse subfossil moa feathers from Holocene New Zealand rockshelter sites and demonstrate that both ancient DNA and plumage information can be recovered from their upper portion, allowing species identification and a means to reconstruct the appearance of extinct taxa. These ancient DNA sequences indicate that the distal portions of feathers are an untapped resource for studies of museum, palaeontological and modern specimens. We investigate the potential to reconstruct the plumage of pre-historically extinct avian taxa using subfossil remains, rather than assuming morphological uniformity with closely related extant taxa. To test the notion of colour persistence in subfossil feathers, we perform digital comparisons of feathers of the red-crowned parakeet (Cyanoramphus novaezelandiae novaezelandiae) excavated from the same horizons as the moa feathers, with modern samples. The results suggest that the coloration of the moa feathers is authentic, and computer software is used to perform plumage reconstructions of moa based on subfossil remains.
Keywords: ancient DNA, feathers, moa, phenotype, plumage reconstruction
1. Introduction
The arrival of humans and their associated mammalian species in New Zealand at approximately AD 1280 (Wilmshurst et al. 2008) resulted in the extinction of 41 per cent of New Zealand's breeding bird species (Tennyson & Martinson 2006). These species are relatively well known osteologically because of New Zealand's rich late Quaternary avifaunal fossil record (Worthy & Holdaway 2002). While most are known only from their bones, a few partially mummified remains have also been found (Anderson 1989; Worthy 1989; Vickers-Rich et al. 1995), while isolated feathers have been recovered from a range of late Holocene rockshelter sediments (Wood 2008; Wood et al. 2008). The majority of subfossil feathers found in New Zealand have been attributed to the extinct palaeognathus (ratite) moa (Aves: Dinornithiformes), although none has been confirmed genetically. If DNA could be recovered routinely from subfossil feathers, it would create many opportunities for genetic studies of extinct taxa and populations, as well as providing an important insight into the appearance of extinct species. However, the survival of DNA in subfossil feathers has not yet been demonstrated, and there is little evidence about the quality or location of DNA in different parts of feathers.
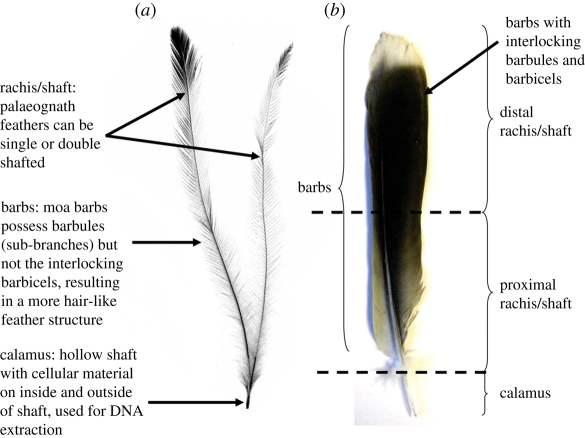
DNA has been extracted previously from the base, or calamus, of feathers from modern and historical museum specimens (Payne & Sorenson 2002; Sefc et al. 2003; Horváth et al. 2005). Extraction protocols have generally ignored the distal components of the feather, such as the rachis and barbs (including barbules and barbicels) (figure 1), which comprises the bulk of the feather structure, because it has been thought that there is no amplifiable DNA in these structures. The distribution of mitochondrial DNA (mtDNA) throughout paralogous structures like hair (Gilbert et al. 2007) and reptilian scales (Fetzner 1999; Feldman & Spicer 2002) raises the possibility that mtDNA might also be present in all parts of the feather structure. Because the calamus is commonly absent from subfossil feathers (owing to breakage before, or during, deposition), the recovery of DNA from distal feather components would have major implications for historical and ancient DNA research.
Figure 1.
(a) Morphological structure of a palaeognathus feather, here shown by emu (Dromaius novaehollandiae). The main shaft (rachis) supports side branches (barbs) and together makes up the distal component of feathers. In vaned feathers, barbs are held together by small sub-branches termed barbules, which in turn are held together by interlocking hooks or barbicels. (b) A schematic representation of the distal feather components sampled in this study using a neo-avian feather to illustrate the role of barbules and barbicels in forming a vaned structure characteristic of flighted birds.
Ancient DNA can be a powerful tool when reconstructing the phenotype of extinct and extant species. It has been used previously to reconstruct mammoth and horse coat colour (Rompler et al. 2006; Ludwig et al. 2009) and to suggest that Neanderthals had differing degrees of skin and hair pigmentation (Lalueza-Fox et al. 2007). However, these studies used ancient DNA to identify colour genes, including the melanocortin-1 receptor (MCR1) gene, rather than directly linking hair samples of known colour to an extinct species of unknown external appearance. Recently, it has been demonstrated that even fossilized feathers from the Cretaceous and Eocene (Vinther et al. 2008) can be preserved as carbonaceous traces of melanosomes (pigment containing organelles). When compared with modern taxa, such fossils provide information on the colour pattern of the original feather (Vinther et al. 2008). However, reconstructions of specific taxa using this methodology are only possible if the fossilized feathers can be identified positively to species. When there is no fossil feather information, reconstruction attempts have relied on the phenotype of closely related extant taxa (e.g. Vickers-Rich et al. 1985; Gill & Martinson 1991; Flannery & Schouten 2001; Murray & Vickers-Rich 2003; Tennyson & Martinson 2006). In the rare cases in which subfossil feathers have been preserved, it has generally been assumed that the colours reflect accurately their original appearance (White 1885; Hamilton 1894). However, historical museum and subfossil feathers are prone to fading from exposure to sunlight or other factors (Oliver 1955; G. Pohland 2007, unpublished data), and it is conceivable that feathers excavated from cave sediment may have also altered. One approach to investigating this issue is to use standardized Munsell colour chips (Villafuerte & Negro 1998) to compare subfossil and modern feathers from the same species and to quantify colour fading for a given site and horizon. The assumption is then made that other feathers in the deposit have been protected similarly from fading and alteration.
To determine whether amplifiable ancient DNA can be extracted from subfossil feathers, we performed trials on nine moa feathers collected from rockshelter sediments in the semi-arid region of Central Otago, South Island, New Zealand. The giant, graviportal moa were the dominant terrestrial herbivores in New Zealand's pre-human terrestrial ecosystems and a striking example of an avian radiation into different niches and habitats. Moa have been the focus of considerable palaeontological (Worthy & Holdaway 2002), palaeoecological (Wood et al. 2008) and evolutionary research (Cooper et al. 1992; Bunce et al. 2003; Huynen et al. 2003; Baker et al. 2005; Lambert et al. 2005). However, relatively little is known about their external phenotype or interspecific plumage variation. Most mummified moa specimens, where skin is present, have just the bases of feathers preserved (e.g. Hutton & Coughtrey 1875; Forrest 1987). As a consequence, it has been difficult to assign isolated moa feathers to species. Ancient DNA analysis offers the potential to link these feathers to known fossil taxa and provides insight into their plumage and appearance. To better understand where DNA is distributed in different parts of a feather, we also tested whether amplifiable DNA was detectable in the rachis and barbs of 10 moa feathers from one of the sites, as well as three modern emu feathers (figure 1). Lastly, to determine whether moa feathers could be used to reconstruct plumage characteristics, we quantified digitally the amount of colour fading by comparing subfossil feathers from another species recovered from the same deposits with living relatives.
2. Material and methods
(a). Materials
We analysed a total of 19 moa feathers excavated from Sawers', Roxburgh Gorge B and Roxburgh Gorge C rockshelters, in Central Otago, South Island, New Zealand, and held in the collections of the Otago Museum, Alexandra Museum and the Australian Centre for Ancient DNA (ACAD) (table S1, electronic supplementary material). The excavated feathers were characteristic of palaeongaths and specimens identified previously as belonging to moa (figure 1; Worthy & Holdaway 2002). The calamus (5 mm) was removed from a set of nine moa feathers (table S1, electronic supplementary material) for genetic analysis, and then each feather was photographed with a Nikon digital camera for digital reconstruction of original colour and plumage.
To investigate the location of DNA in feathers, the rachis and barbs from a further 10 moa feathers from Sawers' rockshelter were examined in a later series of extractions. The two sets of samples were collected at different times from different excavations (table S1, electronic supplementary material). Naturally shed modern emu (Dromaius novaehollandiae) feathers were also used as a positive control (table S1, electronic supplementary material).
(b). Molecular methods
To determine whether DNA could be extracted from subfossil feathers, the calamus from the first nine moa feathers was cut in half longitudinally and further diced with a sterile scalpel blade to help facilitate enzymatic digestion. Ancient DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer's instructions. Multiple negative extraction and amplification controls were included. All DNA extractions and the setup of PCR reactions were performed in the physically remote, isolated and dedicated ancient DNA facility using appropriate logistical and methodological procedures (Cooper & Poinar 2000).
PCR amplifications targeted 31, 180 or 205 bp of the moa mtDNA control region (excluding primers) using the primer pairs 262F/294R (31 bp), 262F/419R (180 bp) and 185F/294R (205 bp; from Cooper et al. 2001; Bunce et al. 2003; Wood et al. 2008) (table S2, electronic supplementary material). Unsuccessful PCR amplifications were subsequently repeated with the following primer pair: 204F/294R (11 bp excluding primers) (Wood et al. 2008; table S2, electronic supplementary material) to determine whether smaller DNA fragments were present. To improve the sequencing results from the short PCR products, the 262F and 294R primers were tagged with M13USP and M13RSP primers, respectively, as described by Wood et al. (2008). PCR reactions were conducted in 25 µl volumes containing a final concentration of 2 mg ml−1 rabbit serum albumen (Sigma), 1× PCR buffer (Invitrogen), 2 mM MgSO4, 200 µM each dNTP, 1 µM each primer, 1 U Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and 1–2 µl template DNA. PCR conditions were as follows: 94°C for 3 min, 55 cycles of 94°C for 30 s, 55°C for 30 s and 68°C for 45 s, with a final extension of 68°C for 10 min. PCR amplification reactions and all downstream post-PCR procedures were carried out in a modern molecular biology laboratory at the University of Adelaide.
To test whether amplifiable DNA was present in the distal portion of both modern and subfossil feathers, an additional 10 moa and 3 modern emu feathers were divided into sections for separate DNA extractions. The entire distal portion (rachis and barbs) of the moa feathers was examined, while for emus, separate DNA extractions were performed on subsamples comprising: the calamus; the distal; and proximal halves of the rachis (only); and all of the barbs (figure 1). Each subsample was minced with a scalpel blade and soaked in 1 : 10 bleach solution for 30 min to remove potential contamination from exterior surfaces, and then rinsed three times with Millipore ultrapure water. DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer's instructions, with the addition of 20 µl of dithiothreitol to the ATL lysis buffer to aid digestion of keratin and left overnight at 55°C on a rotary mixer to completely dissolve the samples. PCR amplifications targeting 31, 180 or 205 bp of the moa mtDNA control region (excluding primers) were performed as described earlier. For emu feathers, a 108 bp fragment of the mtDNA 12S ribosomal RNA gene was amplified following the PCR method outlined earlier using the primer pair 12SE/12SH2 reported in Cooper et al. (2001) (table S2, electronic supplementary material).
PCR products were visualized on a 2–3.5% 1× TBE agarose gel. If primer dimers were present after PCR amplification, PCR products were purified using the AMPure magnetic bead system (Agencourt), otherwise PCR products were purified using 4 U Exo1 and 0.6 U SAP (Fermentas) by incubation at 37°C for 30 min and 80°C for 15 min. Both template strands were sequenced for each mtDNA fragment, using independent PCR reactions for each strand in ancient samples. All sequences were determined using Big Dye Terminator v. 3.1 chemistry and an ABI 3130XL capillary sequencer. All sequences have been deposited in GenBank (GQ253938–GQ253945) or table S3 (electronic supplementary material) if less than 50 bp in length.
(c). Data analysis
All sequences were imported into Sequencher (Genecodes) and consensus sequences assembled. Sequences derived from emu feathers were identified using GenBank BLAST searches. mtDNA sequences obtained from moa feathers were aligned to a database of moa control region sequences (77 published from Cooper et al. 2001; Bunce et al. 2003; Huynen et al. 2003, 2008; Baker et al. 2005 and 300 sequences encompassing the entire geographical range of each moa species in New Zealand; unpublished data) using the ClustalW algorithm implemented in MEGA 4.0 (Kumar et al. 2004). Moa species were identified through similarity to known moa sequences in the database, with matches of 98–100%. To verify the initial identifications, a reference dataset of previously published long (2914 bp) moa sequences (Baker et al. 2005) was used to construct a robust maximum parsimony strict consensus bootstrap phylogeny (10 random sequence addition replications and 500 bootstrap replicates) with PAUP*4.0b10 (Swofford 2000). The reference moa dataset comprised concatenated CR, 12S, COIII, Cyt-b, ND3, ND4, ND5 and tRNA Lys sequences from 25 specimens and an additional 379 bp CR data from 29 specimens lacking the full 2914 bp. The moa feather sequences were identified to species using the bootstrap phylogeny as a backbone constraint by placing the partial control region sequences onto the tree using a full heuristic maximum parsimony search.
(d). Preservation of colour in subfossil feathers
The degree of colour fading in moa feathers was tested using subfossil feathers of the extant red-crowned parakeet (Cyanoramphus novaezelandiae novaezelandiae) excavated from moa feather-bearing sediment horizons within Roxburgh Gorge C rockshelter. These were compared with similar-sized feathers collected recently from captive birds following the digital methodology of Villafuerte & Negro (1998). Feathers were photographed on a white background using the auto colour setting on a Nikon digital camera, at a standard distance (300 mm) and focal length (35 mm), with fluorescent lighting positioned 300 mm above the feathers. Three Munsell colour chips (10YR7/4, 10YR6/6 and 10YR5/4) were placed beside all the photographed feathers. Raw images (NEF format) were opened in Adobe Photoshop 7.0 using the Camera Raw 3.7 plug-in. The magic wand tool in Photoshop was used to select each colour chip, and mean red, green and blue (RGB) values were obtained for each. These were plotted against theoretical RGB values for each chip obtained using Munsell Conversion v. 7.0.1, to calculate a linear regression for each primary colour. This was then used to shift colour values in each photograph, to permit comparison between images. Feather colours were measured by selecting a bright portion of the mid-barb towards the distal end of the feather.
(e). Reconstruction of moa plumage
Standardized digital photographs of moa feathers were opened in Adobe Photoshop 7.0, and the polygonal lasso tool was used to select around the edge of each feather. The clipped feathers were copied as a new layer onto a blank canvas, and the layer was duplicated multiple times to create copies of each feather. Each new layer was then moved so that neighbouring feathers were slightly overlapping, in order to reconstruct the plumage.
3. Results
(a). Ancient DNA from the calamus of subfossil feathers
We successfully amplified and sequenced 31–361 bp of the mtDNA control region from the calamus samples of seven moa feathers (electronic supplementary material, tables S1 and S3). A single unambiguous sequence was obtained from each of the seven feathers. Extractions from the remaining two moa feathers failed to amplify, despite attempts using multiple primer combinations.
The percentage uncorrected sequence divergences for pairwise comparisons of moa taxa for the 31, 180 and 205 bp control region sequences ranged approximately between 3–35%, 3–14% and 5–21%, respectively, demonstrating that there is enough genetic variation within each fragment length to distinguish the various moa species. All seven feather sequences were between 98 and 100 per cent identical to a reference moa sequence, and four moa species were identified: upland moa (Megalapteryx didinus, n = 4), South Island giant moa (Dinornis robustus, n = 1), stout-legged moa (Euryapteryx gravis, n = 1) and heavy-footed moa (Pachyornis elephantopus, n = 1). The species identifications from the moa feathers matched the known Holocene distributions of these taxa based on the analysis of fossil bone (Worthy 1998) and coprolite (Wood et al. 2008) deposits.
(b). DNA from the rachis and barbs of feathers
It was also possible to amplify and sequence mtDNA from the combined rachis and barbs of moa feathers (excluding the calamus) (electronic supplementary material, tables S1 and S3). We amplified and sequenced 31–180 bp of mtDNA control region from four rachis/barb samples of ten moa feathers from Sawers’ rockshelter. All four moa sequences were 98–100% identical to a reference moa sequence. Three moa species were identified: South Island giant moa (n = 1), heavy-footed moa (n = 1) and stout-legged moa (n = 2). For the emu feathers, mtDNA was found to be amplifiable from both the rachis and isolated barbs, respectively. For all three emu feathers, a 108 bp fragment of 12S was amplified and sequenced from each of the four sections (figure 1, electronic supplementary material, table S1). A single unambiguous sequence was obtained from all 12 PCR products, which matched 100 per cent to emu sequences on GenBank (e.g. AF338711.1). There were no PCR products in the extraction and PCR negatives. It is important to note that the precise location of the mtDNA within the feather structure (e.g. within the keratin cells of the rachis and barbs) was not precisely determined. There is also the possibility that despite the bleach treatment, exogenous DNA on the feather might contribute a significant part of the amplifiable DNA. A similar possibility exists for analogous studies of ancient DNA from mammoth (Gilbert et al. 2007) and thylacine hair (Miller et al. 2009), and historical and modern DNA from reptile (Fetzner 1999; Feldman & Spicer 2002) and fish scales (Yue & Orban 2001).
(c). Preservation of colour in subfossil feathers
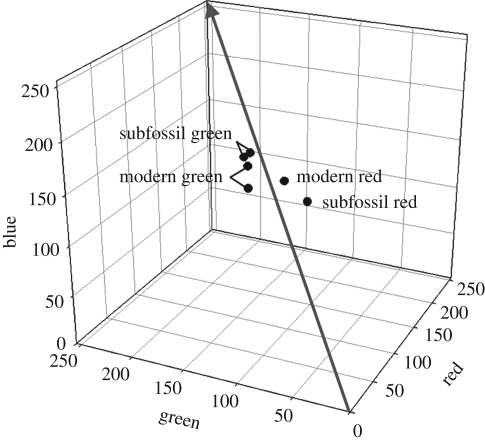
The digital colour comparison illustrated that the colour of subfossil red-crowned parakeet feathers reflected accurately that of the modern feathers (figure 2) indicating that any fading of the subfossil feathers was minor. For green contour feathers, the variation between modern specimens was greater than that observed between some modern and subfossil specimens. We assumed that the colours of moa feathers from the same deposits are also likely to be relatively unmodified, and therefore provide accurate data for plumage reconstructions.
Figure 2.
Quantification of the amount of colour fading in subfossil red facial and green contour feathers of red-crowned parakeet (C. novaezelandiae novaezelandiae) from the Late Holocene Roxburgh Gorge rockshelter B with modern red-crowned parakeet feathers in RGB colour space. The graph indicates that the amount of colour fading in subfossil parakeet feathers is minimal. The colours of moa feathers from the same deposits are also likely to be relatively unmodified.
(d). Reconstruction of moa plumage
Two different morphological and colour types were apparent among the moa feathers (figure 3). The first, present in South Island giant moa, stout-legged moa, upland moa and some heavy-footed moa consisted of slender, medium to long, single or double shafted feathers. These feathers were tan to light brown at the base, grading into dark brown to black at the tip (figure 3a). They were suggestive of a relatively plain, but slightly streaky plumage (figure 4d) similar to brown kiwi (Apteryx australis) which has similarly patterned feathers (figure 4a). The second feather type was present in heavy-footed moa and was a short feather that was dark brown to black for the basal two-thirds and white at the tip (figure 3b). In situ, these feathers would create a speckled plumage pattern (figure 4e, f). Similar-patterned light-tipped feathers are seen in great-spotted kiwi (Apteryx haastii) and little-spotted kiwi, Apteryx oweni (figure 4b,c).
Figure 3.
Characteristic morphology and colour of moa feathers identified from ancient DNA sequences. (a) Feathers identified as upland moa (M. didinus), South Island giant moa (D. robustus), stout-legged moa (E. gravis) and heavy-footed moa (P. elephantopus) exhibited overlapping morphology and colour. The three best examples are shown. From left to right: upland moa (OM Av10793.1), upland moa (OM Av10791.1), South Island giant moa (OM Av10793.2). (b) White-tipped feather (A 06.49.18) identified as heavy-footed moa. Scale bar, 10 mm.
Figure 4.
(a) Plumage of brown kiwi (A. australis); (b) plumage of great-spotted kiwi (A. haastii); (c) plumage of little-spotted kiwi (A. oweni); (d) reconstruction of upland moa (M. didinus), South Island giant moa (D. robustus), stout-legged moa (E. gravis) and heavy-footed moa (P. elephantopus) plumage based on a dark feather; (e and f) reconstruction of upland moa and heavy footed moa plumage based on white-tipped feathers, (e) densely and (f) sparsely spaced.
4. Discussion
The discovery that mtDNA can be amplified from isolated subfossil feathers has allowed the identification of four species of New Zealand moa and provided some important information about the plumage characteristics of this extinct group. Previous studies of historical museum feather specimens (Payne & Sorenson 2002; Sefc et al. 2003; Horváth et al. 2005) have been able to recover genetic information, but we have demonstrated that with a suitably stringent approach (appropriate protocols, targeted primer design for short fragments and low-contamination facilities), it is possible to obtain taxonomically informative sequences both from subfossil feathers and notably from parts of feathers not considered previously to be of use for genetic analysis. There appears to be great potential for genetic studies of birds, both extinct and extant, using feathers where the calamus is not preserved or present, such as from subfossil sites or discarded material in nests. The ability to use the distal portions of feathers also has considerable implications for genetic research on museum specimens, as destructive sampling could be minimized and soft tissue samples such as toe pads could be left intact.
In order to use subfossil feathers to reconstruct the external appearance of extinct avian taxa, we must be confident that the amount of colour fading is minimal. The results reported here show that the colour of subfossil feathers has not faded significantly in the relatively cool, dry conditions of some New Zealand rockshelters. This is potentially aided by the protection from UV exposure in sediments. The genetic identification of four species of moa using isolated feathers provides unique insights into the appearance of these species. Perhaps the most striking is that some heavy-footed moa appear to have had a speckled appearance (figures 3b and 4e,f). This pattern (figure 3b) has been shown previously to be characteristic only for upland moa based on feathers attached in situ to the mummified remains of this species (Hamilton 1894). The results reported here suggest that speckled patterning may have been present in both the heavy-footed and upland moa. In contrast to the situation above, the other feather type analysed in this study was similar in upland moa, South Island giant moa, stout-legged moa and some heavy-footed moa, indicating a plain or slightly streaky appearance (figures 3a and 4d). These feathers could not be separated on either morphological or colour characteristics. This also suggests that upland moa and heavy-footed moa had feathers of differing morphology and colour on different parts of the body or that they might have varied between sexes. This variation among species and plumage patterns would not have been revealed without genetic data, and the analysis of further samples are likely to lead to improved reconstructions. It may also be possible to reconstruct more accurate representations of moa plumage by comparing moa feathers with other extant palaeognathus taxa with differing feather morphology and colour such as the emu (patterned) and cassowary (Casuarius spp.) (non-descript). In addition, because of the similarity of moa and kiwi plumage (figure 4), comparisons with known kiwi colour morphs (Morris & Smith 1988) can be made.
It is likely that the convergent colouring of some moa feathers has been driven by selection on plumage to avoid predation by aerial predators such as Haasts' eagle (Harpagornis mooreii). This concept is supported by the drab camouflage plumage of several other endemic avian New Zealand taxa with similar terrestrial habits (e.g. species of kiwi and kakapo, Strigops habroptilus). It is also probable that moa plumage differed between open and closed-canopy habitat types, sexes and ages—with more disruptive plumage patterns present in the open habitats. The overlapping feather morphology and colour between the identified moa species also raises the possibility that assortative mating in moa was controlled more by call recognition (especially given suggestions of species differences in vocal tract morphology; Worthy & Holdaway 2002), habitat or other unidentified plumage characteristics.
In addition to DNA from subfossil bone (Baker et al. 2005), coprolites (Poinar et al. 1998; Hofreiter et al. 2000), hair (Gilbert et al. 2007) and sediment (Willerslev et al. 2003), ancient DNA from subfossil feathers offer insights into the presence and absence of species on temporal scales. For example, the taxonomic information from moa coprolites (Wood et al. 2008) and subfossil feathers from the same horizons in Roxburgh Gorge B and Sawers' rockshelters indicate that multiple species of moa are present, raising the possibility of competition between moa for prime rockshelter and nesting sites. Interestingly, the success rate for extraction of amplifiable ancient DNA from subfossil moa feathers is much higher than coprolites excavated from the same horizons (4 of 21 coprolites versus 11 of 19 feathers from Central Otago rockshelters had amplifiable ancient DNA; Wood et al. 2008). Furthermore, the size range of amplifiable mtDNA fragments from subfossil feathers (31–205 bp from the calamus, 31–180 bp from distal portions) is comparable to hair (60–130 bp; Gilbert et al. 2007) and coprolites (11–273 bp; Poinar et al. 1998; Hofreiter et al. 2000; Wood et al. 2008), depending on the age and preservation conditions of samples.
Our findings suggest that preserved subfossil feathers from extinct avian taxa in sites around the world are a major potential resource for multi-disciplinary research. Semi-arid sites in Antarctica (Emslie & Patterson 2007), South America (Paabo 1989; Poinar et al. 1998; Hofreiter et al. 2000; authors' personal observation, 2008), North America (Borson et al. 1998; Gilbert et al. 2008), Siberia (Stone 2002; Gilbert et al. 2007), Europe (Loreille et al. 2001) and New Zealand (Worthy & Holdaway 2002; Wood et al. 2008) have preserved bone, skin, fur, hair, coprolites and feathers. In New Zealand, Late Holocene feathers attributed to the extinct Finsch's duck (Chenonetta finschi) have been found in a range of rockshelter deposits. The potential also exists that key enigmatic extinct New Zealand taxa such as the adzebill (Aptornis spp.) and giant goose (Cnemiornis spp.) may also have preserved feathers in subfossil deposits. Until now, subfossil feathers from these palaeontological and archaeological contexts (Borson et al. 1998; Emslie & Patterson 2007) have not been analysed for ancient DNA and this represents an important new area of research.
Subfossil and museum feathers represent a valuable untapped resource of genetic information. In addition, the presence of DNA in the rachis and barbs of moa and modern emu feathers suggests that multiple parts of a feather may contain DNA, providing a method for genetic analysis requiring minimal destruction of valuable specimens. The combination of ancient DNA approaches with the use of appropriate tests for colour fading such as that illustrated here promises to improve our reconstructions of extinct avian taxa.
Acknowledgements
We are very grateful to the following: Otago Museum (Cody Fraser) and the Alexandra Museum (Brian Patrick) for allowing sampling of moa feathers; members of the Australian Centre for Ancient DNA, especially Jessica Metcalf and Jeremy Austin for extensive laboratory and editing advice; Trevor Worthy for discussions on moa and provision of samples and the Australian Research Council and the New Zealand Foundation for Research, Science and Technology for financial support.
References
- Anderson A.1989Prodigious birds Cambridge, UK: Cambridge University Press [Google Scholar]
- Baker A. J., Huynen L. J., Haddrath O., Miller C. D., Lambert D. M.2005Reconstructing the tempo and mode of evolution in an extinct clade of birds with ancient DNA: the giant moas of New Zealand. Proc. Natl Acad. Sci. USA 102, 8257–8262 (doi:10.1073/pnas.0409435102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borson N., Berdan F., Stark E., States J., Wettstein P. J.1998Origin of an Anasazi scarlet macaw feather artifact. Am. Antiquity 63, 131–142 (doi:10.2307/2694780) [Google Scholar]
- Bunce M., Worthy T. H., Ford T., Hoppitt W., Willerslev E., Drummond A., Cooper A.2003Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 425, 172–175 (doi:10.1038/nature01871) [DOI] [PubMed] [Google Scholar]
- Cooper A., Poinar H. N.2000Ancient DNA: do it right or not at all. Science 289, 1139 (doi:10.1126/science.289.5482.1139b) [DOI] [PubMed] [Google Scholar]
- Cooper A., Mourer-Chauvire C., Chambers G. K., von Haeseler A., Wilson A. C., Paabo S.1992Independent origins of New Zealand moas and kiwis. Proc. Natl Acad. Sci. USA 89, 8741–8744 (doi:10.1073/pnas.89.18.8741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A., Fox-Lalueza C., Anderson A., Rambaut A., Austin J., Ward R.2001Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409, 704–707 (doi:10.1038/35055536) [DOI] [PubMed] [Google Scholar]
- Emslie S. D., Patterson W.2007Abrupt recent shift in delta13C and delta15N values in Adelie penguin eggshell in Antarctica. Proc. Natl Acad. Sci. USA 104, 11 666–11 669 (doi:10.1073/pnas.0608477104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman C. R., Spicer G. S.2002Mitochondrial variation in sharp-tailed snakes (Contia tenuis): evidence of a cryptic species. J. Herpetol. 36, 648–655 [Google Scholar]
- Fetzner J. W.1999Extracting high quality DNA from shed reptile skin: a simplified method. Biotechniques 26, 1052–1054 [DOI] [PubMed] [Google Scholar]
- Flannery T., Schouten P.2001A gap in nature: discovering the world's extinct animals Australia: The Text Publishing Company [Google Scholar]
- Forrest R. M.1987A partially mummified skeleton of Anomalopteryx didiformis from Southland. J. R. Soc. N. Z. 17, 399–408 [Google Scholar]
- Gilbert M. T. P., et al. 2007Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science 317, 1927–1930 (doi:10.1126/science.1146971) [DOI] [PubMed] [Google Scholar]
- Gilbert M. T. P., et al. 2008DNA from pre-clovis coprolites in Oregon, North America. Science 320, 786–789 (doi:10.1126/science.1154116) [DOI] [PubMed] [Google Scholar]
- Gill B., Martinson P.1991New Zealand's extinct birds New Zealand: Random Century [Google Scholar]
- Hamilton A.1894On the feathers of a small species of moa (Megalapteryx) found in a cave at the head of the Waikaia River, with a notice of a moa-hunters camping place on the Old Man Range. Trans. Proc. N. Z. Inst. 27, 232–238 [Google Scholar]
- Hofreiter M., Poinar H. N., Spaulding W. G., Bauer K., Martin P. S., Possnert G., Paabo S.2000A molecular analysis of ground sloth diet through the last glaciation. Mol. Ecol. 9, 1975–1984 (doi:10.1046/j.1365-294X.2000.01106.x) [DOI] [PubMed] [Google Scholar]
- Horváth M. B., Martínez-Cruz B., Negro J. J., Kalmár L., Godoy J. A.2005An overlooked DNA source for non-invasive genetic analysis in birds. J. Avian Biol. 36, 84–88 (doi:10.1111/j.0908-8857.2005.03370.x) [Google Scholar]
- Hutton F. W., Coughtrey M.1875Description of some moa remains from the Knobby Ranges. With anatomical notes. Trans. Proc. N. Z. Inst. 7, 266–273 [Google Scholar]
- Huynen L., Miller C. D., Scofield R. P., Lambert D. M.2003Nuclear DNA sequences detect species limits in ancient moa. Nature 425, 175–178 (doi:10.1038/nature01838) [DOI] [PubMed] [Google Scholar]
- Huynen L., Lissone I., Sawyer S., Lambert D.2008Genetic identification of moa remains recovered from Tiniroto, Gisborne. J. R. Soc. N. Z. 38, 231–235 [Google Scholar]
- Kumar S., Tamura K., Nei M.2004MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (doi:10.1093/bib/5.2.150) [DOI] [PubMed] [Google Scholar]
- Lalueza-Fox C., et al. 2007A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science 318, 1453–1455 (doi:10.1126/science.1147417) [DOI] [PubMed] [Google Scholar]
- Lambert D. M., Baker A., Huynen L., Haddrath O., Herbert P. D. N., Miller C. D.2005Is a large-scale DNA-based inventory of ancient life possible? J. Hered. 96, 279–284 (doi:10.1093/jhered/esi035) [DOI] [PubMed] [Google Scholar]
- Loreille O., Roumat E., Verneau O., Bouchet F., Hänni C.2001Ancient DNA from Ascaris: extraction amplification and sequences from eggs collected in coprolites. Int. J. Parasitol. 31, 1101–1106 (doi:10.1016/S0020-7519(01)00214-4) [DOI] [PubMed] [Google Scholar]
- Ludwig A., et al. 2009Coat colour variation at the beginning of horse domestication. Science 324, 485 (doi:10.1126/science.1172750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W., et al. 2009The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Res. 19, 213–220 (doi:10.1101/gr.082628.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R., Smith H.1988Wild south: saving New Zealand's endangered birds TVNZ in Association with Century Hutchinson: New Zealand [Google Scholar]
- Murray P., Vickers-Rich P.2003Magnificent mihirungs: the colossal flightless birds of the Australian dreamtime USA: Indiana University Press [Google Scholar]
- Oliver W.1955New Zealand birds, 2nd edn Wellington, New Zealand: Reed [Google Scholar]
- Paabo S.1989Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl Acad. Sci. USA 86, 1939–1943 (doi:10.1073/pnas.86.6.1939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R. B., Sorenson M. D.2002Museum collections as sources of genetic data. Bonner Zoologische Beitrage 51, 97–104 [Google Scholar]
- Poinar H. N., Hofreiter M., Spaulding W. G., Martin P. S., Stankiewicz B. A., Bland H., Evershed R. P., Possnert G., Paabo S.1998Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 218, 402–406 (doi:10.1126/science.281.5375.402) [DOI] [PubMed] [Google Scholar]
- Rompler H., Rohland N., Lalueza-Fox C., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M.2006Nuclear gene indicates coat-colour polymorphism in mammoths. Science 313, 62 (doi:10.1126/science.1128994) [DOI] [PubMed] [Google Scholar]
- Sefc K. M., Payne R. B., Sorenson M. D.2003Microsatellite amplification from museum feather samples: effects of fragment size and template concentration on genotyping errors. Auk 120, 982–989 (doi:10.1642/0004-8038(2003)120[0982:MAFMFS]2.0.CO;2) [Google Scholar]
- Stone R.2002Mammoth: the resurrection of an ice age giant New York, NY: Basic Books [Google Scholar]
- Swofford D. L.2000PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4 Sunderland, MA: Sinauer Associates [Google Scholar]
- Tennyson A., Martinson P.2006Extinct birds of New Zealand Wellington, New Zealand: Te Papa Press [Google Scholar]
- Vickers-Rich P., van Tets G. F., Knight F.1985Kadimakara: extinct vertebrates of Australia USA: Princeton University Press [Google Scholar]
- Vickers-Rich P., Trusler P., Rowley M. J., Cooper A., Chambers G. K., Bock W. J., Millener P. R., Worthy T. H., Yaldwyn J. C.1995Morphology, myology, collagen and DNA of a mummified moa, Megalapteryx didinus (Aves: Dinornithiformes) from New Zealand. Tuhinga Rec. Mus. N. Z. Te Papa Tongarewa 4, 1–26 [Google Scholar]
- Villafuerte R., Negro J. J.1998Digital imaging for colour measurement in ecological research. Ecol. Lett. 1, 151–154 (doi:10.1046/j.1461-0248.1998.00034.x) [Google Scholar]
- Vinther J., Briggs D. E. G., Prum R. O., Saranathan V.2008The colour of fossil feathers. Biol. Lett. 4, 522–525 (doi:10.1098/rsbl.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.1885Remarks of the feathers of two species of moa. Trans. Proc. N. Z. Inst. 18, 83–84 [Google Scholar]
- Willerslev E., Hanson A. J., Binladen J., Brand T. B., Gilbert M. T. P., Shapiro D., Bunce M., Wuif C., Gilichinsky D. A., Cooper A.2003Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795 (doi:10.1126/science.1084114) [DOI] [PubMed] [Google Scholar]
- Wilmshurst J. M., Anderson A. J., Higham T. F. G., Worthy T. H.2008Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676–7680 (doi:10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.2008Moa (Aves: Dinornithiformes) nesting material from rockshelters in the semi-arid interior of South Island, New Zealand. J. R. Soc. N. Z. 38, 115–129 [Google Scholar]
- Wood J. R., Rawlence N. J., Rogers G. M., Austin J. J., Worthy T. H., Cooper A.2008Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quat. Sci. Rev. 27, 2593–2602 (doi:10.1016/j.quascirev.2008.09.019) [Google Scholar]
- Worthy T. H.1989Mummified moa remains from Mt Owen, northwest Nelson. Notornis 36, 36–38 [Google Scholar]
- Worthy T. H.1998Quaternary fossil faunas of Otago, South Island, New Zealand. J. R. Soc. N. Z. 28, 421–521 [Google Scholar]
- Worthy T. H., Holdaway R. N.2002The lost world of the moa Christchurch, New Zealand: Canterbury University Press [Google Scholar]
- Yue G. H., Orban L.2001Rapid isolation of DNA from fresh and preserved fish scales for polymerase chain reaction. Mar. Biotechnol. 3, 199–204 (doi:10.1007/s10126-001-0010-9) [DOI] [PubMed] [Google Scholar]