Abstract
Dispersal is a fundamental process in ecology because it influences the dynamics, genetic structure and persistence of populations. Furthermore, understanding the evolutionary causes of dispersal pattern, particularly when they differ between genders, is still a major question in evolutionary ecology. Using a panel of 10 microsatellite loci, we investigated at different spatial scales the genetic structure and the sex-specific dispersal patterns in the common vole Microtus arvalis, a small colonial mammal. This study was conducted in an intensive agricultural area of western France. Hierarchical FST analyses, relatedness and assignment tests suggested (i) that females are strongly kin-clustered within colonies; (ii) that dispersal is strongly male-biased at a local scale; and (iii) long-distance dispersal is not rare and more balanced between genders. We conclude that males migrate continuously from colony to colony to reproduce, whereas females may disperse just once and would be mainly involved in new colony foundation.
Keywords: genetic analyses, dispersal distances, natal and breeding dispersal, colonization, female kin clusters, Microtus arvalis
1. Introduction
Dispersal refers to the movement of an organism from its birth place to its first breeding site (natal dispersal) or from one breeding site to another (breeding dispersal). Dispersal plays a fundamental role in population biology and conservation because it influences the dynamics, the genetic structure and the persistence of populations (Clobert et al. 2001). From a metapopulation perspective, two types of dispersive movement are distinguished: colonization (i.e. settlement in an empty patch) and migration sensu stricto (when an individual immigrates into an occupied patch). These two dispersive behaviours differ in their respective consequences for the metapopulation dynamics (Hanski & Gaggiotti 2004).
Birds and mammals have long been recognized to exhibit strong gender-biased dispersal patterns. In mammals for instance, males usually disperse more frequently than females (Greenwood 1980), and are assumed to be the dispersing sex while females are the colonizing sex (Kerth & Petit 2005). Much recent research has focused on understanding the determinants of sex-biased dispersal (for a recent review, see Lawson Handley & Perrin 2007), and four alternative hypotheses were stressed to account for sex-biased dispersal: avoidance of inbreeding, local mate competition, local resource competition and cooperative behaviour among kin. Dispersal is sex-biased when the balance among these evolutionary forces differs between genders. The direction of sex-biased dispersal is in turn influenced by the mating system (Perrin & Goudet 2001). In particular, when males defend female resources and compete for access to females, dispersal is expected to be male-biased because females benefit more than males from staying on their natal patch (Greenwood 1980).
Dispersal may vary in rate (the proportion of philopatric versus dispersing individuals), distance between two successive settlements and timing (natal versus breeding dispersal). All three aspects may have evolved independently or in concert and may be controlled by different or related proximate factors (Ims & Hjermann 2001). Although most studies so far focused only on dispersal rates, dispersal distances may also differ between genders and provide interesting clues to ultimate causes of sex-biased dispersal (Murrell et al. 2002; Rousset & Gandon 2002) because the reasons for long-distance and short-distance dispersal are likely to be very different (Ronce et al. 2001). Short-distance dispersal is probably sufficient for avoiding inbreeding, whereas dispersal over a longer distance might function to colonize new territory or escape crowding (Lawson Handley & Perrin 2007). In turn, knowledge on the timing of dispersal, whether an animal disperses or remains philopatric and if it disperses, whether it disperses once in its life (i.e. natal dispersal followed by breeding site fidelity) or displays secondary dispersal (i.e. breeding dispersal), should be informative for understanding ultimate and proximate causes of dispersal. Therefore, all three aspects of dispersal (rate, distance and timing) are likely to vary with sex, but to our knowledge, very few studies have actually investigated differences between sexes in dispersal distances (Fontanillas et al. 2004; Douadi et al. 2007), and even less have addressed the issue of sex differences in the timing of dispersal. The development of indirect, genetic methods over the last few years yet gives access to these poorly known aspects of dispersal (Lawson Handley & Perrin 2007).
In this study, we investigated the sex-specific dispersal pattern in the common vole, Microtus arvalis. This Eurasian rodent lives in subterraneous burrows in open agricultural land, grazed pastures and meadows and breeds in colonies (Mitchell-Jones et al. 1999). In agricultural landscapes, colonies are usually distributed in patches owing to the fragmented nature of landscape as well as the heterogeneous distribution of permanent or semi-permanent habitats (i.e. grasslands). Observation of groups of breeding females in burrows indicates a polygynous or promiscuous mating system (Boyce & Boyce 1988a) consistent with the male-biased dispersal patterns reported for other similar Microtus species (e.g. Aars & Ims 2000). However, information on the dispersal pattern is incomplete for the common vole and somewhat contradictory: genetic analyses at a large geographical scale suggest male-biased gene flow (Hamilton et al. 2005), whereas local instantaneous measures of immigration rates are not significantly higher for males than for females (Schweizer et al. 2007). Analyses of genetic structures that were carried out in different parts of its distribution range revealed a structured pattern at a fine spatial scale (Schweizer et al. 2007) on the one hand, but a weakly structured pattern at a large spatial scale (Gauffre et al. 2008) on the other. Our poor understanding of dispersal patterns in natural populations of common vole contrasts sharply with the huge amount of experimental work and knowledge cumulated about dispersal cues on common vole and other related Microtus species (Stenseth & Lidicker 1992).
We thus aimed at describing accurately and quantitatively the dispersal pattern (rate, distance and timing) of both sexes in a natural common vole population, using molecular tools. To this end, we performed a hierarchical sampling design: most individuals within colonies, several colonies per locality and several localities in the landscape. This spatially explicit design allowed us (i) to explore the sex-specific pattern of genetic variation at the different spatial scales at which we suspected dispersal to occur and (ii) to use recently developed state- and sex-specific hierarchical analysis of the genetic variation (Fontanillas et al. 2004) to infer sex-specific dispersal rates at the different spatial scales. Our goal was to determine the evolutionary forces driving these sex-specific dispersal patterns. Moreover, we hypothesize that this knowledge could help to resolve the apparent paradox of contrasting dispersal patterns gained from different and independent studies for the common vole.
2. Material and methods
(a). Study area
The study was conducted in central-western France (Région Poitou-Charentes, 46°11′ N, 0°28′ W) in a 500 km2 intensive agricultural area. A full description of the main habitats comprising the agricultural matrix and of the intensity and timing of agricultural practices is given in Gauffre et al. (2008). Currently, the area is dominated by agricultural plots of cereals, colza and spring-sown crops that are annually disturbed by ploughing, which causes the destruction of vole colonies. Meadows, alfalfa plots and edge habitats such as floral trips support lower disturbance (no yearly ploughing), but account for less than 20 per cent of the surface area. Long-term surveys conducted within the study area, however, show that common voles are abundant in all these habitats (Salamolard et al. 2000).
Sampling was conducted in 10 localities from May to July 2007. Each locality corresponded from a single up to three agricultural plot(s) of meadow or alfalfa. These plots are annually mowed in spring and summer. Mowing does not destroy nest sites (common voles build and inhabit subterraneous burrows) but eliminates part of the local population by direct killing or increased risk of predation (data not shown). In this species, no dispersive movement appears to be associated with mowing or any other agricultural disturbance (Jacob & Hempel 2003).
(b). Sampling strategy
Two sampling strategies were conducted in parallel within each of the 10 localities. First, colony sampling consisted of trapping voles directly into the colonies using baited traps placed in front of the burrows. Second, trap line sampling was performed using 100 m-long trap lines without baits (51 individual traps spaced every 2 m) placed at random within the localities and surveyed for 24 h. The number of captures from each of the two sampling strategies and their use for the different statistical analyses are displayed in table 1.
Table 1.
Details of the two samplings and their use for the different statistical analyses.
| statistical analyses |
||||||
|---|---|---|---|---|---|---|
| sampling | stage | number trapped | sex ratio | relatedness | hierarchical F-stat | assignment |
| trap lines | adults | 174 | 0.55 | x | ||
| juveniles | 36 | 0.44 | x | |||
| colonies | adults | 222 | 0.45 | x | x | x |
| juveniles | 79 | 0.49 | x | x | x | |
The 10 localities were on average separated by 12.7 km (1.5–23.5 km). At the time of the sampling, populations were patchily distributed in the landscape, and the different localities (even when separated by short distances) corresponded to separated aggregates of colonies. Within the 10 localities, a total of 105 colonies were sampled. Colonies were located using a GPS system with a precision of 5 m. Colonies sampled in the same locality were on average separated by 175 m (min. 15 m, max. 600 m, median 72 m). Each colony corresponds to subterraneous burrows with several entrances connected by an intense network of trails in surface. We considered that two colonies were different (even when separated by short distances) when their networks were not connected. Animals were weighed, sexed and their reproductive status checked. A very small piece of ear (less than 1 mm2) was collected for genetic analyses. Individuals heavier than 14 g were considered as adults.
(c). DNA conservation and genotyping
Samples were stored in 95 per cent ethanol. DNA extraction was carried out using a standard Chelex protocol. DNA was amplified using the polymerase chain reaction and genotyped with a multiplex panel of 10 microsatellites (Gauffre et al. 2008) in a genotyper ABI PRISM 310 Genetic Analyser (Applied Biosystems). Genotypes were determined using Genescan and Genotyper (Applied Biosystems). All voles were sexed using molecular typing (Bryja & Konecny 2003).
(d). Statistical analyses
(i). Estimating sex-related differences in local dispersal
Sex- and age-related differences in dispersal rates and timing were first investigated at the local scale (i.e. within localities) using the colony sampling (table 1). We performed relatedness analyses at (i) the intracolony scale (i.e. individuals trapped in the same colony) and (ii) the locality scale (excluding pairs from the same colony). At the colony scale, the mean pairwise relatedness is expected to be higher among individuals of the philopatric and less dispersing sex than between those of the most dispersing sex, because the latter, by definition, are less likely to remain in the vicinity of their relatives. At larger scales, differences in mean relatedness should cancel out.
We applied the program SPAGeDI 1.2.f (Hardy & Vekemans 2002) to calculate interindividual relatedness using the Li coefficient corrected for sample size. This estimator shows a high accuracy (low bias) combined with a high precision (low variance) compared with others (Vekemans & Hardy 2004). Relatedness estimation relies on reference allelic frequencies, and we used those calculated over the entire dataset for these calculations. Using SPAGeDI, we tested in each category whether individuals were more related in the colonies than random by permuting, in each locality independently, individuals among colonies. To combine results across localities, we used Fisher's method of combining probabilities by summing −2 ln(p) across datasets. Because −2 ln(p) is χ2 distributed with two degrees of freedom, the sum is χ2 distributed with 2n degrees of freedom, where n is the number of datasets.
Because the pairwise data are non-independent, we could not directly compare relatedness values between the different categories (male versus female, juvenile versus male, etc.) using usual statistical tests (Prugnolle & de Meeus 2002). We thus applied the procedure of Coulon et al. (2006) that consists in generating 1000 random resampling sets without replacement, such that each individual occurs only once in a given resampled set. The difference of mean relatedness Δi between the two categories tested is then calculated for each resampling set. Under the null hypothesis, Δi should follow a normal distribution centred on 0; under the alternative hypothesis (i.e. category-biased dispersal), Δi should be significantly different from 0. We made no a priori on the sign of the difference and thus computed two-sided tests.
(ii). Sex-specific differences in local versus long-distance dispersal
The relative proportion of local versus long-distance dispersal for each gender separately was quantified using the procedure of Fontanillas et al. (2004). The method consists in comparing juvenile (pre-dispersal state) versus adult (post-dispersal state) genetic structures, and local versus large genetic structures, for each sex separately using hierarchical F-statistics. We based this analysis on the colony sampling (table 1). Colonies, the finest hierarchical level, were nested in the localities (superior hierarchical level) and movements between colonies within localities referred to local dispersal, whereas movements between localities referred to long-distance dispersal. At the time of sampling, colonies were patchily distributed in the landscape. Each locality thus corresponded to one such patch. Each locality extended over several hundred square metres, which roughly corresponds to the scale of genetic similarity between individuals as inferred using autocorrelation analyses (Gauffre et al. 2008). The genetic method of Fontanillas et al. (2004) requires sampling to be as exhaustive as possible at the most local scales. We therefore tried to sample all individuals within each colony and all the colonies within a locality. However, because the signal used in this method is the contrast in gene frequencies between pre- and post-dispersal individuals, we excluded individuals weighing between 12 and 16 g because they are a mix of pre- and post-dispersal individuals (they represent noise in statistical terms). We also excluded colonies in which only one individual was trapped. This amounts to 235 common voles from 70 colonies (on average, seven colonies per locality, range 4–12), including 95 adult females (F), 85 adult males (M) and 55 juveniles (J). Within each colony, an average of 1.34 J (range 0–5), 1.95 F (range 0–7) and 1.57 M (range 0–9) were trapped.
The following notations were used: d1 refers to the dispersal rate between colonies within localities (short-distance dispersal), whereas d2 refers to the dispersal rate between localities (long-distance dispersal); Loc stands for locality; Col refers to colony and Tot to total. Accordingly, FLocTot is the correlation of genes within locality relative to the total, and FColLoc is the correlation of genes within colony relative to locality. When computed in a fully hierarchical setting, FLocTot and FColLoc deliver an estimate for d1 + d2. FLocTot can also be computed without taking into account the colony level (non-hierarchical setting). This yields an estimate for d2 (Fontanillas et al. 2004) and will thus be referred to as FST. The 95 per cent confidence intervals (95% CIs) of the different F-statistics were obtained by bootstrapping over loci. Bootstrapped variance components were also used to compute 95 per cent confidence intervals for dispersal rates. Calculations were performed with the package HierFstat (Goudet 2005) for the statistical software R 2.7.1 (R Development Core Team 2008).
(iii). Detection of first-generation immigrants
Finally, we evaluated the proportion of first-generation immigrants among adults within each locality, and for each gender separately, using assignment tests. Both samplings (colony and trap lines) were used for this purpose (table 1). For each individual, we computed its likelihood of belonging to the population where it was sampled (i.e. the Lh statistic) as recommended, when all sources for immigrants have not necessarily been sampled (Paetkau et al. 2004), using the frequency method of Paetkau et al. (1995). The probability of an individual being resident was then assessed using a Monte Carlo resampling procedure. Computations were performed using the program GeneClass 2.0 (Piry et al. 2004). Individuals with a probability of less than 0.05 were excluded as resident. The difference between males and females in the number of migrants was tested using Fisher's exact tests.
3. Results
A total of 511 common voles was caught (table 1), 59 per cent of which came from colony-trapping scheme; overall, 77 per cent of the trapped animals were adults (73% in the colony sampling and 82% in the trap lines sampling). The adult sex ratio (% of males) of caught animals was female-biased in the colony sampling (0.45, n = 174), but male-biased in the trap line sampling (0.55, n = 222), a difference that was just significant (χ2 test, p = 0.045). In addition, the proportion of juveniles was significantly higher in the colony sampling (χ2 test, p = 0.019).
(a). Sex- and age-related local dispersal: comparison within and between breeding colonies
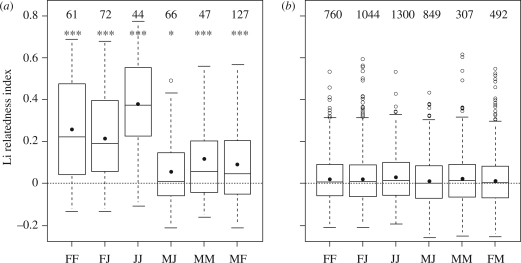
Across the 10 localities, the relatedness among and between M, F and J (figure 1) showed strong variations within colonies (from 0.38 for relatedness among J to 0.07 for M–J relatedness), while it was homogeneous and low at the intercolonies scale (from 0.03 for relatedness among J to 0.011 for M–J relatedness). The combined probabilities of the permutation tests (performed in each locality independently) indicated that whatever the category considered, pairs of individuals originating from the same colony were significantly more related than random (figure 1). However details of these permutation tests showed that in most localities, M were not more related to each other or to J than random (tests were significant at 5% level in 2/7 localities for M, 3/10 for M–J pairs and 4–10 for M–F pairs). Conversely, in almost all localities, F and J were more related in the colonies than expected at random (tests were significant at 5% level in 8/10 localities for F, all for J and 7/9 for F–J pairs). Social structure and sex-biased dispersal were then investigated by comparing within-colony pairwise relatedness for different sex and age categories. Results of these tests indicate that F were significantly more related than M (mean relatedness among F = 0.26, mean relatedness among M = 0.12, p < 0.001) and more related to J than M (mean F–J relatedness = 0.21, mean M–J relatedness = 0.06, p < 0.001).
Figure 1.
Boxplots showing pairwise relatedness values (Li relatedness index, Li et al. 1993) among adult females (FF), between adult females and juveniles (FJ), among juveniles (JJ), between adult males and juveniles (MJ), among adult males (MM) and between adult females and adult males (MF), (a) trapped in the same colony and (b) trapped in the same locality, but not the same colony. Boxes represent the interquartile range, bars within boxes are median values and whiskers indicate the 10th and 90th percentiles. Zero line indicates the theoretical pairwise relatedness value between two individuals taken at random in the population. Dots indicate mean value. Numbers of pairwise data are also indicated. In the first graph, the combined p-value of the permutations tests of individuals among colonies is also provided (*p < 0.05; ***p < 0.001).
(b). Sex-specific differences in local versus long-distance dispersal
The fully hierarchical analysis indicated that the colony level explained the major part of molecular variation. The genetic differentiation was lower between localities (FLocTot = 0.018 and 0.011 for M and F, respectively) than between colonies among localities (FColLoc = 0.047 and 0.097 for M and F, respectively) (table 2). The non-hierarchical analysis showed that FST values (differentiation among localities) did not differ significantly between genders (0.022–0.034 for M and 0.018–0.033 for F) and, as a result, d2 (long-distance dispersal) estimators did not differ significantly between sexes (24% for M versus 27% for F). Conversely, when comparing 95 per cent CI, d1 + d2 (total dispersal) appeared significantly male-biased because in the hierarchical analysis, FColLoc values (differentiation among colonies within localities) were significantly higher in females. Consequently, d1 was also significantly male-biased (table 2). Thus, an estimate of 68 per cent of adult males was immigrant, 44 per cent stemming from other colonies within the same locality (local dispersal) and 24 per cent from other localities (long-distance dispersal). In contrast, 28 per cent of adult females were immigrants. Female dispersal among localities (27%) was the only significant parameter, dispersal among colonies inside a locality being close to zero (1%).
Table 2.
F-statistics and associated dispersal rates in M. arvalis In the non-hierarchical analysis, FST quantifies the genetic differentiation among localities. In the hierarchical analysis, FLocTot quantifies the genetic differentiation between localities within the total and FColLoc the differentiation between colonies within localities. Dispersal rate between localities (d2) and total dispersal rate (d1 + d2) are computed from the non-hierarchical and hierarchical sex- and state-specific analyses, respectively. Dispersal rate between colonies within localities (d1) is derived from the previous estimates. Confidence intervals (95%) are in brackets.
| non-hierarchical analysis |
hierarchical analysis |
||||||
|---|---|---|---|---|---|---|---|
| category | n | FST | d2 | FLocTot | FColLoc | d1 + d2 | d1 |
| juveniles | 55 | 0.047 (0.034–0.058) | 0.021 (0.01–0.033) | 0.161 (0.129–0.19) | |||
| males | 85 | 0.027 (0.022–0.034) | 0.24 (0.20–0.24) | 0.022 (0.017–0.027) | 0.036 (0.016–0.062) | 0.68 (0.53–1) | 0.44 (0.29–0.80) |
| females | 95 | 0.025 (0.018–0.033) | 0.27 (0.25–0.27) | 0.013 (0.004–0.022) | 0.085 (0.054–0.113) | 0.28 (0.24–0.35) | 0.01 (−0.01–0.08) |
(c). First-generation immigrants
Assignment tests conducted over the 396 adult common voles allowed the detection of 54 immigrants (34 M and 20 F) at the 0.05 probability level and 17 immigrants (13 M and 4 F) at the 0.01 probability level. Among them, the relative proportion of M was significantly higher (76% at the 0.01 probability level, Fisher's test, p = 0.026; and 63% at the 0.05 probability level, p = 0.04).
4. Discussion
We demonstrated that dispersal patterns are different for males and females in the common vole and that this sex-biased dispersal is scale- and time-dependent. Dispersal rates are strongly male-biased at short distances, whereas long-distance dispersal is not rare and affects males and females in similar proportions. In addition, males exhibit both natal and breeding dispersal.
(a). Using indirect estimates to detect sex-biased patterns of dispersal
Although dispersal rates and distances are among the most important aspects of an organism's life history, gathering quantitative estimates in natural populations is notoriously difficult. Much has been done in recent years to develop population genetic approaches (Lawson Handley & Perrin 2007). Methods relying on uni-parentally inherited markers (i.e. Y chromosomes for males and mtDNA for females in mammals), although theoretically powerful, are not well suited at small geographical scales because of the low genetic polymorphism of the uni-parentally transmitted genome (see Ishibashi et al. 1997; Aars et al. 1998 for voles). At first glance, bi-parentally inherited markers might seem useless because genes transmitted by one sex are redistributed between sexes in the next generation. However, just after the dispersal phase, the genetic structure assessed from individuals of the more dispersing sex should be slightly less pronounced that assessed from individuals of the less dispersing sex, because the next reproduction event has not yet mixed male and female alleles (Goudet et al. 2002; Prugnolle & de Meeus 2002). Within this framework, powerful methods from different theoretical fields have been developed, such as hierarchical F-statistics (Fontanillas et al. 2004), relatedness comparisons (Coulon et al. 2006) and assignation procedures (Paetkau et al. 2004). All three methods were applied in this study, and they gave consistent results. We wish to emphasize that the three methods measure instantaneous dispersal (Prugnolle & de Meeus 2002), i.e. dispersal of the present adult generation. In that sense, they strongly depart from the traditional F-statistics estimates within the Wright–Fisher model, which are influenced by what occurred within both present and past generations. This is particularly important because most vole populations usually display fluctuating or cyclic population dynamics (Lambin et al. 2006). Moreover, even though underlying assumptions of the hierarchical F-statistics, among which is island structure, are unrealistic in many situations, the method has been found to be robust to departures from this hypothesis (Fontanillas et al. 2004). This is also important because vole populations usually display isolation by distance genetic patterns (Berthier et al. 2006; Gauffre et al. 2008).
Altogether, our data revealed that the dispersal rate of males (68%) was twice that of females (28%). The dispersal bias obtained from the different methods cannot, however, be directly compared unless the sex ratio is balanced because the assignment method counts the number of migrants while the F-statistics analyses estimate dispersal rates. Hence, a male-biased sex ratio could also explain why the number of male migrants exceeds the number of female migrants even if dispersal rates were equal in males and females. Our line-trapping procedure showed male-biased sex ratio, whereas the colony trapping revealed a female-biased sex ratio. This can be explained by differences in mobility between sexes at the time of the sampling and illustrates that sex-ratio differences are difficult to measure in nature (Bryja et al. 2005). Interestingly, a method either depending on sex ratio (the detection of first-generation migrants) or not (hierarchical F-statistics) both supported a male-biased dispersal pattern in the common vole. Yet, when taking into account the spatial scale, a different picture emerged. Dispersal was strongly male-biased at a very local scale, while it appeared to be more balanced at a larger scale. In a previous study on sex-specific dispersal distance in shrews (Fontanillas et al. 2004), an opposite dispersal pattern was inferred (dispersal was found to be female-biased at the local scale and equilibrated among sexes at long distance). Mating systems are different in these two species (monogamy for shrews, polygyny or promiscuity for common voles). Therefore, the difference in sex-specific dispersal pattern of these two models is consistent with the hypothesis that mating system influences the direction of sex-biased dispersal.
(b). Female voles cluster with kin and colonize
In the colonies, females and juveniles were strongly related to each other, indicating that females are organized in matrilines (i.e. groups of related breeding females) also called kin clusters. The building of social bonds among members of one sex is known to act against dispersal in this sex, since individuals benefit from kin cooperation to breed, acquire and defend resources (Lawson Handley & Perrin 2007). Thus kin cooperation among females has the potential to induce female-biased philopatry in polygynous or promiscuous mating systems. Sociality within female kin groups has been described in several species, and kin clusters in females seem to be often associated with defence against aggressive unfamiliar conspecifics, in particular against infanticidal males in Microtus species (Agrell et al. 1998; Ebensperger 1998; Le Galliard et al. 2006). However, although suggested from both theoretical and experimental work (see above references), the reality of female kin clusters in natural populations of voles has seldom been demonstrated (but see Ishibashi et al. 1997). Our work bridges this gap between theoretical, experimental and empirical studies and strengthens the view that kin clusters of philopatric females are a basic component of the social structure in natural Microtus populations.
In our study, females appeared to be essentially philopatric with little or no dispersal at the local scale, although long-distance dispersal was not rare. Relatedness analyses suggest a low occurrence of immigration of females in extant colonies because females are highly related to each other and to the juveniles inside the colonies. This is consistent with field experiments, showing that immigrating microtine females face strong aggressive behaviour from residents when attempting to settle in an occupied colony (Smith & Batzli 2006), but also with experimental studies, in which the presence of immigrant females attempting to settle in colonies depressed the weaning success of resident females and their own fitness (Lambin & Krebs 1993; Lambin & Yoccoz 1998). We thus conclude that female dispersal between extant colonies is rare and/or ineffective because it is very difficult for a female to settle down in a patch or colony, which is already occupied by clusters of related females. We suggest that female dispersal should mostly translate into the colonization of empty patches through new colony foundation and probably results from local resource competition among relatives. Hence, it leads necessarily to longer movements than in males, in accordance with theoretical expectations, which predict that distances travelled to colonize new habitats are generally large (Ronce et al. 2001). Perturbations owing to agricultural practices (e.g. ploughing destroys colonies in annual crops, Jacob 2003) generate heterogeneous common vole densities between agricultural plots and many plots with underexploited resources. Given the patchy structure of the agricultural landscape, this allows colonizing females to reach vacant patch easily. Our hypothesis is also supported by Boyce & Boyce (1988b), who argue that common vole females may eventually disperse after their first fecundation to colonize empty habitats without the stress of finding a mate.
(c). Males disperse continuously for mating
Contrasting with females, males appeared highly mobile at the local scale as revealed by a weak genetic differentiation among males at this scale. Relatedness analyses show that (i) males are not related to females from the colony where they were captured, suggesting that they all display natal dispersal and (ii) even more striking, males trapped in a colony at the peak of the breeding season are not the fathers of the progeny present in the colony at the same time. Given the fast lifecycle of this species (gestation lasts three weeks and individuals reach sexual maturity after few weeks of age), the absence of relatedness between males and juveniles suggests intense breeding dispersal by males. In contrast with birds (Paradis et al. 1998), breeding dispersal has hitherto been rarely documented in mammals. In the common vole, evidence for breeding dispersal is in agreement with the idea that males are thought not to maintain territories but to move as so-called ‘floaters’ between several female territories (Stein 1958). No general theory explaining breeding dispersal has yet been formalized (Berteaux & Boutin 2000). Shifting place just after mating with all receptive females (that may have synchronous oestrus within colony, Poikonen et al. 2008) would maximize males fitness and could explain the breeding dispersal behaviour of males without the need to invoke any other cause. Alternatively, one or a few dominant male(s) may be the father(s) of a synchronous cohort in a colony, and if being dominant is detrimental regarding predation risk, the possibility of rapid death of fathers could not be ruled out to explain the absence of relatedness between males and juveniles captured in the same colony. This hypothesis is, however, difficult to accommodate with the fact that males and females have similar predation risks and that dominance seems to provide protection against predation in some vole species (Koivunen et al. 1998; Banks et al. 2000). Male natal and breeding dispersal acting together lead to the avoidance of brother–sister and mother–son matings. Hence, inbreeding avoidance could be seen in this species as a mere consequence of these behaviours. However, the combination of male dispersal and strict female philopatry at the local scale suggests that inbreeding avoidance by itself could foster the male-biased natal dispersal observed in the common vole (Gandon 1999; Perrin & Mazalov 2000; Ronce et al. 2001). Sex-biased dispersal patterns can also result from unbalanced local mate competition, with males dispersing more than females in species with polygynous or promiscuous mating systems because of more intense male–male competition. In line with previous conclusions drawn by examining mating patterns in American microtines (Boonstra et al. 1987), our inferences about dispersal point to a combined role of local mate competition and inbreeding avoidance in the evolution of mating and dispersal patterns in this species. Disentangling their respective roles will require more direct approaches such as manipulative experiments (Cockburn et al. 1985) or inbreeding depression estimation (Duarte et al. 2003).
In agricultural landscapes, cultivation practices may directly affect the ecology of animals and therefore may influence common vole dispersal: first, by generating plots with underexploited resources that can enhance the success of colonizing females (discussed earlier) and second, by inducing mechanically part of the local common vole population to move outside its plot. Because disturbance is homogeneous at the scale of an agricultural plot (i.e. a locality in our study) and affects both males and females, this could explain the similar dispersal rates observed for males and females at the larger scale. Yet, this hypothesis is lowly supported by the study of Jacob & Hempel (2003) who did not observe evasive movement of radio-tracked common voles following agricultural disturbances. Moving large distances allows individuals to escape crowded conditions or to colonize vacant patches (Ronce & Kirkpatrick 2001), for which selective pressures are unlikely to differ between sexes. This gives little opportunity for a sex bias in dispersal to develop at this scale (Perrin & Goudet 2001).
Acknowledgements
We thank Julien Foucaud, Audrey Sternalski, Olivier Hardy, Xavier Lambin and Oscar Gaggiotti for helpful comments on draft of the manuscript and Maxime Galan, Alexandre Villers and Olivier Fontaine for help in trapping voles. This work was supported by INRA (ECOGER national program), and B.G. received a grant from ACI ECCO.
References
- Aars J., Ims R. A.2000Population dynamic and genetic consequences of spatial density-dependent dispersal in patchy populations. Am. Nat. 155, 252–265 (doi:10.1086/303317) [DOI] [PubMed] [Google Scholar]
- Aars J., Ims R. A., Liu H. P., Mulvey M., Smith M. H.1998Bank voles in linear habitats show restricted gene flow as revealed by mitochondrial DNA (mtDNA). Mol. Ecol. 7, 1383–1389 (doi:10.1046/j.1365-294x.1998.00487.x) [DOI] [PubMed] [Google Scholar]
- Agrell J., Wolff J. O., Ylonen H.1998Counter-strategies to infanticide in mammals: costs and consequences. Oikos 83, 507–517 (doi:10.2307/3546678) [Google Scholar]
- Banks P. B., Norrdahl K., Korpimaki E.2000Nonlinearity in the predation risk of prey mobility. Proc. R. Soc. Lond. B 267, 1621–1625 (doi:10.1098/rspb.2000.1187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux D., Boutin S.2000Breeding dispersal in female North American red squirrels. Ecology 81, 1311–1326 [Google Scholar]
- Berthier K., Charbonnel N., Galan M., Chaval Y., Cosson J. F.2006Migration and recovery of the genetic diversity during the increasing density phase in cyclic vole populations. Mol. Ecol. 15, 2665–2676 (doi:10.1111/j.1365-294X.2006.02959.x) [DOI] [PubMed] [Google Scholar]
- Boonstra R., Krebs C. J., Gaines M. S., Johnson M. L., Craine I. T. M.1987Natal philopatry and breeding systems in voles (Microtus spp.). J. Anim. Ecol. 56, 655–673 (doi:10.2307/5075) [Google Scholar]
- Boyce C. C. K., Boyce J. L.1988aPopulation biology of Microtus arvalis. I. Lifetime reproductive success of solitary and grouped breeding females. J. Anim. Ecol. 57, 711–722 (doi:10.2307/5088) [Google Scholar]
- Boyce C. C. K., Boyce J. L.1988bPopulation biology of Microtus arvalis. II. Natal and breeding dispersal of females. J. Anim. Ecol. 57, 723–736 (doi:10.2307/5089) [Google Scholar]
- Bryja J., Konecny A.2003Fast sex identification in wild mammals using PCR amplification of the Sry gene. Folia Zool. 52, 269–274 [Google Scholar]
- Bryja J., Nesvadbova J., Heroldova M., Janova E., Losik J., Trebaticka L., Tkadlec E.2005Common vole (Microtus arvalis) population sex ratio: biases and process variation. Can. J. Zool. Revue Canadienne De Zoologie 83, 1391–1399 (doi:10.1139/z05-133) [Google Scholar]
- Clobert J., Danchin E., Dhondt A. A., Nichols J. D.2001Dispersal Oxford, UK: Oxford University Press [Google Scholar]
- Cockburn A., Scott M. P., Scotts D. J.1985Inbreeding avoidance and male-biased natal dispersal in Antechinus spp. (Marsupialia, Dasyuridae). Anim. Behav. 33, 908–915 (doi:10.1016/S0003-3472(85)80025-7) [Google Scholar]
- Coulon A., Cosson J. F., Morellet N., Angibault J. M., Cargnelutti B., Galan M., Aulagnier S., Hewison A. J. M.2006Dispersal is not female biased in a resource-defence mating ungulate, the European roe deer. Proc. R. Soc. B 273, 341–348 (doi:10.1098/rspb.2005.3329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douadi M. I., Gatti S., Levrero F., Duhamel G., Bermejo M., Vallet D., Menard N., Petit E. J.2007Sex-biased dispersal in western lowland gorillas (Gorilla gorilla gorilla). Mol. Ecol. 16, 2247–2259 (doi:10.1111/j.1365-294X.2007.03286.x) [DOI] [PubMed] [Google Scholar]
- Duarte L. C., Bouteiller C., Fontanillas P., Petit E., Perrin N.2003Inbreeding in the greater white-toothed shrew Crocidura russula. Evolution 57, 638–645 [DOI] [PubMed] [Google Scholar]
- Ebensperger L. A.1998Strategies and counterstrategies to infanticide in mammals. Biol. Rev. 73, 321–346 (doi:10.1017/S0006323198005209) [Google Scholar]
- Fontanillas P., Petit E., Perrin N.2004Estimating sex-specific dispersal rates with autosomal markers in hierarchically structured populations. Evolution 58, 886–894 [DOI] [PubMed] [Google Scholar]
- Gandon S.1999Kin competition, the cost of inbreeding and the evolution of dispersal. J. Theor. Biol. 200, 345–364 (doi:10.1006/jtbi.1999.0994) [DOI] [PubMed] [Google Scholar]
- Gauffre B., Estoup A., Bretagnolle V., Cosson J. F.2008Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol. Ecol. 17, 4619–4629 (doi:10.1111/j.1365-294X.2008.03950.x) [DOI] [PubMed] [Google Scholar]
- Goudet J.2005HierFstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (doi:10.1111/j.1471-8286.2004.00828.x) [Google Scholar]
- Goudet J., Perrin N., Waser P.2002Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 11, 1103–1114 (doi:10.1046/j.1365-294X.2002.01496.x) [DOI] [PubMed] [Google Scholar]
- Greenwood P. J.1980Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 28, 1140–1162 (doi:10.1016/S0003-3472(80)80103-5) [Google Scholar]
- Hamilton G., Currat M., Ray N., Heckel G., Beaumont M., Excoffier L.2005Bayesian estimation of recent migration rates after a spatial expansion. Genetics 170, 409–417 (doi:10.1534/genetics.104.034199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I., Gaggiotti O.2004Ecology, genetics and evolution of metapopulations Amsterdam, The Netherlands: Elsevier Academic Press [Google Scholar]
- Hardy O. J., Vekemans X.2002Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620 (doi:10.1046/j.1471-8286.2002.00305.x) [Google Scholar]
- Ims R. A., Hjermann D. O.2001Condition dependent dispersal. In Dispersal (eds Clobert J., Danchin E., Dhondt A. A., Nichols J. D.). Oxford, UK: Oxford University Press [Google Scholar]
- Ishibashi Y., Saitoh T., Abe S., Yoshida M. C.1997Sex-related spatial kin structure in a spring population of grey-sided voles Clethrionomys rufocanus as revealed by mitochondrial and microsatellite DNA analyses. Mol. Ecol. 6, 63–71 (doi:10.1046/j.1365-294X.1997.00152.x) [DOI] [PubMed] [Google Scholar]
- Jacob J.2003Short-term effects of farming practices on populations of common voles. Agricult. Ecosyst. Environ. 95, 321–325 (doi:10.1016/S0167-8809(02)00084-1) [Google Scholar]
- Jacob J., Hempel N.2003Effects of farming practices on spatial behaviour of common voles. J. Ethol. 21, 45–50 [Google Scholar]
- Kerth G., Petit E.2005Colonization and dispersal in a social species, the Bechstein's bat (Myotis bechsteinii). Mol. Ecol. 14, 3943–3950 (doi:10.1111/j.1365-294X.2005.02719.x) [DOI] [PubMed] [Google Scholar]
- Koivunen V., Korpimaki E., Hakkarainen H.1998Refuge sites of voles under owl predation risk: priority of dominant individuals? Behav. Ecol. 9, 261–266 (doi:10.1093/beheco/9.3.261) [Google Scholar]
- Lambin X., Krebs C. J.1993Influence of female relatedness on the demography of Townsend's vole populations in spring. J. Anim. Ecol. 62, 536–550 (doi:10.2307/5203) [Google Scholar]
- Lambin X., Yoccoz N. G.1998The impact of population kin-structure on nestling survival in Townsend's voles Microtus townsendii. J. Anim. Ecol. 67, 1–16 (doi:10.1046/j.1365-2656.1998.00181.x) [Google Scholar]
- Lambin X., Bretagnolle V., Yoccoz N. G.2006Vole population cycles in northern and southern Europe: is there a need for different explanations for single pattern? J. Anim. Ecol. 75, 340–349 (doi:10.1111/j.1365-2656.2006.01051.x) [DOI] [PubMed] [Google Scholar]
- Lawson Handley L. J., Perrin N.2007Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 16, 1559–1578 (doi:10.1111/j.1365-294X.2006.03152.x) [DOI] [PubMed] [Google Scholar]
- Le Galliard J. F., Gundersen G., Andreassen H. P., Stenseth N. C.2006Natal dispersal, interactions among siblings and intrasexual competition. Behav. Ecol. 17, 733–740 (doi:10.1093/beheco/arl002) [Google Scholar]
- Li C. C., Weeks D. E., Chakravarti A.1993Similarity of DNA finger prints due to chance and relatedness. Hum. Hered. 43, 45–52 [DOI] [PubMed] [Google Scholar]
- Mitchell-Jones A. J., Amori G., Bogdanowicz W.1999The atlas of European mammals London, UK: T. & A.D. Poyser [Google Scholar]
- Murrell D. J., Travis J. M. J., Dytham C.2002The evolution of dispersal distance in spatially-structured populations. Oikos 97, 229–236 (doi:10.1034/j.1600-0706.2002.970209.x) [Google Scholar]
- Paetkau D., Calvert W., Stirling I., Strobeck C.1995Microsatellite analysis of population-structure in Canadian polar bears. Mol. Ecol. 4, 347–354 (doi:10.1111/j.1365-294X.1995.tb00227.x) [DOI] [PubMed] [Google Scholar]
- Paetkau D., Slade R., Burden M., Estoup A.2004Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 13, 55–65 (doi:10.1046/j.1365-294X.2004.02008.x) [DOI] [PubMed] [Google Scholar]
- Paradis E., Baillie S. R., Sutherland W. J., Gregory R. D.1998Patterns of natal and breeding dispersal in birds. J. Anim. Ecol. 67, 518–536 (doi:10.1046/j.1365-2656.1998.00215.x) [Google Scholar]
- Perrin N., Goudet J.2001Inbreeding, kinship, and the evolution of natal dispersal. In Dispersal (eds Clobert J., Danchin E., Dhondt A. A., Nichols J. D.), pp. 123–141 Oxford, UK: Oxford University Press [Google Scholar]
- Perrin N., Mazalov V.2000Local competition, inbreeding, and the evolution of sex-biased dispersal. Am. Nat. 155, 116–127 (doi:10.1086/303296) [DOI] [PubMed] [Google Scholar]
- Piry S., Alapetite A., Cornuet J. M., Paetkau D., Baudouin L., Estoup A.2004GeneClass2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539 (doi:10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- Poikonen T., Koskela E., Mappes T., Mills S. C.2008Infanticide in the evolution of reproductive synchrony: effects on reproductive success. Evolution 62, 612–621 (doi:10.1111/j.1558-5646.2007.00293.x) [DOI] [PubMed] [Google Scholar]
- Prugnolle F., de Meeus T.2002Inferring sex-biased dispersal from population genetic tools: a review. Heredity 88, 161–165 (doi:10.1038/sj.hdy.6800060) [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2008R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Ronce O., Kirkpatrick M.2001When sources become sinks: migrational meltdown in heterogeneous habitats. Evolution 55, 1520–1531 [DOI] [PubMed] [Google Scholar]
- Ronce O., Olivieri I., Clobert J., Danchin E.2001Perspectives on the study of dispersal evolution. In Dispersal (eds Clobert J., Danchin E., Dhondt A. A., Nichols J. D.), pp. 314–357 Oxford, UK: Oxford University Press [Google Scholar]
- Rousset F., Gandon S.2002Evolution of the distribution of dispersal distance under distance-dependent cost of dispersal. J. Evol. Biol. 15, 515–523 (doi:10.1046/j.1420-9101.2002.00430.x) [Google Scholar]
- Salamolard M., Butet A., Leroux A., Bretagnolle V.2000Responses of an avian predator to variations in prey density at a temperate latitude. Ecology 81, 2428–2441 [Google Scholar]
- Schweizer M., Excoffier L., Heckel G.2007Fine-scale genetic structure and dispersal in the common vole (Microtus arvalis). Mol. Ecol. 16, 2463–2473 (doi:10.1111/j.1365-294X.2007.03284.x) [DOI] [PubMed] [Google Scholar]
- Smith J. E., Batzli G. O.2006Dispersal and mortality of prairie voles (Microtus ochrogaster) in fragmented landscapes: a field experiment. Oikos 112, 209–217 (doi:10.1111/j.0030-1299.2006.13431.x) [Google Scholar]
- Stein G. H. W.1958Die Feldmaus, Die neue Brehm Bücherei Wittemberg, Germany: Ziemsen Verlag [Google Scholar]
- Stenseth N. C., Lidicker W. Z., Jr1992Animal dispersal London, UK: Chapman & Hall [Google Scholar]
- Vekemans X., Hardy O. J.2004New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 13, 921–935 (doi:10.1046/j.1365-294X.2004.02076.x) [DOI] [PubMed] [Google Scholar]