Abstract
Hierarchical properties characterize elephant fission–fusion social organization whereby stable groups of individuals coalesce into higher order groups or split in a predictable manner. This hierarchical complexity is rare among animals and, as such, an examination of the factors driving its emergence offers unique insight into the evolution of social behaviour. Investigation of the genetic basis for such social affiliation demonstrates that while the majority of core social groups (second-tier affiliates) are significantly related, this is not exclusively the case. As such, direct benefits received through membership of these groups appear to be salient to their formation and maintenance. Further analysis revealed that the majority of groups in the two higher social echelons (third and fourth tiers) are typically not significantly related. The majority of third-tier members are matrilocal, carrying the same mtDNA control region haplotype, while matrilocality among fourth-tier groups was slightly less than expected at random. Comparison of results to those from a less disturbed population suggests that human depredation, leading to social disruption, altered the genetic underpinning of social relations in the study population. These results suggest that inclusive fitness benefits may crystallize elephant hierarchical social structuring along genetic lines when populations are undisturbed. However, indirect benefits are not critical to the formation and maintenance of second-, third- or fourth-tier level bonds, indicating the importance of direct benefits in the emergence of complex, hierarchical social relations among elephants. Future directions and conservation implications are discussed.
Keywords: kin selection, social behaviour, population genetics, Loxodonta africana
1. Introduction
Structure in social networks influences population viability and disease transmission as well as evolutionary processes driving speciation and sociality (Krause et al. 2007). The understanding of factors influencing population structuring continues to advance; however, the causes of differential association or avoidance among individuals are complex and as a result remain an important area for research (Couzin 2006). Genetic relatedness can enhance the benefits of social grouping through indirect fitness benefits associated with kin selection (Hamilton 1964; Alexander 1974). Therefore, it is not surprising that many social species aggregate with their kin (Hughes 1998; Clutton-Brock 2002). Non-kin-based aggregations, however, are not uncommon among social species (Lukas et al. 2005; Metheny et al. 2008) and are testament to the direct benefits (e.g. predator vigilance, increased foraging efficiency, resource defence) of social relations (Trivers 1971; Axelrod & Hamilton 1981). Sociality is mediated by the interaction between these benefits (both direct and indirect) and the costs of association (e.g. resource competition and disease or parasite transmission). As a result, the sizes of groups are limited in relation to the cost/benefit ratio of aggregating (Krause & Ruxton 2002).
The most complex societies recognized today, found among some primates, cetaceans and elephants, are flexible fission–fusion organizations with a hierarchical structure (Kummer 1995; Connor 2000; Wittemyer et al. 2005b). The malleable nature of social relationships in fission–fusion societies allows individuals to reposition themselves in the social landscape by coalescing with or separating from conspecifics relative to the shifting cost/benefit ratio of aggregating. Where fission–fusion dynamics have evolved hierarchically, primary groups coalesce to create secondary groups that may be further nested within additional structuring. Groups can form or dissolve across various hierarchical levels in relation to conditions. Such established hierarchical relationships can help optimize behaviour by minimizing the interference costs of social interactions (e.g. Wittemyer & Getz 2007). Complexity in grouping behaviour of this nature may result from the different degrees and time scales at which factors influence aggregations (Krause & Ruxton 2002; Wittemyer et al. 2005b). Kin selection, for example, is generally modulated by the time individuals spend in close proximity (Hamilton 1964; Smith 1964).
African elephants (Loxodonta africana) provide a unique system to investigate the basis of population social properties. Elephants maintain extensive social networks in which they are able to individually discern vocalizations from numerous (100+) individuals (McComb et al. 2001). Their fission–fusion sociality is structured hierarchically with at least four nested levels (Douglas-Hamilton 1972; Moss & Poole 1983; Wittemyer et al. 2005b); a breeding female and her sexually immature offspring are the base social unit (termed tier 1 associates), multiple first-tier units (mother–calf) units compose second-tier groups (containing multiple breeding females and their calves), which coalesce to form third-tier social groups that are nested within fourth-tier groups (figure 1). Female choice occurs within the polygamous mating system (Moss 1983), which can impact group kin structuring (Ross 2001). Previous research has demonstrated that core social groups are resoundingly matrilineal, leading to the conclusion that hierarchical structuring among elephants may be a function of core group expansion and fission processes occurring over long periods of time (Moss 1988; Archie et al. 2006b). Complete analysis of the genetic basis for the range of hierarchical structures in this species, however, has not been conducted until now. We assess the degree to which quantitatively defined, hierarchical social levels are genetically based by testing the predictions that: (i) second-tier (core) group members are direct relatives in the form of mother–calf or sibling pairs (first- or second-order relatives)—i.e. members of core groups are more related than members of higher order groups; (ii) stability of second-tier (core) groups, based on measures of cohesion over time, is positively correlated with group relatedness—i.e. groups with lower relatedness values are more prone to fissions (a precursor for genetically based hierarchical structure); and (iii) higher order structuring (third- and fourth-tier groupings) are matrilineally based and more related than random—i.e. a function of the fission process of social groups lower on the hierarchy. In addition, we investigate the impact of human predation (poaching) on the genetic integrity of elephant society by comparing the results presented here with those from a neighbouring, highly protected population where human predation rates are low.
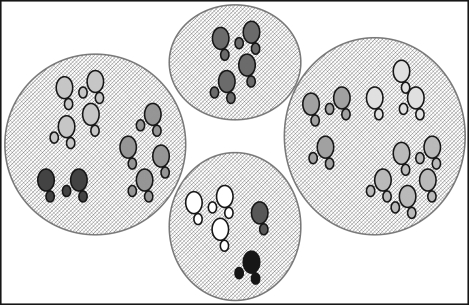
Figure 1.
Elephant hierarchical social structure is characterized by four social levels: basic, first-tier mother–calf units (displayed here as discrete circles); second-tier core groups comprised regularly associating mother–calf units (similarly shaded clusters of discrete first-tier units); third-tier or bond groups (separated clusters of second-tier groups with shaded background); and fourth-tier or clan groups (all units). See Wittemyer et al. (2005b) for more details.
2. Material and methods
(a). Study area and population
The study area lies within the 220 km2 Samburu and Buffalo Springs National Reserves in northern Kenya (37.5° E, 0.5° N). These semiarid parks are located along the Ewaso N'giro River, the major permanent water source in the region, and dominated by Acacia–Commiphora savanna and scrub bush (Barkham & Rainy 1976). As a result of the permanent water, these reserves are a focal area for wildlife. The study area is drought prone and rainfall is highly variable, averaging approximately 350 mm per year and occurring during biannual rainy seasons generally taking place in April/May and November/December.
The elephant population using these reserves numbers approximately 900 individuals (Wittemyer et al. 2005a) and constitutes part of the larger Samburu/Laikipia elephant population comprising over 5000 individuals (Omondi et al. 2002). All individuals observed within the reserves are individually identified, following well-established methods (Moss 1996, 2001). The behaviour and reproductive activity of the entire population has been closely monitored since 1997 (Wittemyer 2001; Wittemyer et al. 2005a).
(b). Quantification of social relationships
Observational records of the study elephants’ social context were collected over a 5-year period beginning in 1998, as described in Wittemyer et al. (2005b), focusing on the most frequently observed ‘resident’ (fifth tier) subpopulation comprising 112 breeding females and their calves, totalling 382 elephants (Wittemyer 2001). The social network of these 112 breeding females was established using 2889 observations of aggregations (averaging 132 observations per individual) for which all breeding females present were registered. Each individual's social group was recorded only once per day to avoid non-independence of observations. The probability of aggregating was calculated from these data as standardized, simple association indices (AIs) (Ginsberg & Young 1992).
Fine-scale social delineations in the study population were defined quantitatively from cluster analysis of AIs, statistically delineating four hierarchical social tiers: first-tier units (mother–calf groups), second-tier groups (core groups), third-tier groups (bond/kinship groups) and, finally, fourth-tier groups (clans) (figure 1). The most dominant individual, as described in analyses of dominance relationships (Wittemyer & Getz 2007), in second-tier groups was defined as that group's matriarch, which was used in subsequent analyses of intergroup relatedness.
(c). Microsatellite genotyping
Genetic variation at 20 microsatellite loci was assessed for 102 of the 112 breeding females used in the original social structure study (Wittemyer et al. 2005a). Genetic extraction was from fresh dung (mucosal portions) or tissue samples, as described in an earlier non-invasive genotyping study (Okello et al. 2005). Further details regarding genotyping protocols and success rates are provided in the electronic supplementary material. Focusing on these 102 breeding females, analysis of the genetic basis for 29 second-tier groups, 12 third-tier groups and 5 fourth-tier groups was conducted in this study—the details of which are described in the electronic supplementary material.
(d). Mitochondrial DNA genotyping
In addition to nuclear markers, we amplified the 5′ hypervariable segment of the mtDNA control region with primers Laf CR1 and Laf CR2 (Nyakaana & Arctander 1999) for 87 of the 112 breeding females, comprising 47 of the 50 second-tier group matriarchs. Fifteen of the adult females known to be offspring of matriarchs (from observations confirmed with relatedness indices estimated using microsatellite variation) were not sampled to avoid redundancy in analysis. The cycling and sequencing parameters followed the protocol described in detail elsewhere (Okello et al. 2008)—a summary of which is provided in the electronic supplementary material.
Seven haplotypes were identified among the 87 breeding females’ mtDNA control region sequences (accession nos GQ357171–GQ357177). The two most common haplotypes, which occurred in 64 (accession no. GQ357171) and 23 per cent (accession no. GQ357173) of the analysed individuals, differed from each other by eight mutational steps. The other five were found in 5 per cent or less of individuals and varied by one or two mutations from the two more common haplotypes.
(e). Analyses of genetic relatedness and pedigrees
Pairwise genetic relatedness from microsatellite genotypic data was estimated using both Relatedness (Queller & Goodnight 1989; Goodnight & Queller 1999) and ML-Relate (Kalinowski et al. 2006). The latter uses a maximum-likelihood-based approach (Thompson & Guo 1991; Marshall et al. 1998; Blouin 2003) that controls for the potential impact of null alleles. In order to minimize bias introduced by averaging pairwise relatedness values within a group (Altmann et al. 1996), we used Relatedness to estimate genetic relatedness within and between known (behaviourally defined) groups or those randomly constructed for comparative purposes through randomization tests (as described below).
The variance and accuracy of Goodnight & Queller (1999) and Kalinowski et al. (2006) relatedness indices (Q&G and ML r, respectively) were assessed by calculating r-values for dyads with known relationships, including mother–calf and half-sibling pairs. Using these known relationships, the accuracy of pedigree assignments by ML-Relate was also assessed. The average relatedness values and pedigree assignments of known relatives were well matched with the following hypothesized values: first, being parent–offspring or full siblings (r = 0.5); second, half siblings or grandmother to grandchild (r = 0.25); and third, aunt to niece/nephew or first cousins (r = 0.125); see electronic supplementary material, table S1. To assess the degree to which social groups in elephants are matrifilial or semisocial (Emlen 1995), the pedigree relationships of the two oldest females in each second-tier group and pairs of matriarchs comprising third- and fourth-tier groups were estimated using ML-Relate. Semisocial groups were categorized as those groups in which the oldest females were half siblings.
(f). Permutation and statistical analyses
Mantel tests (Mantel 1967) were used to assess the correlations between genetic relatedness (r) and AIs among the total samples of breeding females (n = 102) and second-tier group matriarchs (n = 47). The degree to which matriarch associations were correlated with matrilines was assessed similarly, comparing mtDNA haplotype matches with pairwise AIs among second-tier matriarchs (where matrix elements for pairs with matching haplotypes were coded as 1 and non-matching pairs coded as 0). Significance was determined using Monte Carlo simulations. Linear correlation coefficients of the relationship between AIs and r-values were assessed separately within and outside second-, third- and fourth-tier groups.
To determine whether second-tier group members were more closely related to each other than random, we compared r-values of known groups with the distribution of r-values created from 1000 simulated groups of randomly drawn breeding females in the population. The sizes (number of individuals) of randomized groups matched those of known groups, and r-values for these randomized groups were estimated using Relatedness 5.8 in the same manner as for known groups. Groups whose r-values were greater or equal to the 95th percentile of those of randomized groups were considered more closely related than random. Similarly, a randomization test was conducted to assess whether breeding females comprising third- and fourth-tier groups were more closely related than random. For this test, we created 1000 higher order groups from random draws of actual second-tier groups, as relatedness among random draws of breeding females is likely to be low relative to that of multiple second-tier groups comprising third- and fourth-tier groups. Because decision-making in elephants is typically driven by group matriarchs (Moss 1988), we also compared the r-values of second-tier group matriarchs comprising third- and fourth-tier groups with those of randomized groups of matriarchs (consisting of the same number of individuals) following the same procedure.
Because proportions of the seven haplotypes found in the population differed substantially, with the two most common haplotypes carried by approximately 85 per cent of individuals, a randomization test was performed to assess the likelihood that groups possess the same haplotype by chance. Haplotypes from breeding females were shuffled 100 times across groups matching the sizes to second-, third- or fourth-tier groups, and the number of groups sharing the same haplotype was registered for each run. The upper 95th percentile of the number of randomized groups sharing haplotypes was considered the cutoff above which groups were considered to share haplotypes more frequently than expected at random. The same algorithm was performed for matriarchs comprising third- and fourth-tier groups.
The relationship between group stability, defined as changes in group cohesion between wet and dry seasons (as calculated in Wittemyer et al. 2005b), and group relatedness values was assessed using a generalized linear model (GLM). Additional explanatory variables of group size, calf ratio 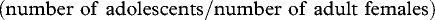 and matriarch age class (Wittemyer & Getz 2007) were also included in the analysis, as these were thought to be the most likely covariates to group stability. Step-wise elimination of non-significant variables was conducted and the full model was statistically compared with the reduced model containing only significant variables using the F-statistic as calculated in the model comparison function of S-PLUS (Venables & Ripley 1999). Model residuals were assessed for homoscedasticity.
and matriarch age class (Wittemyer & Getz 2007) were also included in the analysis, as these were thought to be the most likely covariates to group stability. Step-wise elimination of non-significant variables was conducted and the full model was statistically compared with the reduced model containing only significant variables using the F-statistic as calculated in the model comparison function of S-PLUS (Venables & Ripley 1999). Model residuals were assessed for homoscedasticity.
3. Results
(a). Population level pair-wise relatedness and association indices
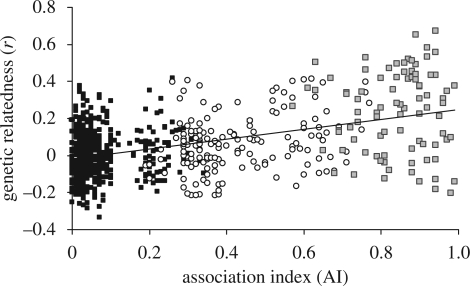
At a coarse level, results from Mantel tests between matrices of r and AI values demonstrate a significant positive correlation among the 102 focal breeding females (Mantel test: ρ = 0.247, p < 0.001) as well as among second-tier group matriarchs (Mantel test: ρ = 0.133, p < 0.001). Similarly, Mantel tests demonstrated that haplotypes were significantly correlated with AIs among matriarchs (ρ = 0.144, p < 0.001). Correlations were weak, however, demonstrating that most of the variation in AI values was not explained by r-values (table 1, figure 2).
Table 1.
Average pair-wise AI and relatedness values (r) calculated using both Queller & Goodnight (1989) and Kalinowski et al. (2006) maximum-likelihood approach for pairs of breeding females within the three hierarchical social tiers are presented along with tier-specific correlation coefficients. Pairs are only represented in one-tier or categorized as non-affiliated if not assigned to any of the three social units.
| social group | N | average AI (s.d.) | method | average r | s.e. | correlation r to AI | p-value |
|---|---|---|---|---|---|---|---|
| second tier | 87 | 0.839 (0.099) | Q&G | 0.234 | 0.023 | <0.001 | 0.941 |
| ML | 0.261 | 0.023 | 0.005 | 0.502 | |||
| third tier | 155 | 0.428 (0.140) | Q&G | 0.067 | 0.012 | 0.078 | <0.001 |
| ML | 0.084 | 0.008 | 0.099 | <0.001 | |||
| fourth tier | 608 | 0.062 (0.065) | Q&G | 0.003 | 0.005 | 0.008 | 0.024 |
| ML | 0.041 | 0.003 | 0.031 | 0.001 | |||
| non-affiliated | 4200 | 0.025 (0.024) | Q&G | −0.003 | 0.002 | <0.001 | 0.480 |
| ML | 0.037 | 0.001 | <0.001 | 0.607 |
Figure 2.
The relationship between relatedness (r) values and AIs across different social tiers shows a significantly positive correlation between the two metrics (R2 = 0.21, p < 0.001; dashed regression line in graph). Among second-tier associates, however, AI and r-values are not significantly correlated (R2 = 0.01, p = 0.90; table 1). Black squares, fourth tier; open circles, third tier; grey squares, second tier.
(b). Genetic basis for core, second-tier groups
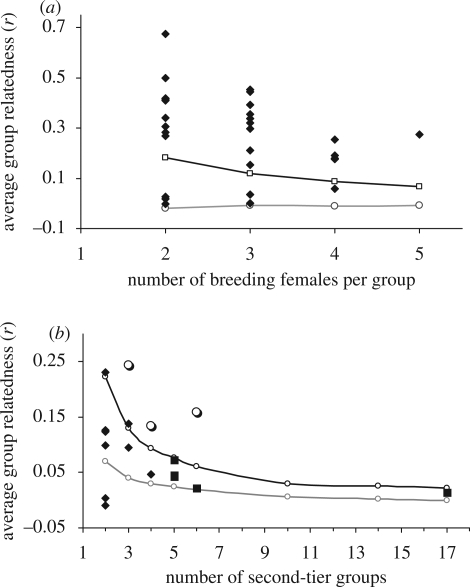
In support of our first hypothesis, our analysis showed that 79 per cent of second-tier groups (23 of 29) were more related than random, i.e. the 95th percentile relatedness value of randomized groups (figure 3a, table 2). The six remaining groups were related more than the median simulated r-value, though less than the 95th percentile. While baseline relatedness values were typically high, averaging r = 0.23 or about the level of second-order relatives (figure 4 and table 1), relatedness and AI values within second-tier groups were uncorrelated, indicating that closer relatedness did not result in tighter associations.
Figure 3.
Average relatedness values of second-, third- and fourth-tier groups compared with simulated relatedness values for groups with the same structure (simulated second-tier groups were derived through a combination of randomly drawn breeding females and simulated third- and fourth-tier groups from randomly drawn second-tier groups; see §2 for further description). (a) Twenty-three of 29 second-tier groups (black diamond) were more related than expected at random (95% of random simulation values). Six of the groups were less related than random, caused by one or two unrelated females joining the group. Open circles, grey line, median simulated r-values; open squares, black line, 95 per cent simulated r-values; black diamonds, average group r. (b). Group relatedness values of the three groups whose third-tier composition did not change at the fourth-tier level (open circles) were significantly greater than random and consistently more related than other groups of similar composition. Only two additional third-tier groups and no fourth-tier groups were significantly related. All group-wise values have been corrected following the Goodnight & Keller (1989) method. Open circle, black line, 95 per cent simulated values; open circle, grey line, median simulated values; filled diamonds, third tier; filled squares, fourth tiers; large open circles, third/fourth tier.
Table 2.
The proportion of groups (actual number are in parentheses) with relatedness values greater than random (the 95th percentile of randomized values) and matching haplotypes were calculated across social levels for both breeding females and second-tier group matriarchs (the latter allowing analysis at higher social levels only).
| breeding females |
matriarchs |
|||
|---|---|---|---|---|
| social level | r-values greater than random | matching haplotypes | r-values greater than random | matching haplotypes |
| second | 79% (23 of 29) | 84% (21 of 23) | n.a. | n.a. |
| third | 42% (5 of 12) | 75% (9 of 12) | 25% (3 of 12) | 83% (10 of 12) |
| fourth | 0% (0 of 5) | 20% (1 of 5) | 20% (1 of 5) | 40% (2 of 5) |
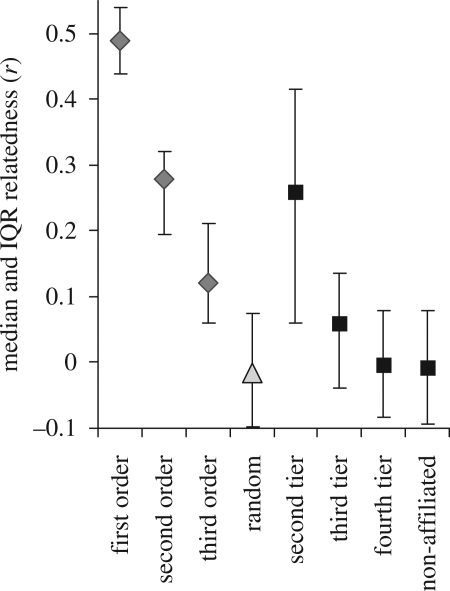
Figure 4.
Average pair-wise relatedness values for the three social tiers analysed are compared with average relatedness values from known first-, second- and third-order relatives (i.e. mother–calf, half sibling and first cousins). Relatedness declines rapidly up the social hierarchy, from approximately the level of second order (half siblings) among second-tier group members to levels no different than random among fourth-tier group associates. Interestingly, relatedness values from randomly selected and unaffiliated pairs were not distinguishable from levels calculated for fourth-tier associates, showing the latter are not genetically based.
Analysis of mitochondrial DNA haplotypes demonstrated that the breeding females in 83 per cent of second-tier groups (19 of 23) descended from the same matriline (table 2), significantly more than that expected at random (p < 0.01). Three of the four groups containing different haplotypes had lower relatedness values than expected at random. Among these four non-matrilocal groups, one comprised three breeding females with differing haplotypes, while the remaining three groups had a single female with a different haplotype from the majority of group members. Three of the second-tier groups with r-values below significant levels shared the same haplotype.
Pedigree relationships among the oldest two breeding females in second-tier groups were typically matrifilial (mother–calf; table 3). Semisocial (sibling) group structure, however, was also observed with a quarter of groups led by half siblings. Overall, group relatedness values were not different between these two structural types (Wilcoxon Z = 0.289, p = 0.773).
Table 3.
Pedigree assignments for the oldest pair of second-tier breeding females and matriarch members of third- and fourth-tier social groups assessed using ML-Relate.
| relatives | r-values | second-tier breeding females | third-tier matriarchs | fourth-tier matriarchs |
|---|---|---|---|---|
| first-order: parent–offspring | 0.5 | 16 (55%) | 1 (2%) | 0 (0%) |
| second-order: half sibling | 0.25 | 6 (21%) | 18 (34%) | 12 (9%) |
| unrelated | 0 | 7 (24%) | 34 (64%) | 118 (91%) |
(c). Social group cohesion and genetic relatedness
Contrary to our second prediction that less related second-tier groups would be less cohesive across seasons (i.e. more prone to fissions), results from a multivariate GLM demonstrated that seasonal stability of second-tier groups was not correlated with their group r-values (t = −0.04, p > 0.05). Group size (t = −0.37, p > 0.05) and calf ratio (t = 0.18, p > 0.05) were also not significant predictors of group stability across seasons. Only matriarch age class provided significant explanatory power (t = 2.97, p = 0.007) in the model, demonstrating that groups led by older matriarchs experienced starker seasonal cohesive changes. The variance explained by the full and reduced GLMs did not differ significantly (likelihood ratio test: Deviance = 0.057, F = 1.495, p = 0.227); the full model contained the explanatory variables of matriarch age, group size, calf ratio, relatedness and associated interaction variables, while the reduced model had the explanatory variable of matriarch age only. Additionally, no difference in stability over seasonal changes was found between matrifilial and non-matrifilial groups (Wilcoxon Z = −0.853, p = 0.394), though matrifilial groups tended to be marginally more cohesive (Z = −1.84, p = 0.062). On a pair-wise basis for all second-tier dyads, AIs did not differ significantly across the three pedigree categories of parent–offspring, half siblings and unrelated dyads (Z = 0.228, p = 0.409).
(d). Genetic basis for hierarchical social tiers
As with the proportion of time spent associating, genetic relatedness declined up the social hierarchy. In contrast to our third prediction, higher order social groups were not strictly genetically based. Among the 12 third-tier groups analysed, group relatedness values in less than half of third-tier groups (five of 12) were greater than expected at random (figure 3b and table 2). Pair-wise relatedness values among third-tier members were typically less than levels among third-order relatives, but greater than random background rates (figure 4). Contrary to second-tier groups, relatedness and AI values were significantly correlated within third-tier groups (table 1). Interestingly, a larger than random number of third-tier groups (nine of 12; p < 0.01) shared the same haplotype, indicating the probable matrilocal origination of such groups despite the relatively low group r-values (table 2). Haplotypes differed among multiple females in two of the three non-matrilocal third-tier groups, while one female differed from all other members in the remaining group; all three had non-significant group relatedness values.
Group relatedness values in the five unique fourth-tier groups were not greater than random (figure 3b and table 2). Neither were fourth-tier pairwise relatedness values (figure 4). However, relatedness values were significantly correlated with AI values among fourth-tier pairs, after excluding social group pairs from second- and third-tiers (table 1). Haplotypes differed between members in four of these five groups, with only one containing the same haplotype across its component second-tier groups (table 2), which is not significantly different from what would be expected at random (p = 0.22).
(e). Relatedness among matriarchs in third- and fourth-tier groups
Analysis of the genetic relatedness among second-tier matriarchs that form third-tier groups demonstrated that 75 per cent of matriarchs (nine of 12) were not related more than expected at random. Among the three third-tier groups whose matriarchs were significantly related, group relatedness values were also greater than random. Approximately, one third of matriarch pairs among third-tier groups were half siblings, with the remainder being categorized as unrelated (table 3). Interestingly, the haplotypes of second-tier matriarchs were the same in significantly more third-tier groups (10 of 12) when compared with random (p < 0.01).
The second-tier matriarchs in one of the 5 fourth-tier groups were related more than random; in contrast to the result for group relatedness where none was related more than random. Pedigree analysis indicated that less than 10 per cent of matriarch pairs were half siblings (table 3). Matriarchs in two of the 5 fourth-tier groups shared the same haplotype, which was marginally significant based on randomization tests (p = 0.08).
4. Discussion
(a). Second-tier groups are not always genetically based units
In support of our first prediction that second-tier core groups were familial in nature, a strong majority of second-tier groups were significantly related. These groups primarily comprised first- and second-order relatives of matrilineal descent (table 3), and the majority had matrifilial (mother–calf) organizations. However, approximately 20 per cent of second-tier groups studied were not significantly related, demonstrating that kinship was not a prerequisite for social affiliation at this fundamental social level and direct benefits alone can elicit strong bonding among elephants (table 2). While the majority of second-tier groups were matrifilial (the organization that hypothetically maximizes group relatedness values), groups containing mother–calf subunits were also led by siblings or even less related pairs—group organizations that according to kin selection theory may be less selectively advantageous. In addition, AIs and relatedness were uncorrelated among second-tier dyads (table 1), and no significant differences were found between the degree of association in mother–calf, half siblings or unrelated pairs; results that demonstrate elephants do not selectively direct interactions based on the degree of relatedness in order to maximize indirect fitness benefits within core groups.
Furthermore, the propensity to fission was not correlated with second-tier group relatedness levels, contrary to our second prediction. Inclusive fitness benefits theoretically influence group stability (Hamilton 1964; Alexander 1974), and the fission of core groups as a function of group size has been posited as the origination of hierarchical social structuring (Douglas-Hamilton 1972; Moss & Poole 1983; Archie et al. 2006b), leading to our prediction of the genetic basis for higher order social groups. But, here, the greatest changes in group cohesion were found among families led by older matriarchs rather than those least genetically related, the potential drivers of which are discussed in the electronic supplementary material. These results indicate that direct benefits play a strong role in facilitating the formation and maintenance of cohesive bonding among females even at fine levels of elephant social organization.
It is important to recognize that inclusive fitness benefits may still be salient to group formation where structure is not optimized to enhance the degree of relatedness among group members. Owing to the generally high association and relatedness metrics among second-tier relatives, inclusive benefits are probably derived from investment in group members even if beneficial interactions are distributed randomly. Additionally, fissions that occur along non-genetic lines in second-tier groups are likely to result in genetic units more related than random. The degree to which interactions are focused on kin potentially varies across groups or age and sex classes for social reasons not apparent in this analysis.
(b). Genetic lineages do not predict hierarchical social tiers
In light of the results for second-tier groups, it is not surprising that higher order social relationships were not strictly genetically based. In contrast to our third prediction, relatedness values among the majority of third- and fourth-tier groups (as well as the pair-wise values between group matriarchs) were below levels distinguishable from background noise (figure 3b). Social affiliation at the third-tier level, however, appears to be for the most part matrilocal, with significantly more third-tier breeding females (75 and 83% of matriarchs) sharing the same haplotype than expected at random (table 2). One or two immigrants appear to cause the non-matrilocality of two of the 3 third-tier groups. This result, as well as the fact that the matriarchs of component second-tier groups in a few third-tier groups were siblings or mother–calf relatives, indicates that the formation of third-tier groups typically originated from familial bonds.
The origination of bonds among fourth-tier group members is less obvious, as significantly fewer members (only one group) than expected at random retained the same haplotype, with clearly non-matrilocal groups making bonds at this level. The numbers of matriarchs within fourth-tier groups sharing the same haplotype, however, were nearly greater than random. As such, matriarchs may choose to maintain matrilocal fourth-tier affiliations where possible (see discussion on the impact of human predation subsequently). While our results indicate that genetic relatedness in higher order groups is not greater than random, matrilocality was present in the highest echelons of elephant society in the study population. In general, the correlation between AIs and relatedness decreased with increasing relatedness (table 1), indicating bonds among relatives tend to be stronger than bonds among non-relatives in the higher social echelons.
(c). Impact of predation on social structure
The Samburu elephant population experienced exceptionally high rates of illegal killing during the 1970s, when the population is purported to have declined by approximately 85 per cent (Poole et al. 1992) and pressure from illegal killing persists for current generations (Wittemyer et al. 2005a). During the same period, the Amboseli elephant population in southern Kenya was relatively undisturbed (Moss 1988, 2001); a population similar to Samburu ecologically, genetically and in terms of density. Both populations have been the focus of long-term behavioural research, including assessment of the genetic basis for multiple-year social relationships, providing a unique opportunity to investigate the impact of human predation on the social properties of this species (methodological differences between the two studies are discussed in the electronic supplementary material).
The genetic patterns among Amboseli elephant groups provide strong evidence of the importance of inclusive fitness benefits in shaping elephant sociality, and the probable kin-based origination of hierarchical structuring (Archie et al. 2006b). In contrast, the well-defined hierarchical structure found in Samburu was not strictly genetically based, with non-relatives comprising groups across social tiers, including the second-tier groups characterized by high associative levels (figure 1 and table 2). The differences between the two populations are probably the manifestation of their different demographic histories. Matrilocal second-tier groups with relatively low microsatellite-based relatedness values in Samburu appear to result from the death of central (probably older) females that were the mother or sibling of other group members. First hand observations of such group altering mortality in Samburu have been relatively common, with over 20 per cent of the 50 focal groups having lost their matriarchs or other mature breeding females during 10 years of close monitoring. Among these typically depredated groups, non-sibling primiparous and adolescent females have been observed to maintain strong affinity after the loss of the group's mature females. Other non-genetic-based second-tier group structures appear to result from unrelated females immigrating into a new family after the loss of its core group members, as suggested for two cases in Amboseli (Archie et al. 2006b). Such immigration events have been observed during the course of our behavioural monitoring twice in Samburu, in addition to those discovered through the genetic analysis presented here. These instances followed predation events where older females were killed by humans and the younger surviving group members did not maintain social bonds with their previous higher tier group members. The alternative has also been observed, where a single primiparous female that lost all its core group affiliates remained solitary, never joining another group. In one second-tier group, all three breeding females were unrelated and had distinct haplotypes, probably exemplifying a situation where remnant single females from predation-decimated families joined together. Such disruptive events at the fundamental social level will clearly have implications for the genetic basis of higher order groups.
Comparison between analyses conducted on elephant populations from Amboseli and Samburu indicate that a genetic component may indeed be important for the evolutionary development of hierarchical social organization, but the elephants of Samburu retained the complex hierarchical structure characteristic of elephant sociality even in light of the erosion of its genetic basis. Perhaps when a population is left undisturbed over generations, weak inclusive fitness benefits crystallize the hierarchal structuring along genetic lines. Following a population disturbance, however, the diversification of the underpinnings of this social structure (running along both genetic and non-genetic lines) suggests that direct benefits are adequate to create and maintain the observed structure. Interestingly, a recent study of a heavily depredated population in Tanzania also shows group formation among non-related individuals (Gobush et al. 2009), though analysis of hierarchical structuring was limited because the study was much shorter with less behavioural data than those analysed here.
Alternatively, it is plausible that the social disruption in Samburu caused by the relatively high rates of adult mortality could elicit novel social bonding (potentially the fourth-tier level), where non-related groups join together to facilitate the direct benefit of anti-predator behaviour (Hamilton 1971). Under extreme predation pressure, all surviving elephants in a population have been observed to group together (Nyakaana et al. 2001). Other recognized direct benefits that may facilitate non-relative acceptance by groups are between-group competition (Wittemyer & Getz 2007), information exchange (Foley et al. 2008) and alloparental care by subordinate and adolescent females (Lee 1987). It is possible that aggregating with non-kin simply emerges from non-plasticity in elephant social behaviour, though the high degree of competition within social groups (Archie et al. 2006a) and diversity of social group structure and size indicate otherwise. In respect to our results showing a genetic basis is not a prerequisite for the creation of hierarchical elephant social relations, future work is needed to determine the differential benefits or lack thereof derived from relations among specific social tier affiliates.
(d). Broader implications
The disintegration of animal social systems can have long-term, negative repercussions on population survivorship rates (Milner et al. 2007). While reproductive rates in elephant populations experiencing extreme predation pressure have collapsed (Barnes & Kapela 1991), the erosion of the genetic foundations of social relations among Samburu's elephants does not appear to be impacting recruitment; although predation rates are relatively low compared with those experienced during poaching peaks. In fact, the Samburu population is currently growing faster than Amboseli's (Moss 2001; Wittemyer et al. 2005a). Post-poaching populations have been seen to rebound in other parts of Africa as well, though recruitment varies strongly across groups (Foley 2002; Foley et al. 2008). Determining the relationships among ecological variability, group structure and fitness remains an important direction of research.
Optimizing direct benefits of sociality in complex social and ecological settings is probably best achieved through the fluid nature of hierarchical societies, where individuals can coalesce or separate according to recognized patterns with minimal interference (i.e. avoiding social disruptions like the establishment of dominance relations during initial contact). The echoes of the genetic basis of relationships reflected in the matrilocal nature of third-tier groups and the cohesive strength of certain, closely related groups indicate kin selection does play a role in the emergence of such social complexity. Perhaps the bonds between individuals that necessitate such fluidity in social relation are most easily established through long-term familial relationships, leading to kin-based associations even where inclusive fitness benefits are weak or non-existent. As the maintenance of elephant hierarchical sociality does not necessitate genetic underpinnings, direct benefits can be surmised to be substantive forces driving the formation of complex, hierarchical social structure among elephants.
Acknowledgements
This research was supported by the National Science Foundation IRFP OISE-0502340 (G.W.), the Danish Agency for International Development (DANIDA) and Danish Natural Science Research Council. We thank the Kenyan Office of the President, the Kenya Wildlife Service (KWS) and the Samburu and Buffalo Springs National Reserve's County Councils, wardens and rangers for their support of our work.
References
- Alexander R. D.1974The evolution of social behavior. Annu. Rev. Ecol. System. 5, 325–383 (doi:10.1146/annurev.es.05.110174.001545) [Google Scholar]
- Altmann J., et al. 1996Behavior predicts genetic structure in a wild primate group. Proc. Natl Acad. Sci. USA 93, 5797–5801 (doi:10.1073/pnas.93.12.5797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie E. A., Morrison T. A., Foley C. A. H., Moss C. J., Alberts S. C.2006aDominance rank relationships among wild female African elephants (Loxodonta africana). Anim. Behav. 71, 117–127 (doi:10.1016/j.anbehav.2005.03.023) [Google Scholar]
- Archie E. A., Moss C. J., Alberts S. C.2006bThe ties that bind: genetic relatedness predicts the fission and fusion of social groups in wild African elephants. Proc. R. Soc. B 273, 513–522 (doi:10.1098/rspb.2005.3361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod R., Hamilton W. D.1981The evolution of cooperation. Science 211, 1390–1396 (doi:10.1126/science.7466396) [DOI] [PubMed] [Google Scholar]
- Barkham J. P., Rainy M. E.1976The vegetation of the Samburu-Isiolo Game Reserve. Afr. J. Ecol. 14, 297–329 (doi:10.1111/j.1365-2028.1976.tb00244.x) [Google Scholar]
- Barnes R. F. W., Kapela E. B.1991Changes in the Ruaha elephant population caused by poaching. Afr. J. Ecol. 29, 289–294 (doi:10.1111/j.1365-2028.1991.tb00466.x) [Google Scholar]
- Blouin M. S.2003DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol. Evol. 18, 503–511 (doi:10.1016/S0169-5347(03)00225-8) [Google Scholar]
- Clutton-Brock T.2002Behavioral ecology—breeding together: kin selection and mutualism in cooperative vertebrates. Science 296, 69–72 (doi:10.1126/science.296.5565.69) [DOI] [PubMed] [Google Scholar]
- Connor R. C.2000Group living in whales and dolphins. In Cetacean societies: field studies of dolphins and whales (eds Tyack P. L., Whitehead H.), pp. 199–218 Chicago, IL: University of Chicago Press [Google Scholar]
- Couzin I. D.2006Behavioral ecology: social organization in fission–fusion societies. Curr. Biol. 16, R169–R171 (doi:10.1016/j.cub.2006.02.042) [DOI] [PubMed] [Google Scholar]
- Douglas-Hamilton I.1972. In On the ecology and behaviour of the African elephant: elephants of Lake Manyara. Oxford, UK: Oxford University [Google Scholar]
- Emlen S. T.1995An evolutionary theory of the family. Proc. Natl Acad. Sci. USA 92, 8092–8099 (doi:10.1073/pnas.92.18.8092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley C. A. H.2002The effects of poaching on elephant social systems Princeton, NJ: Department of Ecology and Evolutionary Biology, Princeton University [Google Scholar]
- Foley C. A. H., Pettorelli N., Foley L.2008Severe drought and calf survival in elephants. Biol. Lett. 4, 541–544 (doi:10.1098/rsbl.2008.0370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg J. R., Young T. P.1992Measuring association between individuals or groups in behavioral studies. Anim. Behav. 44, 377–379 (doi:10.1016/0003-3472(92)90042-8) [Google Scholar]
- Gobush K., Kerr B., Wasser S.2009Genetic relatedness and disrupted social structure in a poached population of African elephants. Mol. Ecol. 18, 722–734 (doi:10.1111/j.1365-294X.2008.04043.x) [DOI] [PubMed] [Google Scholar]
- Goodnight K. F., Queller D. C.1999Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol. Ecol. 8, 1231–1234 (doi:10.1046/j.1365-294x.1999.00664.x) [DOI] [PubMed] [Google Scholar]
- Hamilton W. D.1964Genetical evolution of social behaviour 1 & 2. J. Theor. Biol. 7, 1–56 (doi:10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- Hamilton W. D.1971Geometry for the selfish herd. J. Theor. Biol. 31, 295–311 (doi:10.1016/0022-5193(71)90189-5) [DOI] [PubMed] [Google Scholar]
- Hughes C.1998Integrating molecular techniques with field methods in studies of social behavior: a revolution results. Ecology 79, 383–399 [Google Scholar]
- Kalinowski S. T., Wagner A. P., Taper M. L.2006ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579 (doi:10.1111/j.1471-8286.2006.01256.x) [Google Scholar]
- Krause J., Ruxton G. D.2002Living in groups Oxford, UK: Oxford University Press [Google Scholar]
- Krause J., Croft D. P., James R.2007Social network theory in the behavioural sciences: potential applications. Behav. Ecol. Sociobiol. 62, 15–27 (doi:10.1007/s00265-007-0445-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer H.1995Quest of the sacred baboon Princeton, NJ: Princeton University Press [Google Scholar]
- Lee P. C.1987Allomothering among African elephants. Anim. Behav. 35, 278–291 (doi:10.1016/S0003-3472(87)80234-8) [Google Scholar]
- Lukas D., Reynolds V., Boesch C., Vigilant L.2005To what extent does living in a group mean living with kin? Mol. Ecol. 14, 2181–2196 (doi:10.1111/j.1365-294X.2005.02560.x) [DOI] [PubMed] [Google Scholar]
- Mantel N.1967Detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 [PubMed] [Google Scholar]
- Marshall T. C., Slate J., Kruuk L. E. B., Pemberton J. M.1998Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7, 639–655 (doi:10.1046/j.1365-294x.1998.00374.x) [DOI] [PubMed] [Google Scholar]
- McComb K., Moss C., Durant S. M., Baker L., Sayialel S.2001Matriarchs as repositories of social knowledge in African elephants. Science 292, 491–494 (doi:10.1126/science.1057895) [DOI] [PubMed] [Google Scholar]
- Metheny J. D., Kalcounis-Rueppell M. C., Willis C. K. R., Kolar K. A., Brigham R. M.2008Genetic relationships between roost-mates in a fission–fusion society of tree-roosting big brown bats (Eptesicus fuscus). Behav. Ecol. Sociobiol. 62, 1043–1051 (doi:10.1007/s00265-007-0531-y) [Google Scholar]
- Milner J. M., Nilsen E. B., Andreassen H. P.2007Demographic side effects of selective hunting in ungulates and carnivores. Conserv. Biol. 21, 36–47 (doi:10.1111/j.1523-1739.2006.00591.x) [DOI] [PubMed] [Google Scholar]
- Moss C. J.1983Estrous behavior and female choice in the African elephant. Behaviour 86, 167–196 (doi:10.1163/156853983X00354) [Google Scholar]
- Moss C. J.1988Elephant memories: thirteen years in the life of an elephant family New York, NY: William Morrow [Google Scholar]
- Moss C. J.1996Getting to know a population. In Studying elephants (ed. Kangwana K.), pp. 58–74 Nairobi, Kenya: African Wildlife Foundation [Google Scholar]
- Moss C. J.2001The demography of an African elephant (Loxodonta africana) population in Amboseli, Kenya. J. Zool. 255, 145–156 (doi:10.1017/S0952836901001212) [Google Scholar]
- Moss C. J., Poole J. H.1983Relationships and social structure of African elephants. In Primate social relationships: an integrated approach (ed. Hinde R. A.), pp. 315–325 Oxford, UK: Blackwell Scientific Publications [Google Scholar]
- Nyakaana S., Arctander P.1999Population genetic structure of the African elephant in Uganda based on variation at mitochondrial and nuclear loci: evidence for male-biased gene flow. Mol. Ecol. 8, 1105–1115 (doi:10.1046/j.1365-294x.1999.00661.x) [DOI] [PubMed] [Google Scholar]
- Nyakaana S., Abe E. L., Arctander P., Siegismund H. R.2001DNA evidence for elephant social behaviour breakdown in Queen Elizabeth National Park, Uganda. Anim. Conserv. 4, 231–237 (doi:10.1017/S1367943001001275) [Google Scholar]
- Okello J. B. A., Wittemyer G., Rasmussen H. B., Douglas-Hamilton I., Nyakaana S., Arctander P., Siegismund H. R.2005Noninvasive genotyping and Mendelian analysis of microsatellites in African savannah elephants. J. Hered. 96, 679–687 (doi:10.1093/jhered/esi117) [DOI] [PubMed] [Google Scholar]
- Okello J. B. A., et al. 2008Population genetic structure of savannah elephants in Kenya: conservation and management implications. J. Hered. 99, 443–452 (doi:10.1093/jhered/esn028) [DOI] [PubMed] [Google Scholar]
- Omondi P., Bitok E., Kahindi O., Mayienda R.2002Total aerial count of elephants in Samburu/Laikipia. Nairobi, Kenya: Kenya Wildlife Service [Google Scholar]
- Poole J. H., Aggeawal N., Sinange R., Nganga S., Broten M., Douglas-Hamilton I.1992The status of Kenya's elephants Nairobi, Kenya: African Wildlife Foundation [Google Scholar]
- Queller D. C., Goodnight K. F.1989Estimating relatedness using genetic markers. Evolution 43, 258–275 (doi:10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- Ross K. G.2001Molecular ecology of social behaviour: analyses of breeding systems and genetic structure. Mol. Ecol. 10, 265–284 (doi:10.1046/j.1365-294x.2001.01191.x) [DOI] [PubMed] [Google Scholar]
- Smith J. M.1964Group selection and kin selection. Nature 201, 1145–1147 (doi:10.1038/2011145a0) [Google Scholar]
- Thompson E. A., Guo S. W.1991Evaluation of likelihood ratios for complex genetic models. Math. Med. Biol. 8, 149–169 (doi:10.1093/imammb/8.3.149) [DOI] [PubMed] [Google Scholar]
- Trivers R. L.1971Evolution of reciprocal altruism. Q. Rev. Biol. 46, 35–37 (doi:10.1086/406755) [Google Scholar]
- Venables W. N., Ripley B. D.1999Modern applied statistics with S-Plus New York, NY: Springer [Google Scholar]
- Wittemyer G.2001The elephant population of Samburu and Buffalo Springs National Reserves, Kenya. Afr. J. Ecol. 39, 357–365 (doi:10.1046/j.1365-2028.2001.00324.x) [Google Scholar]
- Wittemyer G., Getz W. M.2007Hierarchical dominance structure and social organization in African elephants Loxodonta africana. Anim. Behav. 73, 671–681 (doi:10.1016/j.anbehav.2006.10.008) [Google Scholar]
- Wittemyer G., Daballen D. K., Rasmussen H. B., Kahindi O., Douglas-Hamilton I.2005aDemographic Status of elephants in the Samburu and Buffalo Springs National Reserves, Kenya. Afr. J. Ecol. 43, 44–47 (doi:10.1111/j.1365-2028.2004.00543.x) [Google Scholar]
- Wittemyer G., Douglas-Hamilton I., Getz W. M.2005bThe socioecology of elephants: analysis of the processes creating multitiered social structures. Anim. Behav. 69, 1357–1371 (doi:10.1016/j.anbehav.2004.08.018) [Google Scholar]