Abstract
The establishment of an endosymbiotic relationship typically seems to be driven through complementation of the host's limited metabolic capabilities by the biochemical versatility of the endosymbiont. The most significant examples of endosymbiosis are represented by the endosymbiotic acquisition of plastids and mitochondria, introducing photosynthesis and respiration to eukaryotes. However, there are numerous other endosymbioses that evolved more recently and repeatedly across the tree of life. Recent advances in genome sequencing technology have led to a better understanding of the physiological basis of many endosymbiotic associations. This review focuses on endosymbionts in protists (unicellular eukaryotes). Selected examples illustrate the incorporation of various new biochemical functions, such as photosynthesis, nitrogen fixation and recycling, and methanogenesis, into protist hosts by prokaryotic endosymbionts. Furthermore, photosynthetic eukaryotic endosymbionts display a great diversity of modes of integration into different protist hosts.
In conclusion, endosymbiosis seems to represent a general evolutionary strategy of protists to acquire novel biochemical functions and is thus an important source of genetic innovation.
Keywords: metabolic complementation, prokaryotic endosymbionts, eukaryotic endosymbionts, evolution, integration
1. Introduction
Endosymbiosis, in the sense of endocytobiosis, with one symbiotic partner (the endosymbiont) living intracellularly within the second symbiotic partner (the host), is the most intimate form of symbiosis. The establishment of an endosymbiotic relationship typically seems to be driven through complementation of the host's limited metabolic capabilities by the biochemical versatility of the endosymbiont, thereby enabling the host to thrive in environments or on diets previously inaccessible (Hoffmeister & Martin 2003; Douglas 2009). For the endosymbiont, the symbiosis provides a nutrient-rich, sheltered environment; yet, it is difficult to demonstrate that endosymbionts benefit significantly from interaction with their hosts (Douglas & Smith 1989). Therefore, some authors regard the process of endosymbiosis rather as an enslavement of the endosymbiont than as a mutually beneficial relationship.
As illustrated by the evolutionary acquisition of plastids and mitochondria, in its most extreme case, an endosymbiosis may lead to an inseparable merger of two symbiotic partners to yield a novel chimeric organism in a process termed ‘symbiogenesis’ (Mereschkowsky 1910). In the course of symbiogenesis, genes are transferred from the endosymbiont to the host nuclear genome, a process dubbed ‘endosymbiotic gene transfer’ (EGT); the respective gene products are targeted to the former endosymbiont that is then regarded as a genetically integrated cell organelle (e.g. Timmis et al. 2004).
Owing to the profound impact that acquisition of plastids and mitochondria had for the evolution of life, the term endosymbiosis is often used as an equivalent for the acquisition of these organelles. At the same time, the myriad transient or stable endosymbiotic relationships that have evolved more recently and repeatedly all across the tree of life are often overlooked. Host partners in these relationships are typically eukaryotes, owing to their larger cell size, phagocytosis and restricted metabolic capabilities, whereas the endosymbiont partners may be either pro- or eukaryotes.
In the context of plastid acquisition, endosymbioses involving a prokaryotic endosymbiont (a cyanobacterium) are referred to as ‘primary endosymbioses’ leading to ‘primary plastids’, as opposed to sequential endosymbioses involving photosynthetic eukaryotes that are termed ‘secondary’ and ‘tertiary endosymbioses’ leading to secondary and ‘tertiary plastids’ (Gould et al. 2008; Archibald 2009; Keeling 2009). It is, however, important to note that the same terms are used differently by zoologists: ‘primary endosymbionts’ are here bacterial endosymbionts that live inside specialized animal host cells in mutually obligate associations, whereas ‘secondary endosymbionts’ are facultative bacterial endosymbionts that coexist with a primary endosymbiont and are not essential for the survival of the host (Moya et al. 2008). Here, we will use the terms in the former sense.
Although the scope of this article is restricted to endosymbionts in protists, i.e. unicellular eukaryotes, there are countless fascinating and well-studied examples of endosymbioses in multicellular organisms, particularly animals. Insects such as aphids and tsetse flies have been able to occupy their ecological niches only due to nutritional support of their bacterial endosymbionts (Moya et al. 2008). Eukaryotic life in deep-sea habitats surrounding hydrothermal vents depends to a large extent on thiotrophic (i.e. sulphur oxidizing) bacterial endosymbionts as primary producers (Dubilier et al. 2008).
In multicellular hosts, however, endosymbionts are typically restricted to specialized organs or tissues, whereas in unicellular hosts, the whole organism is directly involved in the symbiosis. Even though in a few cases EGT was described in multicellular organisms, where endosymbionts had access to the host's germ line owing to their presence in developing gametes (Dunning Hotopp et al. 2007; Nikoh et al. 2008; Klasson et al. 2009), most horizontal gene transfers into eukaryotes have been described in phagotrophic, unicellular eukaryotes (Andersson 2005). Because unicellular organisms have no sequestered germ line, integration of DNA from their food organisms or endosymbionts appears to be rather frequent (Doolittle 1998). Thus, genetic integration seems to be more readily established in protists and it was likely not by chance that all organelles evolved in unicellular organisms.
The phenomenon of endosymbiosis (i) is widespread over the tree of life and various ecosystems, (ii) is of great physiological importance in many eukaryotic groups, (iii) represents a general mechanism of cellular evolution in eukaryotes, and (iv) could aid in understanding the evolutionary process of organellogenesis (Keeling & Palmer 2008). However, owing to the large diversity in phylogenetic affiliation of both partners, the different physiological functions performed by the endosymbionts, as well as the various degrees and modes of integration of the endosymbionts into the protist host cell, no exhaustive treatment of endosymbiotic associations is possible given the limited space of a short review article. Here, we will address different conceptual models of endosymbiosis using selected examples from the recent literature comprising both prokaryotic and eukaryotic endosymbionts of protists and highlight their impact on our current view of eukaryotic evolution.
2. Prokaryotic endosymbionts in protists
(a). Photosynthetic endosymbionts
The origin of photosynthesis in the biosphere introduced an energy resource orders of magnitude larger than that available from redox reactions associated with weathering or hydrothermal activity (Des Marais 2000). Through the endosymbiotic acquisition of plastids, this virtually infinite source of energy became directly available to eukaryotes. In the course of evolution, primary photosynthetic eukaryotes diversified; additionally, plastids spread horizontally via secondary and tertiary endosymbioses (Gould et al. 2008; Archibald 2009). Today, photosynthetic eukaryotes exhibit a bewildering diversity of primary producers that provide the Earth with most of its biomass. Despite the large evolutionary success of photosynthetic eukaryotes, plastids had a monophyletic origin within the cyanobacteria and were likely acquired only once, probably more than 1 Gyr ago (Moreira et al. 2000; McFadden & van Dooren 2004; Rodríguez-Ezpeleta et al. 2005). Thus, our knowledge of the evolutionary processes leading from a free-living cyanobacterium to an integrated photosynthetic organelle is highly speculative, as is the question of why the acquisition of primary plastids apparently remained a singular event.
Only recently, evidence for a second, more recent primary endosymbiosis leading to a photosynthetic eukaryote was presented for the amoeba Paulinella chromatophora (Marin et al. 2005). This places P. chromatophora in a centre stage for understanding early events in the evolution of a phototrophic eukaryote.
(i). The amoeba P. chromatophora with photosynthetic cyanobacterial endosymbionts
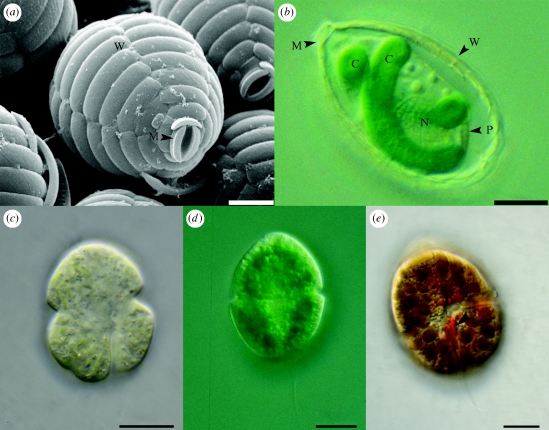
Paulinella chromatophora (figure 1a,b) is a thecate amoeba of cercozoan affiliation (Bhattacharya et al. 1995; Cavalier-Smith & Chao 2003) that occurs ubiquitously in freshwater ponds and lakes. In contrast to their closest marine relatives that feed on cyanobacteria (Johnson et al. 1988; Hannah et al. 1996), P. chromatophora has dispensed with phagotrophic nutrition (Lauterborn 1895; Penard 1905; Kies 1974). Instead, it carries two sausage-shaped photosynthetic entities, termed chromatophores (Lauterborn 1895), that support the phototrophic lifestyle of the amoeba (Kies & Kremer 1979). A high level of symbiotic integration of the chromatophores is implied by the strict synchronization of chromatophore division with the host cell cycle (Hoogenraad 1927 and Nowack, E. C. M. & Melkonian, M. 2006 (unpublished observations)). The chromatophores are enclosed by a membrane of unknown origin and are located in the cytoplasm of the amoeba. They resemble in colour and ultrastructure (possession of carboxysomes, unstacked thylakoids and a peptidoglycan wall) cyanobacteria of the genus Synechococcus (Kies 1974); however, they are roughly 20 times larger than free-living Synechococcus spp. Phylogenetic analyses of the complete ribosomal operon as well as of a concatenated multiple protein dataset revealed that the chromatophores evolved from α-cyanobacteria (i.e. the Prochlorococcus/Synechococcus clade), separating them clearly from plastids that evolved from β-cyanobacteria (Marin et al. 2005, 2007; Yoon et al. 2006). Different isolates of P. chromatophora differ notably in molecular sequence data and morphology of their thecae, suggesting the existence of different species; however, the well-supported monophyly of their chromatophores in phylogenetic analyses of the 16S rRNA strongly suggests that their chromatophores arose from a common cyanobacterial ancestor (Yoon et al. 2009).
Figure 1.
Paulinella chromatophora cells. (a) Scanning electron microscopic image. (b) Light microscopic image (differential interference contrast). C, chromatophore; M, mouth opening; N, nucleus; P, plasma membrane and W, cell wall composed of silica scales. Light microscopic images of dinoflagellates with unusual plastids. (c) L. chlorophorum, (d) G. aeruginosum, and (e) K. foliaceum, arrowhead highlights the eyespot. Images kindly provided by Barbara Surek (c,e) and Karl-Heinz Linne von Berg (d). Scale bar, (a,b) 5 µm; (c–e) 10 µm.
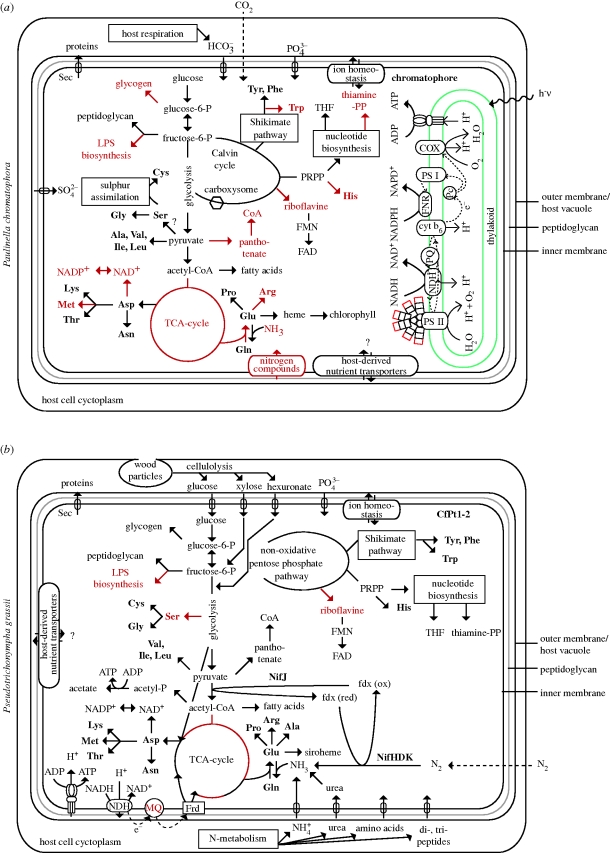
The analysis of the complete chromatophore genome sequence of P. chromatophora strain CCAC 0185 revealed a fundamental genome reduction (Nowack et al. 2008). The single, circular chromosome of 1.02 Mb encodes only 867 protein-coding genes and is, therewith, the smallest cyanobacterial genome reported to date. Although the chromatophore genome contains an almost complete set of photosynthesis genes, it lacks not only genes thought to be dispensable for an intracellular lifestyle but also genes of essential pathways for amino acid and cofactor biosynthesis (figure 2a), characterizing the chromatophore as a photosynthetic entity that is absolutely dependent on its host for growth and survival. The inability to produce several proteinogenic amino acids and critical cofactors strongly implies that the host cell compensates for functions missing in the chromatophore. Intriguingly, the strong metabolic interdependence of host cell and chromatophore contrasts with the paucity of chromatophore-encoded transport systems, raising the question, how metabolite exchange is accomplished.
Figure 2.
Metabolism of bacterial endosymbionts in protists as deduced from their genome sequences. Arrows in black represent pathways largely encoded and arrows in red represent pathways largely missing. (a) Chromatophore of Paulinella and (b) CfPt1-2 Bacteroides-type endosymbiont of the cellulolytic termite gut flagellate P. grassii (modified from Hongoh et al. 2008b). Question marks denote uncertainties in function or identity of a protein. Amino acids are in bold. COX, cytochrome-c oxidase; FNR, ferredoxin : NADP+ reductase; FRD, fumarate reductase; MQ, menaquinone; NDH, NADH dehydrogenase; PQ, plastoquinone and PRPP, 5-phosphoribosyl 1-pyrophosphate.
Furthermore, the chromatophore genome analysis revealed the lack of a few low-molecular weight components of the photosystems and several enzymes essential for DNA replication such as ligases. These findings strongly suggest protein import into the chromatophore, as the functions lacking clearly cannot be compensated at the metabolite level. Recently, the first evidence for gene transfer from the chromatophore to the nuclear genome of P. chromatophora was reported. The psaE gene, which encodes a low-molecular weight photosystem I subunit, is not present on the chromatophore genome but was identified as a P. chromatophora expressed sequence tag (EST) sequence (Nakayama & Ishida 2009). The psaE cDNA contains a typical eukaryotic polyA tail, and the genomic sequence exhibits introns; these features strongly suggest that the gene is located on the nuclear genome. However, although import of the nuclear-encoded PsaE protein into the chromatophore is likely, no direct evidence for protein import was presented yet.
(b). Nitrogen-fixing or recycling endosymbionts
Nitrogen is a common component of numerous biomolecules and accordingly essential for the growth and development of all organisms. Whereas the major part of nitrogen occurs as molecular nitrogen (N2) in the atmosphere, in many terrestrial and aquatic habitats biologically available nitrogen represents a growth-limiting factor. Nevertheless, only some bacteria possess the ability to fix atmospheric N2 by means of the nitrogenase enzyme complex. Eukaryotes, in contrast, are only able to use atmospheric N2 through symbiotic interactions with N2-fixing bacteria. Such kinds of symbioses are well known for multicellular organisms, as exemplified by the legume/rhizobia endosymbioses, but are also frequent in protists.
(i). The diatom Rhopalodia gibba with N2-fixing cyanobacterial endosymbionts
One well-studied example is the pennate freshwater diatom Rhopalodia gibba. Rhopalodia gibba cells host—in addition to a photosynthetically active secondary plastid—one to four cyanobacterial endosymbionts of ellipsoid shape (ca 6 × 5 µm in size) in cytoplasmic vacuoles (Drum & Pankratz 1965; Geitler 1977). The endosymbionts, referred to as ‘spheroid bodies’, are closely related to the N2-fixing cyanobacterium Cyanothece (Prechtl et al. 2004). Fragments of the genome of the spheroid bodies that were recently sequenced support the previous notion that the R. gibba endosymbionts are able to fix N2 (Floener & Bothe 1980), but have dispensed with photosynthesis: a complete set of genes involved in N2 fixation was identified, whereas essential photosynthesis genes were found to be eroded to pseudogenes (Kneip et al. 2008).
An advanced level of symbiotic integration is indicated by the following findings: R. gibba cells were never observed without spheroid bodies; the maximum number of spheroid bodies per cell seems to be restricted; spheroid bodies are vertically transmitted and attempts to cultivate the endosymbiont outside of the host cell have failed (Drum & Pankratz 1965; Prechtl et al. 2004).
(ii). Cellulolytic termite gut flagellates with nitrogen-fixing or recycling Bacteroidales-type endosymbionts
In terrestrial habitats or diets nitrogen also often represents a limiting factor, as exemplified by wood-feeding termites. For the digestion of lignocelluloses, the predominant component of woody plants, lower termites rely on anaerobic flagellates of the Parabasalia and Oxymonadida that are located in their hindgut; the majority of which harbour numerous prokaryotic endosymbionts (Ohkuma 2003). Widespread and often predominating in the gut microbial communities are bacteria belonging to the candidate phylum Termite Group 1 (TG1) of the Bacteroidales (Stingl et al. 2005; Ohkuma et al. 2007). The endosymbiotic bacteria colonize the cytoplasm of their flagellate hosts in large numbers and are surrounded by two membranes; the outermost membrane being either derived from the outer membrane of the Gram-negative bacterium or from the host (Stingl et al. 2005). Since neither endosymbiotic bacteria nor the flagellates are cultivable, these symbiotic associations are not readily accessible for research.
Recently, two complete genome sequences of the TG1 species CfPt1-2 and Rs-D17 were obtained from endosymbiont DNA extracted from two different single flagellate host cells by whole genome amplification (Hongoh et al. 2008a,b). These genome sequences yielded the first insight into the physiological basis of the symbiotic association between cellulolytic termite gut protists and their bacterial endosymbionts.
CfPt1-2, the endosymbiont of Pseudotrichonympha grassii (Parabasalia), accounts for 70 per cent of the bacterial cells in the gut of the termite Coptotermes formosanus. About 105 endosymbionts are housed within a single flagellate cell. A genome analysis of the 1.1 Mb chromosome of the endosymbiont, encoding a meagre 758 protein-coding genes, demonstrated not only its ability to recycle putative host nitrogen waste products, such as ammonium and urea, for biosynthesis of several amino acids and cofactors, but also revealed the unexpected ability to fix N2 (figure 2b).
Rs-D17 is the endosymbiont of Trichonympha agilis (Parabasalia) that thrives in the gut of the termite Reticulitermes speratus. Here, about 4 × 103 endosymbionts are housed within a single flagellate cell. The circular chromosome of 1.1 Mb, encoding 761 protein-coding genes, does not encode enzymes of the nitrogenase complex and, therewith, precludes the ability of the bacterium to fix N2. However, the fact that biosynthetic capabilities for 15 amino acids and various cofactors were not only retained, but some of the respective genes had even been duplicated, implies a function of the endosymbiont in the supply of these essential compounds, deficient in lignocellulose food, to their host protists and eventually to the termite.
These highly complex bacteria/flagellate/termite symbiotic associations probably support the ability of lower termites to survive on recalcitrant plant matter as the sole nutrient source and, thereby, contribute largely to the success of these insects of worldwide distribution.
(c). Methanogenic endosymbionts
(i). Anaerobic ciliates with methanogenic archaeal endosymbionts
Anaerobic ciliates occur worldwide in anoxic aquatic sediments as well as symbiotically in the guts of many animals, in particular, those which rely on fermenting bacteria for digestion of structural plant matter (Fenchel & Finlay 1991a). Many free-living anaerobic ciliates are known to contain endosymbiotic methanogenic archaea, e.g. Metopus contortus, Trimyema sp. and Cyclidium porcatum (Fenchel & Finlay 1991a; van Hoek et al. 2000). These ciliates contain hydrogenosomes, i.e. H2-evolving redox organelles that are thought to have been derived from mitochondria (Boxma et al. 2005). The endosymbionts, distantly related methanogenic archaea (Embley & Finlay 1993), probably use the H2 produced by the hydrogenosomes to reduce CO2 and generate energy and excrete dissolved organics that are used by the host (Fenchel & Finlay 1991a). However, owing to the absence of genomic data for these endosymbionts, the nature of the associations remains speculative.
Apparently, all of these symbioses have a facultative character for both partners (Fenchel & Finlay 1991a); however, for a few ciliate species (e.g. M. contortus), increased growth rates were determined in the presence of the symbionts (Fenchel & Finlay 1991b; Shinzato et al. 2007).
An intriguing aspect of the symbioses is their morphological diversity. In M. contortus, the cell wall of the endosymbiont appears to be stripped away, which goes along with an increase in the size of the symbiont (Finlay & Fenchel 1991; Embley et al. 1992). The endosymbiont of Trimyema retains its cell wall but changes size and shape to form a large stellate structure (Embley & Finlay 1993). In both cases, host hydrogenosomes closely associate with the enlarged symbionts. Cyclidium porcatum cells contain a unique complex of ca 8 µm length containing hydrogenosomes, eubacteria (of unknown affiliation) and methanogens; the latter two tightly integrated into the strongly fissured organelle (Esteban et al. 1993). A recent survey of ESTs in anaerobic rumen ciliates identified archaeal and eubacterial genes that were probably acquired by EGT. Among these, genes involved in the catabolism of complex carbohydrates were over-represented, suggesting that the acquisition of these genes has greatly facilitated the colonization of the rumen by anaerobic ciliates (Ricard et al. 2006).
(d). General remarks on bacterial endosymbionts within protists
The prokaryotic endosymbionts presented here exemplify extremely diverse physiological concepts, encompassing a photosynthetic cyanobacterial endosymbiont in a heterotrophic amoeba, a N2-fixing, non-photosynthetic cyanobacterial endosymbiont in a photosynthetic diatom, nitrogen-fixing and recycling endosymbionts of the Bacteroidales in cellulolytic, anaerobic gut flagellates and methanogenic archaea in anaerobic free-living and rumen ciliates.
The range of prokaryotic endosymbionts within protists in nature is much larger. It has been estimated that about 5 per cent of all algal cells in natural populations are likely to contain endosymbiotic bacteria (Surek & Melkonian 1983). Countless free-living amoebae and ciliates house bacterial endosymbionts (Fokin 2004; Schmitz-Esser et al. 2008). Amoebal hosts have lately attracted considerable scientific interest as they are hypothesized to represent (i) ‘training grounds’ for pathogens, adapting bacteria to intracellular environments (Molmeret et al. 2005), (ii) genetic ‘melting pots’ promoting cross-species conjugation as a result of the co-occurrence of different intracellular bacteria in amoebae (Ogata et al. 2006), and (iii) reservoirs for pathogens enabling their survival outside their specific host species (Greub & Raoult 2004; Horn 2008).
Unfortunately, for the majority of the endosymbiotic bacteria reported in protists, still not much more than a morphological description is available, precluding any conclusions about their physiological role as well as a clear recognition of the bacteria as endosymbionts, pathogens or prey.
Recent advances in genome sequencing technology allowed the rapid determination of complete genome sequences of prokaryotic genomes at reasonable costs. As a result, a better understanding of the physiological basis of many bacterial endosymbioses, mainly in invertebrates, has been achieved. Here, the crucial nutritional role of endosymbionts in diverse lineages has been unveiled (for reviews see Zientz et al. 2004; Baumann 2005; Moya et al. 2008). Common features in genome evolution of endosymbiotic bacteria were discovered such as acquisition of an AT-bias and genome reduction (Moya et al. 2008). Genome reduction is typically biased towards loss of genes that are clearly dispensable in an intracellular environment, and also loss of regulatory functions, biosynthetic pathways for metabolites provided by the host cell, DNA repair capacities and transport functions (e.g. Shigenobu et al. 2000; Akman et al. 2002; Gil et al. 2003; Kuwahara et al. 2007). The same features generally apply also for the bacterial endosymbionts of protists.
The survival of the endosymbionts despite the lack of essential genes is preferentially explained through compensation of the lacking functions by the host at the metabolite level. However, sufficient transport capacities to explain the large-scale metabolite import are consistently missing from endosymbiont genomes, suggesting the insertion of host-derived transport systems into the endosymbiont. Since also in primary plastids the majority of solute transporters are host derived (Tyra et al. 2007), typical early steps in the evolution of a stable endosymbiotic relationship between pro- and eukaryote seem to be the loss of biosynthetic capacity for essential metabolites in the endosymbiont and complementation of the lost functions by the host using host-derived transport systems.
Typical genome sizes of endosymbiotic bacteria range between 0.4 and 1.9 Mb; however, in the extreme case of Carsonella ruddii, an endosymbiotic bacterium present in psyllids, phloem sap-feeding insects, the genome has shrunk to less than 0.16 Mb (Nakabachi et al. 2006). The reductive process was accompanied by the loss of a large number of the genes involved in DNA replication, transcription and translation (Tamames et al. 2007). Since these functions cannot be compensated at the metabolite level, protein import is obviously required for the maintenance of this unit. With the discovery of the extraordinarily reduced genome of C. ruddii, the hitherto clear distinction between endosymbionts and organelles began to blur.
3. Eukaryotic endosymbionts in protists
In contrast to the diversity of functions fulfilled by prokaryotic endosymbionts, the main function performed by eukaryotic endosymbionts is photosynthesis. Many aquatic multicellular organisms live in symbioses with photosynthetic algae as typified by the well-known examples of corals, clams or the cnidarian Hydra. Algal symbioses are particularly advantageous in photic, oligotrophic environments. Even though Foraminifera display a wide variety of algal endosymbionts and secondary plastids evolved from algal endosymbionts in the Excavata (Euglenophyta) and the Rhizaria (Chlorarachniophyceae), the greatest diversity of eukaryotic endosymbionts in protists is clearly found within the Chromalveolates.
According to the higher complexity of eukaryotic cells compared with prokaryotic cells, also the modes of endosymbiotic integration found among eukaryotic endosymbionts spans a much wider array than that found among prokaryotic endosymbionts (reviewed by Schnepf 2004).
(a). Paramecium bursaria
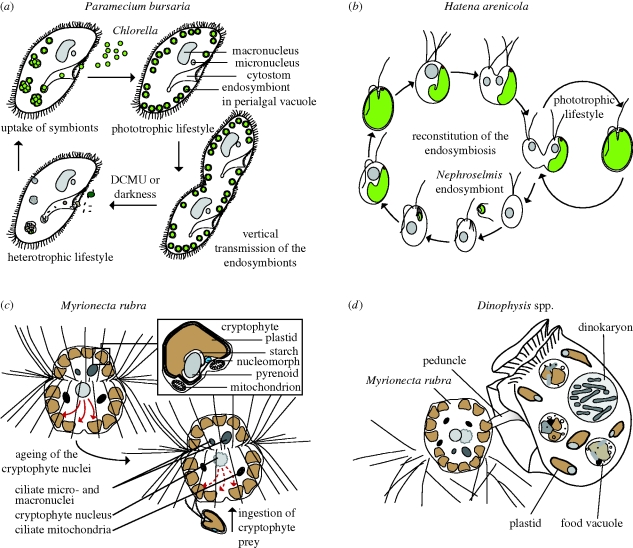
The common freshwater ciliate Paramecium bursaria (figure 3a) hosting green-algal symbionts of the genus Chlorella is a textbook example of endosymbiosis in protists. The ciliate contains hundreds of endosymbionts in individual perialgal vacuoles close to the surface of the cell (Kodama & Fujishima 2009) that are vertically transmitted to daughter cells upon host cell division (Siegel 1960). Endosymbionts supply the host mainly with photosynthetically fixed carbon in the form of maltose, supporting photoautotrophic growth of the ciliate (Brown & Nielsen 1974), while the host provides the endosymbiont with nitrogen compounds (Reisser 1976). Phylogenetic analyses of SSU rDNA and ITS2 revealed four distinct lineages of endosymbionts from the Chlorellaceae and a second green-algal clade containing species of Coccomyxa and Paradoxia multiseta, suggesting multiple origins of the symbiosis (Hoshina & Imamura 2008). Interestingly, despite its apparent stability, the symbiotic association is not obligatory, i.e. both partners can be grown separately and under appropriate conditions, and the symbiosis can be reconstituted (Weis 1983).
Figure 3.
Life history of protists with photosynthetic eukaryotic endosymbionts. (a) In P. bursaria aposymbiosis can be induced by DCMU (DCMU = 3-(3,4-Dichlorophenyl)-1,1-dimethylurea) treatment or growth in the dark. Upon availability of Chlorella cells, the symbiosis is reconstituted in the light. (b) Cell division of H. arenicola yields one symbiont-bearing green and one symbiont-lacking colourless cell, which reconstitutes the phototrophic lifestyle by ingestion of a Nephroselmis cell (modified from Okamoto & Inouye 2006). (c) In M. rubra, performance of the cryptophyte-derived kleptoplastids depends on transcriptional activity of the cryptophyte nuclei (symbolized by red arrows). Ageing nuclei are replaced by the uptake of new cryptophyte prey. (d) Dinophysis cell during myzocytotic uptake of kleptoplastids from Myrionecta rubra.
(b). Hatena arenicola
Hatena arenicola is a katablepharid flagellate recently described from an intertidal beach in Japan that shows a peculiar life history (Okamoto & Inouye 2005, 2006): In natural populations, most H. arenicola cells harbour a green-algal endosymbiont of the genus Nephroselmis bounded by a single membrane of unknown origin. Remarkably, upon cell division, the endosymbiont is inherited only by one of the two daughter cells, resulting in a symbiont-bearing green cell and a symbiont-free colourless cell that re-establishes the phototrophic lifestyle by ingestion of a new Nephroselmis cell (figure 3b). The endosymbiont occupies a distinct position and orientation inside the host cell, with the endosymbiont's eyespot always placed at the apex of the host cell, indicating a closely regulated interaction between both partners.
Interestingly, the uptake of the correct Nephroselmis species or strain within a short time leads to dramatic morphological changes in both symbiotic partners: free-living Nephroselmis cells are about 10 µm in length and kidney shaped. In contrast, in the endosymbiont the plastid is selectively enlarged up to 10-fold its original size, one to four endosymbiont nuclei were observed, other cell components, such as the endomembrane system, mitochondria and cytoskeleton including flagella, are degraded to various degrees. In the colourless H. arenicola cells, a complex apical feeding apparatus forms that degenerates within a short time after uptake of the endosymbiont. Unfortunately, as of yet H. arenicola cannot be cultured, hampering a closer examination of this fascinating symbiotic association.
(c). Myrionecta rubra Jankowski (=Mesodinum rubrum Lohmann = Cyclotrichium meunieri Powers)
A striking example of cellular chimerism was recently discovered in the ubiquitous marine ciliate Myrionecta rubra, which has attracted the interest of the scientific community for decades owing to recurrent red tides caused by its massive blooms (Ryther 1967; Lindholm 1985).
Myrionecta rubra contains cryptophyte plastids (Taylor et al. 1969; Hibberd 1977) that are most closely related to the free-living Geminigera cryophila (Johnson et al. 2006) and enable the phototrophic lifestyle of the ciliate (Smith & Barber 1979). Myrionecta rubra depends on ingestion of cryptophyte prey to sustain growth and a culture of the fragile ciliate was only successfully established when it was provided with the cryptophyte Teleaulax sp. (later determined to be G. cryophila; Johnson et al. 2006) as prey (Gustafson et al. 2000).
The cryptophyte plastids of M. rubra have long been known to be organized in numerous ‘complexes’ that besides plastids also contain cryptophyte cytoplasm and mitochondria (figure 3c), and that are bounded by a host vacuole membrane and two ER membranes (Taylor et al. 1969). Interestingly, cryptophyte nuclei are also observed in M. rubra, but are sequestered independently from the plastidal complexes (Johnson et al. 2007). The cryptophyte nucleus retains its function for up to 30 days, is transcriptionally active and serves plastids derived from multiple cryptophyte cells (Johnson et al. 2007). Thus, the sequestered cryptophyte nuclei appear to regulate the cryptophyte organelle performance. Because the retention time of prey nuclei is shorter than that for plastids, an average M. rubra cell may have eight cryptophyte plastids per single prey nucleus and prey nuclei need to be replaced by continuous feeding on cryptophyte algae (Johnson et al. 2007). Loss of prey nuclei results in the inability of the plastids to divide, leading to a decline in organelle number and biochemical potential. Thus, M. rubra cells apparently depend on recurrent stealing of cryptophyte nuclei, a highly unusual lifestyle, termed ‘karyoklepty’ by Johnson et al. (2007).
(d). Dinoflagellates
Dinoflagellates do not only represent frequent endosymbionts, but also readily form endosymbiotic associations with a variety of algae, in which they function as host partners (Schnepf & Elbrächter 1999). In line with their readiness to accept algal endosymbionts, dinoflagellates display an unusual diversity of stable secondary and tertiary plastids. Besides the peridinin-containing plastids, probably of red-algal origin, that are probably the ancestral state, there are haptophyte-derived 19′-hexanoyloxy- and/or 19′-butanoyloxy-fucoxanthin-containing plastids (e.g. in Karenia spp., Karlodinium maicrum), and green-algal-derived chlorophyll b-containing plastids (e.g. in Lepidodinium viride and Lepidodinium chlorophorum; figure 1c). The finding of non-peridinin-containing plastids implies the repeated replacement of plastids via secondary and tertiary symbiosis. Numerous dinoflagellates harbour algal endosymbionts at different stages of symbiotic integration and are likely in the process of acquisition of novel secondary or tertiary plastids. Interestingly, different host cells evolved a variety of strategies to obtain photosynthetic units.
(e). Dinoflagellates with cryptophyte-derived endosymbionts
In several dinoflagellate lineages, unstable phycobilin-containing plastids occur that originate from cryptophytes.
Amphidinium poecilochroum and Gymnodinium aeruginosum (=G. acidotum) contain cryptophyte-derived plastids that are surrounded by four envelope membranes including a nucleomorph. The plastids are accompanied by varying amounts of further cryptophyte cell material that is separated from the host cytoplasm by a further (probably) host-derived vacuole. Amphidinium poecilochroum pierces cryptophyte prey with its peduncle and ingests their cytoplasm and organelles under formation of a phagocytotic vacuole, a process known as ‘myzocytosis’ (Larsen 1988). Subsequently, ingested cell material including plastids is slowly digested. It is unclear whether the cryptophyte plastids represent food only or whether they remain photosynthetically active for some time and thus, have to be considered kleptoplastids. Kleptoplastidy is the temporary retention of plastids, obtained from ingested algal prey, that remain functional providing photosynthetic products to the predator (host).
Unlike A. poecilochroum, which can ingest several cryptophyte cells, each G. aeruginosum cell (figure 1d) contains only a single cryptophyte cell; this displays besides plastids, mitochondria, endomembrane system, and in 10–30% of the cases, a cryptophyte nucleus (Wilcox & Wedemayer 1984; Schnepf et al. 1989; Farmer & Roberts 1990). The ingested cryptophyte is not restricted to a small phagocytic vacuole, but is strongly lobed throughout the host cell (Farmer & Roberts 1990). Gymnodinium aeruginosum becomes colourless after four to seven cell divisions after isolation of single cells from natural samples and finally dies (Melkonian, M. 2002 (unpublished observations)). It is likely that deterioration of the plastid starts after digestion of the symbiont nucleus. In co-culture with Chroomonas, G. aeruginosum has been maintained over nine months (Fields & Rhodes 1991). Whereas many Chroomonas strains can serve as prey for G. aeruginosum, only few are apparently able to re-establish kleptoplastidy (Melkonian, M. 2006 (unpublished observations)).
A very peculiar case of kleptoplastidy has recently been studied in some detail in Dinophysis spp. Photosynthetic members of the toxin-producing, bloom-forming genus Dinophysis contain plastids of cryptophyte origin (Schnepf & Elbrächter 1988; Takishita et al. 2002). These are surrounded by two membranes only, and a nucleomorph is absent (Lucas & Vesk 1990). Establishment of cultures of Dinophysis spp. had consistently failed, no matter which cryptophyte species was offered as prey, until the ciliate M. rubra (see above) was provided as prey (Park et al. 2006). This method turned out to be useful for further Dinophysis species (Nagai et al. 2008; Nishitani et al. 2008; Park et al. 2008). Apparently, Dinophysis spp. feed myzocytotically on M. rubra, ingesting preferably the cryptophyte-derived kleptoplastids (figure 3d), which they retain for up to two months (Park et al. 2008). However, it was not yet unambiguously demonstrated that M. rubra is the only source of plastids in Dinophysis spp., nor is it understood which role M. rubra plays in preparing the kleptoplastids for operation in Dinophysis spp.
(f). Dinoflagellates with diatom-derived endosymbionts
In several dinoflagellates, peridinin-containing plastids are replaced with diatom-derived fucoxanthin-containing endosymbionts, e.g. Durinskia baltica (=Peridinium balticum), Kryptoperidinium foliaceum (=Peridinium foliaceum = Glenodinium foliaceum), Gymnodinium quadrilobatum, Peridiniopsis spp. and Peridinium quinquecorne (Schnepf & Elbrächter 1999; Takano et al. 2008). A red eyespot surrounded by three membranes, present in the dinoflagellate host cell, probably represents a remnant of the former peridinin plastid. As opposed to the host cells, which appear monophyletic, phylogenies of plastid-encoded rbcL and endosymbiont-encoded 18S rRNA genes reveal multiple origins of the endosymbionts within the diatoms (Takano et al. 2008). Nevertheless, dinoflagellates with diatom-derived endosymbionts represent the most stable endosymbiotic association discussed in this section.
Host cell and endosymbiont are highly interdigitated and are separated by a single membrane of unknown origin (compare Eschbach et al. 1990; Schnepf & Elbrächter 1999). The endosymbiont retains in addition to the four membrane-bounded plastid, the diatom nucleus, cytoplasm, ER, ribosomes and mitochondria (Schnepf & Elbrächter 1999). For K. foliaceum (figure 1e) and D. baltica, the functionality of both endosymbiont and host mitochondria was demonstrated by the expression analysis of essential genes of the electron transport chain (Imanian & Keeling 2007). This is apparently the first example of functional polyphyletic mitochondria within one cell. Photosynthetic products are transferred to the host cell, where they are stored as starch (Dodge 1986).
The cell cycles of endosymbiont and host are synchronized, which enables prolonged photoautotrophic cultures of the cells. Interestingly, division of the diatom nucleus occurs amitotically, and neither chromatin condensation nor spindles are observed (Dodge 1971; Tippit & Pickett-Heaps 1976). The variable size of the resulting daughter nuclei suggests a loss of function of the endosymbiont genome owing to EGT of endosymbiont nuclear genes to the dinokaryon (Figueroa et al. 2009). In the sexual cycle of D. baltica and K. foliaceum plasmogamy and karyogamy of the host were shown to be followed by that of the endosymbiont (Chesnick & Cox 1989; Figueroa et al. 2009). Although the Kryptoperidinium/diatom symbiosis is binucleate in culture, natural blooms of K. foliaceum have been found in which an endosymbiont nucleus could not be detected, suggesting kleptoplastidy may occur as well in this organism (Kempton et al. 2002).
(g). Foraminifera
Larger foraminifera are amoeboid protists of the Rhizaria living in calcified shells that reach giant sizes of 0.01–6 cm and are abundant in the benthos of tropical and semi-tropical marine habitats (Lee 1995). These foraminifera host a polyphyletic array of endosymbiotic algae, such as dinoflagellates, diatoms, chlorophytes, rhodophytes, chrysophytes and haptophytes, that reside within the host cytoplasm enclosed in a symbiont vacuole (Richardson 2001; Holzmann et al. 2006).
Foraminiferal endosymbionts are vertically transmitted during asexual reproduction of the host. Upon gametogenesis, however, the large size of most endosymbionts compared with foraminiferal gametes precludes retention of the endosymbiont; hence, symbionts need to be reacquired after gametogamy, leading to a cyclic nature of the interaction (Röttger et al. 1998). The relationship between foraminiferal host species and particular endosymbionts is not generally fixed, i.e. the same host species may harbour different endosymbiont species, sometimes within one cell (Lee et al. 1989). However, in the soritid foraminifera, which host dinoflagellate endosymbionts of the genus Symbiodinium, 14 out of 22 soritid phylotypes revealed strict symbiont specificity and only one was found to be the host for more than two phylotypes of Symbiodinium (Garcia-Cuetos et al. 2005).
Even though most foraminiferal hosts are mixotrophic and only few can grow without any obvious food source (e.g. Heterostegina depressa), host cells usually cannot survive for longer periods without their endosymbiotic algae (Lee 1995). In contrast, for the algal endosymbionts the symbiosis is not obligate.
(h). General remarks on eukaryotic endosymbionts within protists
In this section, various conceptual models for the integration of eukaryotic endosymbionts into protist cells were presented, ranging from periodic associations of two facultative symbiotic partners, via temporary retention of photosynthetic prey cells or their organelles, to permanent, obligate symbioses. Any model may go along with dramatic morphological changes of one or both partners.
The destruction of prey cells precludes interpreting kleptoplastids as true endosymbionts and the process of kleptoplastidy is best characterized as predation with farming of the prey organelles. An important question is what sustains the long-term stability of the kleptoplastids, when the prey cells are destroyed and the eukaryotic nucleus, sometimes also the nucleomorph, which encodes essential plastid proteins (Douglas et al. 2001) are lost. An interesting solution is presented by the karyoklepty found in M. rubra, i.e. the uptake of transcriptionally active cryptophyte nuclei to operate the cryptophyte kleptoplastids. Dinoflagellates may encode many genes necessary for plastid function on their nuclear genome; a fact that might facilitate the integration of new photosynthetic endosymbionts. This hypothesis is substantiated by the finding of plastid genes in the heterotrophic dinoflagellate Crypthecodinium cohnii (Sanchez-Puerta et al. 2007), suggesting that many (or perhaps all) colourless dinoflagellates may harbour previously undetected leucoplasts, as well as the demonstration of retention and use of many genes for plastid-targeted proteins, originating from the ancestral peridinin-containing plastid in Karlodinium micrum, a dinoflagellate with a tertiary haptophyte-derived plastid (Patron et al. 2006).
Kleptoplastids are found in a wide range of host species such as ciliates (Stoecker et al. 1987; Esteban et al. 2009), foraminifera (Richardson 2001) and also in multicellular organisms (Mujer et al. 1996). Interestingly, EGT was recently demonstrated in the sea slug Elysia chlorotica, which feeds on the xanthophyte Vaucheria litorea, and retains its plastids in functional state for up to nine months (Mujer et al. 1996). Despite the lack of algal nuclei, not only presence, but also expression of the Vaucheria psbO gene, a nuclear-encoded plastid-targeted gene with an essential function in photosystem II, was demonstrated in the sea slug tissue (Rumpho et al. 2008). As argued above, genetic integration should be facilitated in protists as opposed to multicellular organisms, owing to the absence of a sequestered germ line. Thus, kleptoplastids provide their hosts not only temporarily with photosynthetic products and perhaps oxygen in anoxic habitats as was recently suggested for the common freshwater ciliate Histiobalantium natans (Esteban et al. 2009), but might become genetically integrated, either by pre-existing plastid functions encoded on the host nucleus, or by EGT enabled by a simultaneous uptake of prey nuclei. In conclusion, kleptoplastids might represent a useful intermediate step in the acquisition of novel plastids.
Secondary and tertiary plastids evolved several times independently from eukaryotic phototrophs, whereas the origin of photosynthetic organelles by primary endosymbiosis was presumably a unique event and only recently the first case of an independent acquisition of a primary photosynthetic endosymbiont was described. This ratio might reflect the larger difficulties of integrating a prokaryotic system into a eukaryotic cell: gene transfer between two eukaryotic systems might be facilitated by similar genome structures (presence of eukaryotic ribosome binding sites, introns, polyadenylation signals, etc.). Additionally, and maybe more importantly, evolution of a protein import mechanism to target the former prokaryotic compartment, likely the main hurdle for genetic integration of a prokaryote into a eukaryotic cell, is already established in secondary and tertiary endosymbioses. The new eukaryotic endosymbiont can be accessed via the endomembrane system (Gould et al. 2008).
4. Conclusions and future perspectives
The evolutionary success of endosymbioses is evident from the wide range of eukaryote groups that have established endosymbiotic associations. Endosymbionts in protists may be prokaryotes that perform a multitude of new biochemical functions in the host cells, such as photosynthesis, nitrogen fixation, nitrogen recycling, methanogenesis or sulphide oxidation. Moreover, eukaryotic photosynthetic endosymbionts found various modes of integration into different host cells. Thus, endosymbiosis represents a common evolutionary strategy of eukaryotes to acquire novel biochemical potential and novel compartments to fulfil the respective functions.
If EGT (but not necessarily protein import) occurs, endosymbiosis represents, furthermore, a source of genetic innovation in eukaryotes, a requirement that is met in bacteria by frequent inter- or intraspecies horizontal gene transfer (e.g. Zhaxybayeva et al. 2006). Cases of EGT were detected in several multicellular organisms, even though direct proof of protein import is still pending. The stable retention of endosymbionts with enormously reduced genomes such as Carsonella ruddii, as well as the presence of a gene encoding a subunit of photosystem I in the nuclear genome of P. chromatophora, however, imply that the evolution of some kind of protein import mechanism is possible outside canonical organelles. These findings illustrate the difficulties drawing a clear line between endosymbiont and organelle and provide a new view on the classical concept of the eukaryotic cell.
Organisms such as Paulinella, Myrionecta and Dinophysis have been known for decades. Still only recently they started to unveil their secrets, driven by progress in sequencing technologies and culture experiments. Moreover, there are numerous endosymbiotic associations in nature, about which only scarce knowledge is available and that might reveal fascinating new insights into eukaryotic evolution and diversity, which cannot be obtained from studies of the classical model organisms.
Attempts to study additional endosymbiotic associations in laboratory cultures should be encouraged. A culture represents an indispensible base to examine the life cycle of an endosymbiosis and, therewith, determine the stability of the association, its permanent or periodic, obligate or facultative nature. Phylogenetic studies, which do not necessarily depend on the availability of a culture, may help to understand the specificity of a symbiotic relationship and elucidate its evolutionary history. For a broad understanding of the physiological role of an endosymbiont, genome studies are indispensible. As exemplified by the genomes of the endosymbionts of gut-dwelling flagellates, complete genome sequences do not depend any longer on a culture of the organism or symbiosis of interest, but may be obtained from a limited number of cells by whole genome amplification.
Important questions that still need to be addressed in most endosymbiotic relationships are how does the host cell control growth and division of the endosymbiont, which means of communication exist between the symbiotic partners and by which mechanisms are metabolites, or even proteins, exchanged. An answer to these questions could also help to understand the complex process of organelle evolution that shaped life on this planet once and forever.
Footnotes
One contribution of 12 to a Theme Issue ‘Evolution of organellar metabolism in unicellular eukaryotes’.
References
- Akman L., Yamashita A., Watanabe H., Oshima K., Shiba T., Hattori M., Aksoy S.2002Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407 (doi:10.1038/ng986) [DOI] [PubMed] [Google Scholar]
- Andersson J. O.2005Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197 (doi:10.1007/s00018-005-4539-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J. M.2009The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 (doi:10.1016/j.cub.2008.11.067) [DOI] [PubMed] [Google Scholar]
- Baumann P.2005Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189 (doi:10.1146/annurev.micro.59.030804.121041) [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Helmchen T., Melkonian M.1995Molecular evolutionary analyses of nuclear-encoded small-subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyta. J. Eukaryot. Microbiol. 42, 65–69 (doi:10.1111/j.1550-7408.1995.tb01541.x) [DOI] [PubMed] [Google Scholar]
- Boxma B., et al. 2005An anaerobic mitochondrion that produces hydrogen. Nature 434, 74–79 (doi:10.1038/nature03343) [DOI] [PubMed] [Google Scholar]
- Brown J. A., Nielsen P. J.1974Transfer of photosynthetically produced carbohydrate from endosymbiotic Chlorellae to Paramecium bursaria. J. Protozool. 21, 569–570 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T., Chao E. E. Y.2003Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154, 341–358 (doi:10.1078/143446103322454112) [DOI] [PubMed] [Google Scholar]
- Chesnick J. M., Cox E. R.1989Fertilization and zygote development in the binucleate dinoflagellate Peridinium balticum (Pyrrhophyta). Am. J. Bot. 76, 1060–1072 (doi:10.2307/2444528) [Google Scholar]
- Des Marais D. J.2000Evolution—when did photosynthesis emerge on earth? Science 289, 1703–1705 [PubMed] [Google Scholar]
- Dodge J. D.1971Dinoflagellate with both a mesocaryotic and a eucaryotic nucleus. 1. Fine structure of nuclei. Protoplasma 73, 145–157 (doi:10.1007/BF01275591) [DOI] [PubMed] [Google Scholar]
- Dodge J. D.1986A re-examination of the relationship between unicellular host and eucaryotic endosymbiont with special reference to Glenodinium foliaceum. In Endocytobiology, vol. 2 (eds Schenk H. E. A., Schwemmler W.), pp. 1015–1026 Berlin, Germany: de Gruyter [Google Scholar]
- Doolittle W. E.1998You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311 (doi:10.1016/S0168-9525(98)01494-2) [DOI] [PubMed] [Google Scholar]
- Douglas A. E.2009The microbial dimension in insect nutritional ecology. Funct. Ecol. 23, 38–47 (doi:10.1111/j.1365-2435.2008.01442.x) [Google Scholar]
- Douglas A. E., Smith D. C.1989Are endosymbioses mutualistic? Trends Ecol. Evol. 4, 350–352 [DOI] [PubMed] [Google Scholar]
- Douglas S., et al. 2001The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096 (doi:10.1038/35074092) [DOI] [PubMed] [Google Scholar]
- Drum R. W., Pankratz S.1965Fine structure of an unusual cytoplasmic inclusion in the diatom genus Rhopalodia. Protoplasma 60, 141–149 (doi:10.1007/BF01248136) [Google Scholar]
- Dubilier N., Bergin C., Lott C.2008Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740 (doi:10.1038/nrmicro1992) [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp J. C., et al. 2007Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (doi:10.1126/science.1142490) [DOI] [PubMed] [Google Scholar]
- Embley T. M., Finlay B. J.1993Systematic and morphological diversity of endosymbiotic methanogens in anaerobic ciliates. Antonie van Leeuwenhoek 64, 261–271 (doi:10.1007/BF00873086) [DOI] [PubMed] [Google Scholar]
- Embley T. M., Finlay B. J., Brown S.1992RNA sequence analysis shows that the symbionts in the ciliate Metopus contortus are polymorphs of a single methanogen species. FEMS Microbiol. Lett. 97, 57–61 (doi:10.1111/j.1574-6968.1992.tb05439.x) [DOI] [PubMed] [Google Scholar]
- Eschbach S., Speth V., Hansmann P., Sitte P.1990Freeze-fracture study of the single membrane between host cell and endocytobiont in the dinoflagellates Glenodinium foliaceum and Peridinium balticum. J. Phycol. 26, 324–328 (doi:10.1111/j.0022-3646.1990.00324.x) [Google Scholar]
- Esteban G., Guhl B. E., Clarke K. J., Embley T. M., Finlay B. J.1993Cyclidium porcatum n. sp.: a free-living anaerobic scuticociliate containing a stable complex of hydrogenosomes, eubacteria and archaeobacteria. Eur. J. Protistol. 29, 262–270 [DOI] [PubMed] [Google Scholar]
- Esteban G. F., Finlay B. J., Clarke K. J.2009Sequestered organelles sustain aerobic microbial life in anoxic environments. Environ. Microbiol. 11, 544–550 (doi:10.1111/j.1462-2920.2008.01797.x) [DOI] [PubMed] [Google Scholar]
- Farmer M. A., Roberts K. R.1990Organelle loss in the endosymbiont of Gymnodinium acidotum (Dinophyceae). Protoplasma 153, 178–185 (doi:10.1007/BF01354002) [Google Scholar]
- Fenchel T., Finlay B. J.1991aThe biology of free-living anaerobic ciliates. Eur. J. Protistol. 26, 201–215 [DOI] [PubMed] [Google Scholar]
- Fenchel T., Finlay B. J.1991bEndosymbiotic methanogenic bacteria in anaerobic ciliates: significance for the growth efficiency of the host. J. Protozool. 38, 18–22 [Google Scholar]
- Fields S. D., Rhodes R. G.1991Ingestion and retention of Chroomonas spp. (Cryptophyceae) by Gymnodinium acidotum (Dinophyceae). J. Phycol. 27, 525–529 (doi:10.1111/j.0022-3646.1991.00525.x) [Google Scholar]
- Figueroa R. I., Bravo I., Fraga S., Gracés E., Llaveria G.2009The life cycle of Kryptoperidinium foliaceum, a dinoflagellate with two eukaryotic nuclei. Protist 160, 285–300 (doi:10.1016/j.protis.2008.12.003) [DOI] [PubMed] [Google Scholar]
- Finlay B. J., Fenchel T.1991Polymorphic bacterial symbionts in the anaerobic ciliated protozoan Metopus. FEMS Microbiol. Lett. 79, 187–190 (doi:10.1111/j.1574-6968.1991.tb04526.x) [Google Scholar]
- Floener L., Bothe H.1980Nitrogen fixation in Rhopalodia gibba, a diatom containing blue-greenish inclusions symbiotically. In Endocytobiology, endosymbiosis and cell biology (eds Schwemmler W., Schenk H. E. A.), pp. 541–552 Berlin, Germany: Walter de Gruyter & Co [Google Scholar]
- Fokin S. I.2004Bacterial endocytobionts of ciliophora and their interactions with the host cell. Int. Rev. Cytol. 236, 181–249 (doi:10.1016/S0074-7696(04)36005-5) [DOI] [PubMed] [Google Scholar]
- Garcia-Cuetos L., Pochon X., Pawlowski J.2005Molecular evidence for host-symbiont specificity in soritid Foraminifera. Protist 156, 399–412 (doi:10.1016/j.protis.2005.08.003) [DOI] [PubMed] [Google Scholar]
- Geitler L.1977Life history of the Epithemiaceae Epithemia, Rhopalodia and Denticula (Diatomophyceae) and their presumable symbiotic spheroid bodies. Plant Syst. Evol. 128, 259–275 (doi:10.1007/BF00984562) [Google Scholar]
- Gil R., et al. 2003The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl Acad. Sci. USA 100, 9388–9393 (doi:10.1073/pnas.1533499100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. B., Waller R. R., McFadden G. I.2008Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517 (doi:10.1146/annurev.arplant.59.032607.092915) [DOI] [PubMed] [Google Scholar]
- Greub G., Raoult D.2004Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433 (doi:10.1128/CMR.17.2.413-433.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson D. E., Stoecker D. K., Johnson M. D., Van Heukelem W. F., Sneider K.2000Cryptophyte algae are robbed of their organelles by the marine ciliate Mesodinium rubrum. Nature 405, 1049–1052 (doi:10.1038/35016570) [DOI] [PubMed] [Google Scholar]
- Hannah F., Rogerson A., Anderson O. R.1996A description of Paulinella indentata n. sp. (Filosea: Euglyphina) from subtidal coastal benthic sediments. J. Eukaryot. Microbiol. 43, 1–4 (doi:10.1111/j.1550-7408.1996.tb02464.x) [Google Scholar]
- Hibberd D. J.1977Observations on ultrastructure of the cryptomonad endosymbiont of the red water ciliate Mesodinium rubrum. J. Mar. Biol. Assoc. UK 57, 45–61 (doi:10.1017/S0025315400021226) [Google Scholar]
- Hoffmeister M., Martin W.2003Interspecific evolution: microbial symbiosis, endosymbiosis and gene transfer. Environ. Microbiol. 5, 641–649 (doi:10.1046/j.1462-2920.2003.00454.x) [DOI] [PubMed] [Google Scholar]
- Holzmann M., Berney C., Hohenegger J.2006Molecular identification of diatom endosymbionts in nummulitid Foraminifera. Symbiosis 42, 93–101 [Google Scholar]
- Hongoh Y., et al. 2008aComplete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc. Natl Acad. Sci. USA 105, 5555–5560 (doi:10.1073/pnas.0801389105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongoh Y., et al. 2008bGenome of an endosymbiont coupling N2 fixation to cellulolysis within protist cells in termite gut. Science 322, 1108–1109 (doi:10.1126/science.1165578) [DOI] [PubMed] [Google Scholar]
- Hoogenraad H. R.1927Zur Kenntnis der Fortpflanzung von Paulinella chromatophora Lauterb. Zool. Anz. 72, 140–150 [Google Scholar]
- Horn M.2008Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62, 113–131 (doi:10.1146/annurev.micro.62.081307.162818) [DOI] [PubMed] [Google Scholar]
- Hoshina R., Imamura N.2008Multiple origins of the symbioses in Paramecium bursaria. Protist 159, 53–63 (doi:10.1016/j.protis.2007.08.002) [DOI] [PubMed] [Google Scholar]
- Imanian B., Keeling P. J.2007The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evol. Biol. 7, 172 (doi:10.1186/1471-2148-7-172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. W., Hargraves P. E., Sieburth J. M.1988Ultrastructure and ecology of Calycomonas ovalis Wulff, 1919 (Chrysophyceae), and its redescription as a testate rhizopod, Paulinella ovalis n. comb. (Filosea: Euglyphina). J. Protozool. 35, 618–626 [Google Scholar]
- Johnson M. D., Tengs T., Oldach D., Stoecker D. K.2006Sequestration, performance, and functional control of cryptophyte plastids in the ciliate Myrionecta rubra (Ciliophora). J. Phycol. 42, 1235–1246 (doi:10.1111/j.1529-8817.2006.00275.x) [Google Scholar]
- Johnson M. D., Oldach D., Delwiche C. F., Stoecker D. K.2007Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 445, 426–428 (doi:10.1038/nature05496) [DOI] [PubMed] [Google Scholar]
- Keeling P. J.2009Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 56, 1–8 (doi:10.1111/j.1550-7408.2008.00371.x) [DOI] [PubMed] [Google Scholar]
- Keeling P. J., Palmer J. D.2008Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (doi:10.1038/nrg2386) [DOI] [PubMed] [Google Scholar]
- Kempton J. W., et al. 2002Kryptoperidinium foliaceum blooms in South Carolina: a multi-analytical approach to identification. Harmful Algae 1, 383–392 (doi:10.1016/S1568-9883(02)00051-3) [Google Scholar]
- Kies L.1974Electron microscopical investigations on Paulinella chromatophora Lauterborn, a thecamoeba containing blue-green endosymbionts (cyanelles). Protoplasma 80, 69–89 (doi:10.1007/BF01666352) [DOI] [PubMed] [Google Scholar]
- Kies L., Kremer B. P.1979Function of cyanelles in the thecamoeba Paulinella chromatophora. Naturwissenschaften 66, 578–579 (doi:10.1007/BF00368819) [Google Scholar]
- Klasson L., Kambris Z., Cook P. E., Walker T., Sinkins S. P.2009Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10, 9 (doi:10.1186/1471-2164-10-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C., Voss C., Lockhart P. J., Maier U. G.2008The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol. Biol. 8, 16 (doi:10.1186/1471-2148-8-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Fujishima M.2009Timing of perialgal vacuole membrane differentiation from digestive vacuole membrane in infection of symbiotic algae Chlorella vulgaris of the ciliate Paramecium bursaria. Protist 160, 65–74 (doi:10.1016/j.protis.2008.06.001) [DOI] [PubMed] [Google Scholar]
- Kuwahara H., et al. 2007Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr. Biol. 17, 881–886 (doi:10.1016/j.cub.2007.04.039) [DOI] [PubMed] [Google Scholar]
- Larsen J.1988An ultrastructural study of Amphidinium poecilochroum (Dinophyceae), a phagotrophic dinoflagellate feeding on small species of cryptophytes. Phycologia 27, 366–377 [Google Scholar]
- Lauterborn R.1895Protozoenstudien. Z. Wiss. Zool. 59, 537–544 [Google Scholar]
- Lee J. J.1995Living sands—the symbiosis of protists and algae can provide good models for the study of host symbiont interactions. Bioscience 45, 252–261 (doi:10.2307/1312418) [Google Scholar]
- Lee J. J., McEnery M. E., Terkuile B., Erez J., Röttger R., Rockwell R. F., Faber W. W., Lagziel A.1989Identification and distribution of endosymbiotic diatoms in larger foraminifera. Micropaleontology 35, 353–366 (doi:10.2307/1485677) [Google Scholar]
- Lindholm T.1985Mesodinium rubrum—a unique photosynthetic ciliate. Adv. Aquat. Microbiol. 3, 1–48 [Google Scholar]
- Lucas I. A. N., Vesk M.1990The fine structure of two photosynthetic species of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 26, 345–357 (doi:10.1111/j.0022-3646.1990.00345.x) [Google Scholar]
- Marin B., Nowack E. C. M., Melkonian M.2005A plastid in the making: evidence for a second primary endosymbiosis. Protist 156, 425–432 (doi:10.1016/j.protis.2005.09.001) [DOI] [PubMed] [Google Scholar]
- Marin B., Nowack E. C. M., Glöckner G., Melkonian M.2007The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol. Biol. 7, 85 (doi:10.1186/1471-2148-7-85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G. I., van Dooren G. G.2004Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 14, R514–R516 (doi:10.1016/j.cub.2004.06.041) [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. S.1910Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen. Biol. Centralb. 30, 278–303 [Google Scholar]
- Molmeret M., Horn M., Wagner M., Santic M., Abu Kwaik Y.2005Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28 (doi:10.1128/AEM.71.1.20-28.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D., Le Guyader H., Philippe H.2000The origin of red algae and the evolution of chloroplasts. Nature 405, 69–72 (doi:10.1038/35011054) [DOI] [PubMed] [Google Scholar]
- Moya A., Peretó J., Gil R., Latorre A.2008Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9, 218–229 (doi:10.1038/nrg2319) [DOI] [PubMed] [Google Scholar]
- Mujer C. V., Andrews D. L., Manhart J. R., Pierce S. K., Rumpho M. E.1996Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc. Natl Acad. Sci. USA 93, 12 333–12 338 (doi:10.1073/pnas.93.22.12333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Nitshitani G., Tomaru Y., Sakiyama S., Kamiyama T.2008Predation by the toxic dinoflagellate Dinophysis fortii on the ciliate Myrionecta rubra and observation of sequestration of ciliate chloroplasts. J. Phycol. 44, 909–922 (doi:10.1111/j.1529-8817.2008.00544.x) [DOI] [PubMed] [Google Scholar]
- Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H. E., Moran N. A., Hattori M.2006The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267 (doi:10.1126/science.1134196) [DOI] [PubMed] [Google Scholar]
- Nakayama T., Ishida K.2009Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 19, R284–R285 (doi:10.1016/j.cub.2009.02.043) [DOI] [PubMed] [Google Scholar]
- Nikoh N., Tanaka K., Shibata F., Kondo N., Hizume M., Shimada M., Fukatsu T.2008Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 18, 272–280 (doi:10.1101/gr.7144908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani G., Nagai S., Takano Y., Sakiyama S., Baba K., Kamiyama T.2008Growth characteristics and phylogenetic analysis of the marine dinoflagellate Dinophysis infundibulus (Dinophyceae). Aquat. Microb. Ecol. 52, 209–221 (doi:10.3354/ame01233) [Google Scholar]
- Nowack E. C. M., Melkonian M., Glöckner G.2008Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 (doi:10.1016/j.cub.2008.02.051) [DOI] [PubMed] [Google Scholar]
- Ogata H., La Scola B., Audic S., Renesto P., Blanc G., Robert C., Fournier P. E., Claverie J. M., Raoult D.2006Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2, 733–744 (doi:10.1371/journal.pgen.0020076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma M.2003Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbiol. Biotechnol. 61, 1–9 [DOI] [PubMed] [Google Scholar]
- Ohkuma M., Sato T., Noda S., Ui S., Kudo T., Hongoh Y.2007The candidate phylum ‘Termite Group 1’ of bacteria: phylogenetic diversity, distribution, and endosymbiont members of various gut flagellated protists. FEMS Microbiol. Ecol. 60, 467–476 (doi:10.1111/j.1574-6941.2007.00311.x) [DOI] [PubMed] [Google Scholar]
- Okamoto N., Inouye I.2005A secondary symbiosis in progress? Science 310, 287 (doi:10.1126/science.1116125) [DOI] [PubMed] [Google Scholar]
- Okamoto N., Inouye I.2006Hatena arenicola gen. et sp. nov., a katablepharid undergoing probable plastid acquisition. Protist 157, 401–419 (doi:10.1016/j.protis.2006.05.011) [DOI] [PubMed] [Google Scholar]
- Park M. G., Kim S., Kim H. S., Myung G., Kang Y. G., Yih W.2006First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat. Microb. Ecol. 45, 101–106 (doi:10.3354/ame045101) [Google Scholar]
- Park M. G., Park J. S., Kim M., Yih W.2008Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J. Phycol. 44, 1154–1163 (doi:10.1111/j.1529-8817.2008.00579.x) [DOI] [PubMed] [Google Scholar]
- Patron N. J., Waller R. F., Keeling P. J.2006A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 357, 1373–1382 (doi:10.1016/j.jmb.2006.01.084) [DOI] [PubMed] [Google Scholar]
- Penard E.1905Notes sur quelques Sarcodinés—12. Paulinella chromatophora Lauterborn. Rev. Suisse Zool. 13, 585–616 [Google Scholar]
- Prechtl J., Kneip C., Lockhart P., Wenderoth K., Maier U. G.2004Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol. Biol. Evol. 21, 1477–1481 (doi:10.1093/molbev/msh086) [DOI] [PubMed] [Google Scholar]
- Reisser W.1976Metabolic interactions between Paramecium bursaria Ehrbg. and Chlorella spec. in Paramecium bursaria-symbiosis: 1. Nitrogen and carbon metabolism. Arch. Microbiol. 107, 357–360 (doi:10.1007/BF00425352) [DOI] [PubMed] [Google Scholar]
- Ricard G., et al. 2006Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7, 22 (doi:10.1186/1471-2164-7-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. L.2001Endosymbiont change as a key innovation in the adaptive radiation of Soritida (Foraminifera). Paleobiology 27, 262–289 (doi:10.1666/0094-8373(2001)027<0262:ECAAKI>2.0.CO;2) [Google Scholar]
- Rodríguez-Ezpeleta N., Brinkmann H., Burey S. C., Roure B., Burger G., Löffelhardt W., Bohnert H. J., Philippe H., Lang B. F.2005Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330 (doi:10.1016/j.cub.2005.06.040) [DOI] [PubMed] [Google Scholar]
- Röttger R., Dettmering C., Krüger R., Schmaljohann R., Hohenegger J.1998Gametes in nummulitids (Foraminifera). J. Foraminifer. Res. 28, 345–348 [Google Scholar]
- Rumpho M. E., Worful J. M., Lee J., Kannan K., Tyler M. S., Bhattacharya D., Moustafa A., Manhart J. R.2008Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl Acad. Sci. USA 105, 17 867–17 871 (doi:10.1073/pnas.0804968105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryther J. H.1967Occurrence of red water off Peru. Nature 214, 1318–1319 (doi:10.1038/2141318a0) [Google Scholar]
- Sanchez-Puerta M. V., Lippmeier J. C., Apt K. E., Delwiche C. F.2007Plastid genes in a non-photosynthetic dinoflagellate. Protist 158, 105–117 (doi:10.1016/j.protis.2006.09.004) [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S., Toenshoff E. R., Haider S., Heinz E., Hoenninger V. M., Wagner M., Horn M.2008Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl. Environ. Microbiol. 74, 5822–5831 (doi:10.1128/AEM.01093-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf E.2004Protoctists and microalgae: antagonistic and mutualistic associations and the symbiogenesis of plastids. In Progress in botany, vol. 65 (eds Esser K., Lüttge U., Beyschlag W., Murata J.), pp. 3–51 Berlin, Germany: Springer-Verlag [Google Scholar]
- Schnepf E., Elbrächter M.1988Cryptophycean-like double membrane-bound chloroplast in the dinoflagellate, Dinophysis Ehrenb.: evolutionary, phylogenetic and toxicological implications. Bot. Acta 101, 196–203 [Google Scholar]
- Schnepf E., Elbrächter M.1999Dinophyte chloroplasts and phylogeny—a review. Grana 38, 81–97 [Google Scholar]
- Schnepf E., Winter S., Mollenhauer D.1989Gymnodinium aeruginosum (Dinophyta): a blue-green dinoflagellate with a vestigial, anucleate, cryptophycean endosymbiont. Plant Syst. Evol. 164, 75–91 (doi:10.1007/BF00940431) [Google Scholar]
- Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H.2000Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (doi:10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- Shinzato N., Watanabe I., Meng X. Y., Sekiguchi Y., Tamaki H., Matsui T., Kamagata Y.2007Phylogenetic analysis and fluorescence in situ hybridization detection of archaeal and bacterial endosymbionts in the anaerobic ciliate Trimyema compressum. Microb. Ecol. 54, 627–636 (doi:10.1007/s00248-007-9218-1) [DOI] [PubMed] [Google Scholar]
- Siegel R. W.1960Hereditary endosymbiosis in Paramecium bursaria. Exp. Cell Res. 19, 239–252 (doi:10.1016/0014-4827(60)90005-7) [DOI] [PubMed] [Google Scholar]
- Smith W. O., Barber R. T.1979Carbon budget for the autotrophic ciliate Mesodinium rubrum. J. Phycol. 15, 27–33 (doi:10.1111/j.0022-3646.1979.00027.x) [Google Scholar]
- Stingl U., Radek R., Yang H., Brune A.2005‘Endomicrobia’: cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71, 1473–1479 (doi:10.1128/AEM.71.3.1473-1479.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecker D. K., Michaels A. E., Davis L. H.1987Large proportion of marine planktonic ciliates found to contain functional chloroplasts. Nature 326, 790–792 (doi:10.1038/326790a0) [Google Scholar]
- Surek B., Melkonian M.1983Intracellular bacteria in the Euglenophyceae: prolonged axenic culture of an alga—bacterial system. In Endocytobiology, vol. 2 (eds Schenk H. E. A., Schwemmler W.), pp. 475–486 Berlin, Germany: de Gruyter [Google Scholar]
- Takano Y., Hansen G., Fujita D., Horiguchi T.2008Serial replacement of diatom endosymbionts in two freshwater dinoflagellates, Peridiniopsis spp. (Peridiniales, Dinophyceae). Phycologia 47, 41–53 (doi:10.2216/07-36.1) [Google Scholar]
- Takishita K., Koike K., Maruyama T., Ogata T.2002Molecular evidence for plastid robbery (Kleptoplastidy) in Dinophysis, a dinoflagellate causing diarrhetic shellfish poisoning. Protist 153, 293–302 (doi:10.1078/1434-4610-00106) [DOI] [PubMed] [Google Scholar]
- Tamames J., Gil R., Latorre A., Peretó J., Silva F. J., Moya A.2007The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 7, 7 (doi:10.1186/1471-2148-7-181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor F. J. R., Blackbou D. J., Blackbou J.1969Ultrastructure of chloroplasts and associated structures within marine ciliate Mesodinium rubrum (Lohmann). Nature 224, 819–821 (doi:10.1038/224819a0) [Google Scholar]
- Timmis J. N., Ayliffe M. A., Huang C. Y., Martin W.2004Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (doi:10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
- Tippit D. H., Pickett-Heaps J. D.1976Apparent amitosis in the binucleate dinoflagellate Peridinium balticum. J. Cell Sci. 21, 273–289 [DOI] [PubMed] [Google Scholar]
- Tyra H. M., Linka M., Weber A. P. M., Bhattacharya D.2007Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8, R212 (doi:10.1186/gb-2007-8-10-r212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek A. H. A. M., van Alen T. A., Sprakel V. S. I., Leunissen J. A. M., Brigge T., Vogels G. D., Hackstein J. H. P.2000Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 17, 251–258 [DOI] [PubMed] [Google Scholar]
- Weis D.1983Infection in Paramedium bursaria as an inductive process. In Endocytobiology, vol. 2 (eds Schenk H. E. A., Schwemmler W.), pp. 523–532 Berlin, Germany: de Gruyter [Google Scholar]
- Wilcox L. W., Wedemayer G. J.1984Gymnodinium acidotum Nygaard (Pyrrophyta), a dinoflagellate with an endosymbiotic cryptomonad. J. Phycol. 20, 236–242 (doi:10.1111/j.0022-3646.1984.00236.x) [Google Scholar]
- Yoon H. S., Reyes-Prieto A., Melkonian M., Bhattacharya D.2006Minimal plastid genome evolution in the Paulinella endosymbiont. Curr. Biol. 16, R670–R672 (doi:10.1016/j.cub.2006.08.018) [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Nakayama T., Reyes-Prieto A., Andersen R. A., Boo S. M., Ishida K., Bhattacharya D.2009A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol. Biol. 9, 98 (doi:10.1186/1471-2148-9-98) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O., Gogarten J. P., Charlebois R. L., Doolittle W. F., Papke R. T.2006Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 16, 1099–1108 (doi:10.1101/gr.5322306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E., Dandekar T., Gross R.2004Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68, 745–770 (doi:10.1128/MMBR.68.4.745-770.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]