Abstract
Cells need a constant supply of precursors to enable the production of macromolecules to sustain growth and survival. Unlike metazoans, unicellular eukaryotes depend exclusively on the extracellular medium for this supply. When environmental nutrients become depleted, existing cytoplasmic components will be catabolized by (macro)autophagy in order to re-use building blocks and to support ATP production. In many cases, autophagy takes care of cellular housekeeping to sustain cellular viability. Autophagy encompasses a multitude of related and often highly specific processes that are implicated in both biogenetic and catabolic processes. Recent data indicate that in some unicellular eukaryotes that undergo profound differentiation during their life cycle (e.g. kinetoplastid parasites and amoebes), autophagy is essential for the developmental change that allows the cell to adapt to a new host or form spores. This review summarizes the knowledge on the molecular mechanisms of autophagy as well as the cytoplasm-to-vacuole-targeting pathway, pexophagy, mitophagy, ER-phagy, ribophagy and piecemeal microautophagy of the nucleus, all highly selective forms of autophagy that have first been uncovered in yeast species. Additionally, a detailed analysis will be presented on the state of knowledge on autophagy in non-yeast unicellular eukaryotes with emphasis on the role of this process in differentiation.
Keywords: amoebes, Atg proteins, kinetoplastid parasites, macroautophagy, selective autophagy, yeast
1. Introduction
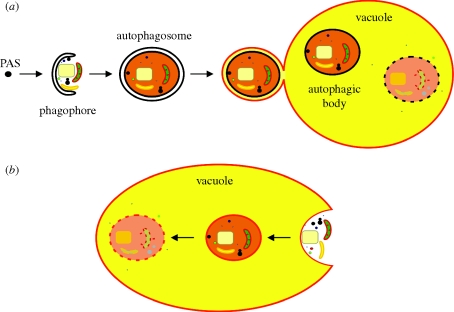
Autophagy is the process by which organisms recycle their intracellular components through the vacuole/lysosome (Yorimitsu & Klionsky 2005). The term autophagy actually covers multiple processes. Non-selective (or macro-) autophagy involves random uptake of portions of the cytoplasm (cytosol and organelles) in the vacuole/lysosome for recycling (Takeshige et al. 1992). This process is essential for cellular survival when the nutrient supply becomes limiting. Additionally, in many eukaryotes, macroautophagy is also essential during differentiation (in baker's yeast also for sporulation). When macroautophagy is induced, a double membrane forms around a portion of cytoplasm, resulting in the formation of a structure termed an autophagosome. After completion, the outer layer of the autophagosome fuses with the vacuolar membrane and a single-membrane structure, referred to as an autophagic body, enters the vacuolar lumen where it is recycled into re-usable components by vacuolar hydrolases (figure 1a). Proteins involved in macroautophagy have been designated Atg proteins (Klionsky et al. 2003).
Figure 1.
Macro- and microautophagy in yeast. (a) Schematic representation of macroautophagy in baker's yeast. Upon induction of (nitrogen) starvation, the PAS (pre-autophagosomal structure or phagophore assembly site) incorporates membrane material and grows out to become the double membrane-layered phagophore that randomly sequesters cytoplasmic components (proteins and organelles). After complete engulfment of the cytoplasmic material, an autophagosome is formed. Subsequently, the outer membrane of the autophagosome fuses with the vacuolar membrane. As a result, this membrane obtains vacuolar characteristics (red colour) and ultimately becomes part of the vacuolar membrane, thereby increasing the size of the organelle. Additionally, a single membrane-bound autophagic body enters the vacuole, where it will become degraded by vacuolar hydrolases. (b) Schematic representation of microautophagy in baker's yeast. During microautophagy, the membrane of the vacuole engulfs a portion of the cytoplasm (including organelles). As a result, a vesicle with a single membrane originating from the vacuolar membrane (red colour) is formed inside the vacuole. This contrasts to macroautophagy, where the membrane of the autophagic body originates from the autophagosome (black colour). After complete engulfment, the vesicle and its contents are degraded by vacuolar hydrolases. Consequently, during microautophagy, the size of the vacuolar membrane is reduced. During selective forms of autophagy, mechanisms similar to macro- and microautophagy are used to selectively package peroxisomes, mitochondria, endoplasmic reticulum, ribosomes and portions of the nucleus (see text for details).
In addition to macroautophagy, random uptake of cellular components can also occur via microautophagy, a process that is characterized by uptake of a portion of cytoplasm by direct invagination of the vacuolar membrane (figure 1b; Kunz et al. 2004). Because the molecular mechanisms of this process do not depend on Atg proteins, microautophagy will not be discussed further in this review.
A multitude of related processes requires Atg proteins for selective uptake of proteins/organelles into the vacuole (table 1). In all cases, a common theme appears to be the presence of a specific receptor-like protein that links the protein/organelle to be transported to the autophagy machinery (for a review, see Kraft et al. 2009).
Table 1.
Types of autophagy and related processes in eukaryotes. Cvt, cytoplasm-to-vacuole-targeting; ER, endoplasmic reticulum; PMN, piecemeal microautophagy of the nucleus. Processes in bold are described in the text. For an overview of all types of autophagy, see Klionsky et al. (2007).
| process | selective | direct involvement of Atg proteins |
|---|---|---|
| aggrephagy | yes | yes |
| chaperone-mediated autophagy | yes | no |
| crinophagy | yes | no |
| Cvt pathway | yes | yes |
| ER-phagy | yes | yes |
| heterophagy (endocytosis) | no/yes | no |
| macroautophagy | no | yes |
| macropexophagy | yes | yes |
| microautophagy | no | no |
| micropexophagy | yes | yes |
| mitophagy | yes | yes |
| PMN | yes | yes |
| ribophagy | yes | yes |
| vid pathway | yes | no |
| xenophagy | yes | yes |
In this review, first macroautophagy and related processes in Saccharomyces cerevisiae will be discussed. Subsequently, I will summarize the knowledge on autophagy in other unicellular eukaryotes.
2. Proteins involved in macroautophagy in yeast
Invariably, Atg proteins have been first identified in yeast species. However, for many Atg proteins, homologues have been found encoded by the genomes of other eukaryotes. In general, the core macroautophagy machinery is composed of conserved components, while the proteins essential for selective forms of autophagy have usually not been identified in eukaryotes other than yeast species (e.g. Meijer et al. 2007). In the next sections, I will term those homologues arising from a speciation event orthologues, while proteins with weak similarity that nevertheless (are thought to) perform the same function will be designated functional counterparts.
(a). Atg proteins required for the initial stages of autophagy
The protein kinase Tor regulates cell growth in response to nutrient availability and cellular stress (Schmelzle & Hall 2000). During nutrient-rich conditions, a complex containing Tor (TORC1) inhibits macroautophagy, but allows the selective, autophagy-related cytoplasm-to-vacuole-targeting (Cvt) pathway to proceed. The principal downstream effectors of Tor are thought to be members of the type 2A protein phosphatase family, which regulate both pathways via the phosphorylation state of the so-called Atg1 complex (reviewed by Cebollero & Reggiori 2009). This complex is essential for all forms of autophagy and its core is composed of the Ser/Thr protein kinase Atg1 and Atg13. Under nutrient-rich conditions, TORC1 blocks macroautophagy by allowing hyperphosphorylation of Atg13. Upon nutrient limitation, the phosphorylation state of Atg13 is reduced, which enhances its affinity for Atg1. The resulting lowly phosphorylated Atg1/Atg13 complex initiates macroautophagy. Thus, the Atg1 complex represents an important switch between macroautophagy and the selective Cvt pathway. Recent data indicate that the kinase activity of Atg1 is essential for both autophagy and the Cvt pathway (Kabeya et al. 2005). Unfortunately, the physiological protein substrate for the Atg1 kinase remains unknown. Atg1 and Atg13 are not the sole components of the Atg1 complex. A multitude of other Atg proteins can either directly or indirectly associate with the complex (Atg11, Atg17, Atg19, Atg20, Atg24, Atg29, Atg31 and Vac8). Some of these are exclusively required for macroautophagy, while others are exclusively required for the Cvt pathway (electronic supplementary material, table S1). It is unlikely that all these components interact with the Atg1/Atg13 complex simultaneously, but as yet no separate complexes have been isolated. Notably, Atg11 is only essential for the selective forms of autophagy, suggesting a function as a general scaffold protein that links the various cargoes to Atg1 (Cebollero & Reggiori 2009). Nevertheless, Atg11 has so far only been identified in fungi (Meijer et al. 2007).
(b). Formation of a double membrane-layered vesicle
Many of the autophagy-related pathways require a cascade of reactions that leads to the formation of the double-layered membrane (the phagophore), which sequesters proteins/organelles from the cytosol. This process is initiated by the recruitment of proteins and lipids to a presumed membranous structure known as the pre-autophagosomal structure or phagophore assembly site (PAS; Kim et al. 2002; Suzuki et al. 2002). A complex that controls the early stages of all autophagy-related pathways is the class III phosphatidylinositol 3-kinase (PI 3-K) complex, which functions at the PAS. This complex consists of the Ser/Thr protein kinase Vps15, the PI 3-K Vps34 as well as Atg6 and Atg14 (Kihara et al. 2001). The activity of the PI 3-K complex on membrane-located PI molecules allows recruitment of PI 3-P binding proteins including Atg18, Atg21, the previously mentioned Atg20 and Atg24 and the integral membrane protein Atg27 (Nice et al. 2002; Wurmser & Emr 2002; Strømhaug et al. 2004).
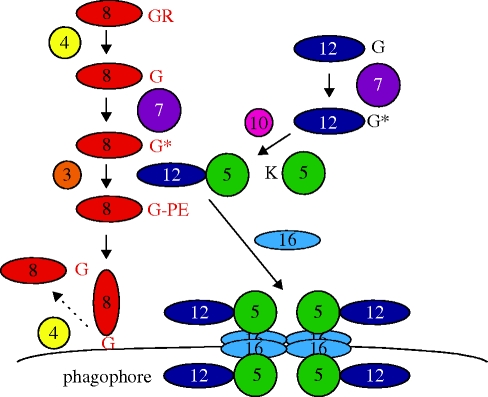
The second step of vesicle formation requires two sets of proteins that participate in two ubiquitin (Ub) conjugation-like reactions (figure 2; Ohsumi 2001). In the first conjugation reaction, the protease Atg4 processes the Ub-like protein Atg8 at its C-terminus, whereupon the protein becomes covalently attached via its C-terminal glycine residue to phosphatidylethanolamine (PE) at the PAS/phagophore. In analogy to the Ub conjugation machinery, this conjugation step requires the activities of an E1 enzyme (Atg7) and an E2 enzyme (Atg3). In addition, binding of Atg8 to PE also depends on the product of a second conjugation reaction, involving another Ub-like protein, Atg12, which becomes covalently attached via its C-terminal glycine residue to a conserved lysine residue of Atg5. This conjugation step is also catalyzed by the E1 enzyme Atg7, but requires another E2 enzyme, namely Atg10. Recent data indicate that the Atg5=Atg12 conjugate functions analogous to an E3 enzyme in the conjugation of Atg8 to PE (Hanada et al. 2007). Finally, the coiled-coil protein Atg16 becomes non-covalently attached to the Atg5=Atg12 conjugate. Membrane attachment of Atg8 to PE and the formation of the Atg5=Atg12/Atg16 complex are essential to drive vesicle enlargement. The latter complex has also been suggested to form a transient coat, thereby shaping the phagophore membrane. During this process, also the integral membrane proteins Atg9 and Atg27 and the membrane-associated protein Atg23 (Reggiori et al. 2004; Yen et al. 2007; see below) are brought to the phagophore, presumably in membrane vesicles that become incorporated in the growing sequestering membrane. During membrane expansion, Atg9, Atg23 and Atg27 continuously cycle between the phagophore and other not well-defined membranous structures.
Figure 2.
The core autophagy machinery. Schematic representation of the two Ub-like conjugation systems that drive phagophore enlargement by forming Atg8-PE and the Atg5=Atg12/Atg16 complex. The numbers refer to Atg proteins. Specific amino acid residues required for the conjugation reactions are indicated in the one-letter code. The asterisks indicate the Atg7 activated forms of the Ub-like proteins Atg8 and Atg12. PE, phosphatidylethanolamine. For details, see text.
(c). Recycling of components from autophagosomes
Not all proteins involved in phagophore expansion remain localized to the double membrane structure. Many proteins, including Atg8 and the Atg5=Atg12/Atg16 complex that are on the outside of the membranous structure, are released upon completion of sequestration. As indicated above, also the membrane proteins Atg9, Atg23 and Atg27 are continuously being retrieved from the growing phagophore, a process that depends on the activity of Atg1, Atg2 and Atg18 (Reggiori et al. 2004). Release of the covalently attached Atg8 protein from the completing autophagosome involves its cleavage from PE by the protease Atg4 (figure 2). However, a significant portion of Atg8 remains trapped inside the vesicle and will accompany the cargo into the vacuole, where it will be degraded.
(d). Degradation of engulfed material in the vacuole, and release of breakdown products
As soon as the autophagosomal membrane has been completed, fusion of its outer membrane occurs with the vacuolar membrane. This step uses the same components as homotypic vacuole fusion (e.g. SNAREs and the HOPS complex; Klionsky 2005). Because of their general function in membrane fusion events, these proteins have not obtained an Atg nomenclature and they will not be discussed further. Fusion results in incorporation of sequestered material in the vacuole matrix, where it will be degraded. This is followed by a release of the breakdown products from the vacuole. The baker's yeast vacuole contains a multitude of hydrolases, including the proteases PrA, PrB, CPS and CPY. In addition, two Atg proteins have been identified that are required for degradation of some autophagy cargo (electronic supplementary material, table S1). The integral membrane protein Atg15, a putative lipase thought to lyse the membrane of the autophagic body, is transported via the secretory pathway to the late Golgi and subsequently enters the vacuole lumen via the multi-vesicular body pathway (Epple et al. 2001, 2003). Atg22 is an integral vacuolar membrane protein with similarity to permeases of the major facilitator superfamily that has been suggested to function in export of regenerated material from the vacuole to the cytosol (Suriapranata et al. 2000; Yang et al. 2006).
(e). Selective forms of autophagy
During the last years, a number of selective autophagy-related processes have been uncovered requiring the function of Atg proteins: the biogenetic Cvt pathway and the degradative pexophagy, mitophagy, ER-phagy, ribophagy and piecemeal microautophagy of the nucleus (PMN) pathways (table 1; reviewed by Kraft et al. 2009). In these processes, cargo recognition is an essential step that involves specific sets of proteins (electronic supplementary material, table S1). Often, an intact actin cytoskeleton also appears to be essential for these selective autophagy pathways, which is not the case for macroautophagy. Nevertheless, once recognition has taken place, the Atg machinery described above is required to sort the tagged material to the vacuole lumen.
(i). The cytoplasm-to-vacuole-targeting pathway
The Cvt pathway occurs during nutrient-rich conditions and involves the selective transport of the hydrolases aminopeptidase I (Ape1) and α-mannosidase (Ams1) to the vacuole (reviewed by Teter & Klionsky 2000). So far, the Cvt pathway has only been identified in the yeast species S. cerevisiae and Pichia pastoris (Kim et al. 1997; Farré et al. 2007). In this pathway, precursor Ape1 molecules form dodecamers in the cytosol that assemble into larger Ape1 complexes. Subsequently, a Cvt-specific receptor protein, Atg19, binds to the N-terminus of pre-Ape1, thereby forming the Cvt complex. This complex also contains Ams1, the second Cvt cargo protein. Atg11 functions as the adaptor between Atg19 and the PAS, and enables tight packaging of cargo-receptor molecules into a double-membrane structure, designated the Cvt vesicle. This process requires all the components of the core autophagy machinery as well as the actin cytoskeleton (Monastyrska et al. 2008). Subsequently, the outer layer of the Cvt vesicle fuses with the vacuolar membrane to allow the release of a Cvt body in the vacuole lumen, which will lyse and release its contents. Similar to Atg8 (see above), the Atg11 and Atg19 molecules that are bound to the inner membrane of the Cvt vesicle enter the vacuole where they become degraded, while the precursor Ape1 molecules become activated. Remarkably, Cvt vesicle formation from the PAS is not initiated when both the cargo proteins or either Atg11 or Atg19 is absent, implying that the vesicle is only formed when cargo is available and properly attached to the PAS (Shintani & Klionsky 2004). In addition, Atg19 becomes ubiquitinated, which may enhance its binding capacity to Cvt cargo proteins (Baxter et al. 2005; see below).
(ii). Pexophagy
In many organisms, peroxisomes are inducible organelles. Conversely, when the induced peroxisomes become metabolically redundant, or are damaged, they are removed from the cytosol by autophagy-related pathways, termed macro- and micropexophagy. These processes have been studied in detail in the methylotrophic yeast species Hansenula polymorpha and P. pastoris (reviewed by Sakai et al. 2006). During macropexophagy, multiple membrane layers sequester a single peroxisome resulting in the formation of a pexophagosome of which the outer membrane layer fuses with the vacuole where the peroxisome becomes hydrolysed. Micropexophagy involves the uptake of a cluster of peroxisomes through direct engulfment by the vacuolar membrane.
For recognition of peroxisomes during macropexophagy two peroxisomal membrane proteins were shown to be essential, namely Pex3 and Pex14 (Bellu et al. 2001, 2002). Both proteins are peroxins—i.e. also required for peroxisome biogenesis (reviewed by Kiel et al. 2006). Atg30 represents a candidate receptor protein that can bind both Pex3 and Pex14 as well as Atg11, thereby possibly linking peroxisomes to be degraded to the autophagy machinery (Farré et al. 2008). Atg11 and Atg30 are essential for both macro- and micropexophagy. During the latter process, a specifically formed membrane structure designated the micropexophagy-specific membrane apparatus (MIPA, presumably derived from the PAS; Mukaiyama et al. 2004) contains many of the core Atg proteins. But unlike macropexophagy, where tagged peroxisomes become completely surrounded by a sequestering membrane derived from the PAS, during micropexophagy the MIPA is exclusively required to fuse the two sides of the vacuole that engulf a peroxisome cluster.
(iii). Mitophagy
Similar to peroxisomes, mitochondrial function and morphology in baker's yeast depend on the growth conditions. During growth on glucose, fermentation can occur and mitochondria are poorly differentiated. In contrast, when yeast cells are grown on carbon sources requiring respiration, mitochondrial biogenesis is robust. This implies that turnover of mitochondria (mitophagy) can be easily induced in yeast by changing the carbon source for growth. For this several distinct mechanisms are available. In addition to non-selective macroautophagy, these include selective micro- and macromitophagy (reviewed by Tolkovsky 2009). So far, a few proteins specific for mitophagy have been identified in S. cerevisiae, but their role is not well understood (Priault et al. 2005; electronic supplementary material, table S1) The role of Atg genes in mitophagy was investigated by Kanki & Klionsky (2008). These authors concluded that mitophagy is indeed a selective process that appears not to be just an adaptation of the Cvt pathway. Recently, a putative receptor protein, designated Atg32, was identified that is exclusively required for mitophagy (Kanki et al. 2009).
(iv). Degradation of portions of the endoplasmic reticulum (ER-phagy)
In addition to sequestration by random macroautophagy, portions of the endoplasmic reticulum (ER) can also be selectively packaged into autophagosomes upon induction of the unfolded-protein response (UPR) or during starvation (Hamasaki et al. 2005; Bernales et al. 2006). Under strong UPR-inducing conditions, ER-phagy depends on Atg proteins (electronic supplementary material, table S1). When induced by starvation, ER-phagy is only partly Atg dependent (Hamasaki et al. 2005; Mijaljica et al. 2006). This suggests that during starvation selective uptake of portions of the ER by autophagosomes may not use the entire phagophore assembly machinery. Also, actin seems to be required for ER-phagy, indeed implying that ER-phagy is a selective process (Hamasaki et al. 2005).
(v). Ribophagy
For a long time it was thought that the presence of ribosomes in autophagosomes was caused by random macroautophagy. However, under certain conditions ribosomes are more rapidly degraded than cytosolic proteins, implying a selective mechanism (Kraft et al. 2008). As might be expected, ribophagy of both the 60S and 40S ribosomal subunits requires the core Atg machinery as well as specific proteins (electronic supplementary material, table S1). Intriguingly, the specific degradation of the 60S ribosomal subunit, but not that of the 40S subunit, was affected when the Ub protease Ubp3 or its cofactor Bre3 was absent. Remarkably, both proteins have been shown to interact with the Cvt receptor Atg19 (Baxter et al. 2005). This suggests that Ub conjugation may be important for efficient transport of proteins/organelles to the vacuole, reminiscent of the role of Ub during aggrephagy, the transport of protein aggregates via the receptor SQSTM1/p62 to lysosomes in mammalian cells (Seibenhener et al. 2007).
(vi). Piecemeal microautophagy of the nucleus
Even the nucleus is not excluded from degradation by the vacuole. In baker's yeast, starvation also results in pinching-off and degradation of portions of the nucleus by PMN. Morphologically, this pathway resembles microautophagy, which is largely independent of Atg proteins. Nevertheless, it was demonstrated that PMN not only requires the core Atg machinery, but also Atg11, implying a selective mode of autophagy (Krick et al. 2008; electronic supplementary material, table S1). During PMN, nuclear–vacuolar junctions (NVJs) are formed via an interaction between the vacuolar membrane protein Vac8 and the outer nuclear membrane protein Nvj1 (Roberts et al. 2003). Nutrient starvation increases Nvj1 levels, and recruits the ankyrin repeat protein Osh1 and the enoyl reductase Tsc13 to the NVJ (Kvam & Goldfarb 2004; Kvam et al. 2005). As a consequence, part of the NVJ buds into the vacuole resulting in a bleb that is pinched off into the vacuole lumen, where it will be degraded. During starvation, PMN is thought to be mainly involved in regulating certain nuclear processes and is probably not a major source of nutrients.
3. Autophagy in other unicellular eukaryotes
Although autophagy in unicellular eukaryotes has mainly been studied in S. cerevisiae, this process has been observed in many other species, e.g. in trichomonads (Benchimol 1999), kinetoplastid parasites and amoebes. However, only in few of these species were autophagy-related processes studied in some detail. In many cases, this was triggered by the observation that autophagy correlated with cell differentiation. In the following sections, the focus will be on describing autophagy processes in those species where studies included some molecular analysis.
(a). Kinetoplastid parasites
Trypanosomatids are the causative agents of chronic and potentially fatal diseases that occur mainly in developing countries. Leishmania spp. cause different forms of leishmaniasis in various parts of the world, Trypanosoma brucei is responsible for human sleeping sickness and a similar cattle disease known as nagana in large parts of sub-Saharan Africa and Trypanosoma cruzi causes Chagas disease in Latin America (reviewed by Barrett et al. 2003). These parasites all have a life cycle that alternates between an insect vector and a mammalian host, requiring profound metabolic and morphological changes.
(i). The role of autophagy in Leishmania
Leishmania are most commonly parasites of humans and dogs that occur in different developmental forms in sandflies and mammals (reviewed by Bates & Rogers 2004). In the alimentary tract of the insect vector, Leishmania exist in two main forms of motile cells known as promastigotes: the multiplicative, non-infective procyclic promastigotes and the non-multiplicative, but infective metacyclic promastigotes. Upon injection into the mammalian host, Leishmania promastigotes are taken up by macrophages in which the metacyclic forms differentiate into small multiplicative, non-motile amastigotes that live in a lysosomal compartment known as the parasitophorous vacuole.
Recent data have shown that (macro)autophagy is essential for the differentiation of procyclic into metacyclic promastigotes and of metacyclic promastigotes into amastigotes (reviewed by Besteiro et al. 2007). During differentiation, an increase in the abundance of autophagosomes and an increased level of protein degradation were observed (Besteiro et al. 2006; Williams et al. 2006). In these parasites, the lysosome has multiple shapes that reflect the different functions in the various developmental forms (reviewed by Waller & McConville 2002). Leishmania mexicana promastigotes contain a tubular-vesicular compartment termed the multi-vesicular tubule (MVT) that accumulates FM4-64 and other endocytosed markers and also harbours lysosomal cysteine and serine peptidases. In contrast, amastigotes are characterized by the presence of a large membrane-bound compartment termed the megasome, which has an acidic pH and contains many peptidases. These lysosome-like structures have highly varying enzyme contents depending on the stage of parasite differentiation. Importantly, two lysosomal cysteine peptidases, CPA and CPB, have been directly implicated in both autophagy and development. In mutant parasites lacking these enzymes, promastigotes have an enhanced number of autophagosome-containing MVTs. Although mutant cells survived, cell remodelling into amastigotes appeared to be largely prevented (Williams et al. 2006). Additionally, Besteiro et al. (2006) demonstrated that a block at a late stage of endosomal sorting by inactivation of the Leishmania major VPS4 gene resulted in an accumulation of cytosolic autophagosomes that could not be processed further. As a consequence, Lm-vps4 mutants were less able to survive nutrient deprivation and were unable to differentiate into metacyclic promastigotes.
Genomics studies (Besteiro et al. 2006; Williams et al. 2006, 2009) have revealed that in L. major all Atg proteins of the two ubiquitination cascades are present. Remarkably, 26 Atg8-related proteins were identified that fall into four families, ATG8 (one member), ATG8A (three members), ATG8B (nine members) and ATG8C (13 members). Of these, only Lm-ATG8 shows high similarity to yeast and mammalian Atg8, and this protein has been successfully used to label autophagosomes (Besteiro et al. 2006). The other Atg8-related proteins also appear to localize to puncta in promastigotes, with members of the ATG8A family possibly localizing to autophagosomes as well (Williams et al. 2009). Two Atg4 proteins were identified in L. major (Lm-ATG4.1 and Lm-ATG4.2), while single orthologues were found for Atg3 and Atg7. Distant Atg5, Atg12 and Atg16 homologues only showed weak similarity to their yeast and human counterparts.
A deletion of Lm-ATG4.2 was shown to have a defect in autophagy and resulted in promastigotes that were less able to withstand starvation and unable to differentiate (Besteiro et al. 2006). Although in this mutant GFP · Lm-ATG8 puncta were still present during starvation, highly increased levels of the lipidated form of the protein were observed. This suggests that the Lm-atg4.2 mutant is not blocked in the activation of Lm-ATG8 prior to its conjugation to PE, but rather in the release of Lm-ATG8 from PE at the autophagosomal membrane (cf. figure 2). This view is consistent with the observation that in vitro Lm-ATG4.1 had proteolytic activity towards Lm-ATG8, whereas Lm-ATG4.2 showed only minor activity towards this protein (Williams et al. 2009). Apparently, the two functions combined in baker's yeast Atg4 are present in two different forms of the peptidase in L. major.
Functional complementation in S. cerevisiae atg mutants demonstrated that members of all Lm-ATG8 families could complement Sc-atg8. Similarly, the L. major homologues of Atg5 and Atg10 could replace their yeast counterparts, despite their weak similarity. Unlike yeast and mammalian Atg12, the protein designated Lm-ATG12 has a C-terminal extension after the key glycine residue that is essential for conjugation to Atg5. Lm-ATG12 also shows significant similarity to Atg8 proteins (see below). Both Lm-ATG4 proteins were unable to cleave the C-terminal extension from Lm-ATG12, implying that an alternative protease must be present to free the glycine residue. It was demonstrated that only a mutant Lm-ATG12 protein with a free C-terminal glycine could replace Sc-Atg12. Moreover, an RFP · Lm-ATG12 fusion protein localized to punctate structures in starving L. major promastigotes, which occasionally contained also GFP · Lm-ATG8. Biochemical data indicated the presence of an RFP · Lm-ATG12 complex with a higher molecular mass, pointing towards the presence of a possible Lm-ATG12=Lm-ATG5 conjugate. Based on these data, Williams et al. (2009) concluded that the putative Lm-ATG12 protein was indeed the functional counterpart of Sc-Atg12. Whether in L. major this novel type of Atg12 has displaced a close relative of yeast Atg12 during evolution or whether L. major and relatives have never contained anything closely resembling yeast Atg12 remains to be investigated.
(ii). Autophagy in Trypanosoma cruzi
The life cycle of T. cruzi involves four major developmental stages (for a review, see De Souza 2002). The infective stage of the parasite, the metacyclic trypomastigote, enters the mammalian host from the insect faeces through wound openings or mucous membranes. In the mammalian host, the metacyclic trypomastigote differentiates into a replicative form designated amastigote. After several rounds of replication in the host's cells, the amastigote differentiates into the bloodstream trypomastigote, which can enter new cells and perpetuate the infection. When the insect bites an infected host, the bloodstream trypomastigote differentiates into the replicative epimastigote that lives in the insect's gut. Finally, in the rectum of the insect, the epimastigote differentiates into the infective metacyclic trypomastigote that is ready to infect its host again.
It was noted that differentiation of the epimastigote in the insect's rectum was triggered by nutritional stress caused by the very low prevalent nutrient content. The insect's epimastigote stage harbours a lysosome-like organelle designated the reservosome (Cunha-e-Silva et al. 2006), which contains endocytosed material, lipids and a major cysteine proteinase, cruzipain. Apparently, massive proteolysis occurs during differentiation into metacyclic trypomastigotes. Indeed, it was demonstrated that cysteine proteinase inhibitors blocked differentiation (Franke de Cazzulo et al. 1994). Recently, it was observed that differentiating epimastigotes have very intense Tc-Atg8.1 staining, while metacyclic trypomastigotes have almost no signal. Furthermore, the Tc-Atg8.1 signal co-localized in part with carboxypeptidase in reservosomes, suggesting that the epimastigotes were undergoing massive autophagy (Alvarez et al. 2008).
Genome analysis has identified candidate ATG genes in the T. cruzi genome (Herman et al. 2006; Alvarez et al. 2008). From the key Atg proteins, single orthologues of Atg7 and Atg3 and two candidate Atg8 (Tc-Atg8.1 and Tc-Atg8.2) and Atg4 (Tc-Atg4.1 and Tc-Atg4.2) proteins were found as well as a distant Atg16.
In an in vitro cleavage assay, Tc-Atg4.1 was able to rapidly process both Tc-Atg8.1 and Tc-Atg8.2, while Tc-Atg4.2 showed little activity towards these proteins, reminiscent of the situation observed in L. major (see above). Nevertheless, both Tc-ATG4 genes could complement an Sc-atg4 mutant. Tc-Atg8.1 displayed the highest similarity to Sc-Atg8 and could partially replace Sc-Atg8, while Tc-Atg8.2 could not. Moreover, only HA-tagged Tc-Atg8.1 localized to large spots, thought to represent autophagosomes, and this localization was dependent on the key glycine residue at the C-terminus of the protein. In contrast, HA-tagged Tc-Atg8.2 only showed a weak punctate localization during starvation. Based on these results, the authors concluded that Tc-Atg8.1 was probably the functional homologue of yeast and mammalian Atg8. The function of Tc-Atg8.2 remained unclear (see below).
(iii). Pexophagy in Trypanosoma brucei
Also T. brucei undergoes a series of developmental changes during its life cycle in the insect vector and the mammalian host (for a review, see Matthews 2005). The parasite is transmitted by the tsetse fly when it takes its blood meal, and a non-dividing metacyclic form present in the insect's salivary gland enters the bloodstream of the mammalian host. This quickly develops into a rapidly dividing slender form, resulting in a parasitaemia in the host's bloodstream. Subsequently, the slender form undergoes cell cycle arrest and develops into short-stumpy forms. When the tsetse fly bites an infected host, only the short stumpy parasites survive in the insect's midgut and develop into a procyclic form. Finally, the parasite undergoes multiple developmental phases on its way to the insect's salivary gland that finally culminate in the infective metacyclic form.
So far, the bloodstream form of the parasite has been studied in most detail. This form of the parasite lives in an environment where glucose is present at a fairly constant level, and consequently has developed a simple, but special, way to benefit most from its surroundings. It obtains all its energy (ATP) via glycolysis in peroxisome-related organelles called glycosomes where uniquely virtually all enzymes of the glycolytic pathway are concentrated, while mitochondrial pathways are highly repressed (reviewed by Michels et al. 2006). The glycosome is essential for the viability of the bloodstream form of the parasite, and mutants that lack glycosomes die as a result of accumulation of high levels of glycolytic intermediates (see Haanstra et al. 2008 and the references therein). In contrast, the procyclic (insect) form of the parasite has a much more complex metabolism with a normally developed mitochondrion. This form relies for growth more on amino acids like proline that is abundantly present in the midgut of the insect (Michels et al. 2006). Consequently, the number, size and enzymatic contents of the glycosomes undergo large variations during the developmental changes of the parasite, which is reminiscent of the situation observed in methylotrophic yeasts, where peroxisomes are synthesized and degraded by pexophagy dependent on their requirement (reviewed by Sakai et al. 2006). This has initiated an investigation into the autophagic degradation of glycosomes in T. brucei during its various stages of differentiation. Indeed, the first results are consistent with the notion that during differentiation ‘old’ glycosomes are degraded while new organelles become synthesized, which contain an enzyme repertoire better adapted to the lifestyle of the new form of the parasite (Herman et al. 2008).
Like autophagy, pexophagy uses all Atg proteins required to build the autophagosome. Indeed, an in silico analysis of the genomes of T. brucei and the related T. cruzi (see above) has identified many of the core autophagy components (Herman et al. 2006). However, the components of the second conjugation machinery (Atg5, Atg10, Atg12 and Atg16) were not found, leading the authors to conclude that both parasites may harbour only a minimal form of autophagy (see below).
(b). Amoebes
(i). Autophagy in the slime mould Dictyostelium discoideum
Dictyostelium discoideum is a soil amoeba that in nature feeds on bacteria. When grown on rich media or bacterial lawns, D. discoideum exists as a unicellular organism. However, upon starvation, chemotaxis results in aggregation of many thousands of amoebae forming a mound that undergoes development to form a multi-cellular fruiting body, in which stalks support balls of spores. In this structure, the stalk cells become highly vacuolated and undergo developmental cell death (reviewed in Kessin 2001). This has led to the speculation that autophagy may be required for development. Analysis of the D. discoideum genome has identified (most of) the core components required for macroautophagy (Otto et al. 2003, 2004; Tekinay et al. 2006; Rigden et al. 2009). The study of D. discoideum atg mutants has demonstrated that the development of the amoebae into the fruiting body requires autophagy (Otto et al. 2003, 2004). However, whether and how far development proceeds depend on the Dd-atg mutant analysed. The Dd-atg1-1 mutant has the most severe phenotype and does not produce any spores, while conversely Dd-atg6A and Dd-atg8 mutants can make fruiting bodies, which contain fewer and immature spores (Otto et al. 2004). Also, a mutant lacking a mammalian-style Dd-Atg16L (initially identified as TipD) has a severely reduced fruiting body and spore production (Stege et al. 1999). In addition to a block in development, Dd-atg mutants are hypersensitive to amino acid starvation and degrade significantly less protein than wild-type cells. Furthermore, during starvation, Dd-atg mutants retain many organelles in the cytoplasm, while starved wild-type cells have an almost empty cytoplasm with few organelles.
Molecular studies have demonstrated that in D. discoideum Atg5 · GFP and Atg12 · GFP molecules are present in higher molecular mass complexes, which depend on the presence of Dd-Atg7 (Otto et al. 2003). This is reminiscent of the Atg5=Atg12 conjugate that is found in yeast and mammalian cells. Indeed, sequence analysis has confirmed that the lysine residue on Atg5, which is the site of isopeptide bond formation with Atg12, is fully conserved in Dd-Atg5 (Lys186; electronic supplementary material, figure S1). As in yeast and mammalian cells, GFP · Dd-Atg8 localized to spots in wild-type D. discoideum cells in a Dd-Atg5-dependent manner.
A detailed study of Dd-Atg1 (Tekinay et al. 2006) has revealed that the kinase activity and the conserved C-terminal region of the protein are essential during autophagy and development. A kinase-dead mutant (K36A), unable to complement the Dd-atg1-1 mutant, had a dominant-negative phenotype in wild-type cells, thus phenocopying the Dd-atg1-1 mutant. Deletion of the C-terminal 40 amino acids of Dd-Atg1, a highly conserved region, abrogated the co-localization of Dd-Atg1K36A with Dd-Atg8 at a presumed PAS. Unexpectedly, the Dd-atg1-1 mutant accumulated immature phagophores as well as vesicles thought to represent membrane material required for autophagosome completion, a phenotype not observed in Dd-atg5 cells. This suggests a role for the Dd-Atg1 protein at a late stage of autophagosome formation, consistent with a possible role in Dd-Atg9 recycling. In yeast and human atg1 mutants, phagophores are usually absent, which has led to the speculation that in these organisms Atg1 might be involved in a very early step of autophagy as well. This might not to be the case in D. discoideum.
Although many proteins from yeast origin have orthologues or functional counterparts in mammals including man, in certain cases, mammalian proteins have been identified that have no counterpart in fungi, but are present in the unicellular D. discoideum. This makes this amoebe the simplest model system to study these mammalian proteins. One such protein is Vmp1, an ER-localized membrane protein associated with pancreatitis and cancer. A Dd-vmp1 mutant has many phenotypes consistent with problems in the secretory pathway, including the formation of aberrant autophagosomes and a compromised growth and development (Calvo-Garrido et al. 2008). In mammalian cells, Vmp1 has been localized to the ER, but also to autophagosomes, where it co-localizes with LC3/Atg8 (Ropolo et al. 2007). Furthermore, it interacts with Beclin-1/Atg6. Whether Dd-Vmp1 also has a direct role in autophagosome formation will require further investigation.
(ii). Autophagy in parasitic amoebes
Entamoeba histolytica and Entamoeba invadens are enteric protozoans that cause amoebiasis in humans and reptiles, respectively (Petri 2002). The virtually anaerobic eukaryote E. histolytica has a simple lifestyle that comprises two forms. The active/feeding stage is the mobile and proliferative trophozoite. This stage is responsible for the pathology of amoebiasis in the human host. The other form is the dormant but highly infective cyst that is essential for transmission of the disease. E. histolytica is a highly unusual eukaryote that appears to lack many of the organelles observed in other eukaryotic species like peroxisomes, Golgi apparatus and normal mitochondria. Instead, a single mitochondrion-related organelle designated the mitosome has been observed in this species. As a consequence, E. histolytica is considered one of the earliest branches of the eukaryote tree (Hasegawa et al. 1993; Tovar et al. 1999).
Since the cyst stage is the transmissible form of the organism, interfering with the transition of the active trophozoite into the cyst (a process designated encystation) has been the focus of investigations for many years. So far, encystation can only be induced in vitro in the reptile-specific E. invadens, making this organism the model of choice to study molecular processes in Entamoeba sp. The observation that in many eukaryotes autophagy plays a major role in differentiation and spore formation has triggered an investigation into the role of autophagy during vegetative growth and encystation in Entamoeba sp. Genome analysis has uncovered some of the core proteins involved in autophagy, three proteins of the PI 3-K complex (Vps15, Vps34 and Atg6) and the proteins of one of the Ub-related conjugation systems (Atg3, Atg4, Atg7 and Atg8; Picazarri et al. 2008a). From their analysis, the authors concluded that the proteins of the second Ub-like conjugation system, Atg5, Atg10, Atg12 and Atg16, were absent in E. histolytica and E. invadens. This has strengthened the notion that many unicellular eukaryotes may have a primitive form of autophagy (see below).
So far, the occurrence of Atg8-punctate structures and analysis of Atg8 lipidation have been the major tools in investigating the presence and formation of autophagosomes in Entamoeba sp. (Picazarri et al. 2008a,b). It was observed that Atg8-positive structures were present in vegetatively growing E. histolytica and E. invadens trophozoites, but were not induced by nutrient deprivation or under-stress conditions. Moreover, growth of trophozoites was inhibited by PI 3-K inhibitors. These data suggest that autophagy is important for proliferation of Entamoeba sp. and thus may play a housekeeping role. Autophagosome formation increased significantly in E. invadens during in vitro-induced encystation, and was preceded by the emergence of the lipidated form of Ei-Atg8. Since PI 3-K inhibitors inhibited the formation of autophagosomes as well as encystation, there appears to be a direct connection between the two processes.
4. Identification of Atg proteins in unicellular eukaryotes
Via genome analysis, a number of Atg orthologues has been identified in the above-mentioned unicellular eukaryotes. In most cases, in silico genome searches have used exclusively S. cerevisiae Atg protein sequences as queries. Such a strategy can be successful when the query protein is highly conserved or the target organism is evolutionarily close (e.g. another yeast species). However, in many of the instances described above, this is not the case. Additionally, many researchers have been very cautious in their data interpretation to avoid over-prediction. Although such caution is justified, this may also lead to incomplete datasets that may significantly affect the conclusions drawn. From their studies, several authors have concluded that a number of the core autophagy proteins, specifically the second Ub-like conjugation system consisting of Atg5, Atg10, Atg12 and Atg16, are not present in certain parasites, which has fuelled the idea that these parasites may only have a primitive form of autophagy.
A reanalysis of genome databases demonstrates that this notion may be incorrect. Many additional candidate Atg proteins can be identified when a query sequence is used from an organism that is evolutionarily distinct from baker's yeast (e.g. animal or plant; cf. Rigden et al. 2009). Figures S1–S3 in the electronic supplementary material show alignments that include candidate Atg5, Atg10 and Atg16 orthologues from L. major, T. cruzi, T. brucei, D. discoideum and E. histolytica. Remarkably, all these organisms appear to have a mammalian-style Atg16L protein with C-terminal WD40 repeats. Furthermore, the crucial lysine residue in Atg5 required for Atg12 conjugation seems not conserved in Atg5 orthologues in Trypanosomatidae, although two other conserved lysines are present in close vicinity. At first sight, sequence analysis suggests that in most parasites an Atg12 homologue is absent (electronic supplementary material, figure S4). However, it was recently shown that a protein designated ATG12 in L. major (see above), which shows significant similarity to Atg8 proteins, is the functional counterpart of S. cerevisiae Atg12 (Williams et al. 2009). Since both Ub-related proteins are thought to originate from a common ancestor, a protein related to Atg8 may therefore also represent an Atg12 counterpart. Such a candidate Atg12 protein can also be identified in T. brucei and T. cruzi (Tb-Atg8.2 and Tc-Atg8.2). Additionally, the E. histolytica genome encodes two almost identical Atg8 proteins (Eh-Atg8a and Eh-Atg8b), which are evolutionarily rather distant from known Atg8 and Atg12 proteins (electronic supplementary material, figure S4). Possibly, these E. histolytica proteins represent a novel type of Atg protein that actually fulfils both functions. However, experimental proof for this hypothesis is currently lacking. Nevertheless, it would appear that also these protozoan parasites, which according to phylogenetic analysis diverged early in evolution from the main branch of the eukaryotic tree, may contain the entire set of core autophagy proteins.
5. Conclusions and perspectives
Initially, autophagy was thought to be a random process involving degradation of portions of the cytoplasm. During the last years it has become clear that autophagy actually comprises multiple forms of related processes, with the majority being involved in highly selective cargo transport to the vacuole/lysosome. Most of these pathways have been identified in yeast models and some (like aggrephagy, the degradation of ubiquitinated protein aggregates via SQSTM1/p62) in mammalian systems. Only during the last few years has the study of autophagy in other unicellular eukaryotes been initiated, mainly because it was realized that this process is required for cellular differentiation, which is an essential part of the lifestyle of these organisms. Nevertheless, so far (non-selective) macroautophagy has been the focus of research in these species. Whether also selective modes of autophagy take place during differentiation is largely unknown. Only in T. brucei selective degradation of peroxisomes (glycosomes) has been observed, implying that some selective form of autophagy can occur in kinetoplastid parasites, but molecular details are lacking. Nevertheless, considering the fact that many selective autophagy pathways in yeast have only very recently been uncovered, it is likely that also in kinetoplastid parasites and amoebes selective pathways will be identified. Based on the low conservation of selective Atg proteins in yeast species, it can be expected that such pathways use proteins distinct from those identified in yeast species and mammals.
The recent availability of genome sequences for many unicellular eukaryotes has enabled the identification of orthologues and functional counterparts of Atg proteins in these species. So far, molecular studies have shown that a number of these proteins can indeed replace yeast Atg proteins. Thus, it can be assumed that a basic form of autophagy is conserved in all eukaryotes. Nevertheless, many candidate Atg proteins might have been missed, and others have not been studied yet. Clearly, much additional research is required to answer the question whether some unicellular eukaryotes might have a more primitive form of autophagy.
Finally, many of the described unicellular eukaryotes cause human or animal diseases, with differentiation being essential for infection. In most cases, autophagy was shown to be required for cellular differentiation, and most probably precedes this process. This implies that interfering with autophagy in these parasites/amoebes may be a promising venue to fight the diseases they cause in humans. Candidate target proteins for such a scheme may be those proteins that are involved in the more selective autophagy pathways, since these are expected to show little conservation enabling a more specific drug targeting.
Footnotes
One contribution of 12 to a Theme Issue ‘Evolution of organellar metabolism in unicellular eukaryotes’.
References
- Alvarez V. E., Kosec G., Sant'Anna C., Turk V., Cazzulo J. J., Turk B.2008Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283, 3454–3464 (doi:10.1074/jbc.M708474200) [DOI] [PubMed] [Google Scholar]
- Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S.2003The trypanosomiases. The Lancet 362, 1469–1480 (doi:10.1016/S0140-6736(03)14694-6) [DOI] [PubMed] [Google Scholar]
- Bates P. A., Rogers M. E.2004New insights into the developmental biology and transmission mechanisms of Leishmania. Curr. Mol. Med. 4, 601–609 (doi:10.2174/1566524043360285) [DOI] [PubMed] [Google Scholar]
- Baxter B. K., Abeliovich H., Zhang X., Stirling A. G., Burlingame A. L., Goldfarb D. S.2005Atg19p ubiquitination and the cytoplasm to vacuole trafficking pathway in yeast. J. Biol. Chem. 280, 39 067–39 076 (doi:10.1074/jbc.M508064200) [DOI] [PubMed] [Google Scholar]
- Bellu A. R., Komori M., van der Klei I. J., Kiel J. A. K. W., Veenhuis M.2001Peroxisome biogenesis and selective degradation converge at Pex14p. J. Biol. Chem. 276, 44 570–44 574 (doi:10.1074/jbc.M107599200) [DOI] [PubMed] [Google Scholar]
- Bellu A. R., Salomons F. A., Kiel J. A. K. W., Veenhuis M., van der Klei I. J.2002Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 277, 42 875–42 880 (doi:10.1074/jbc.M205437200) [DOI] [PubMed] [Google Scholar]
- Benchimol M.1999Hydrogenosome autophagy: an ultrastructural and cytochemical study. Biol. Cell 91, 165–174 (doi:10.1016/S0248-4900(99)80039-2) [DOI] [PubMed] [Google Scholar]
- Bernales S., McDonald K. L., Walter P.2006Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423 (doi:10.1371/journal.pbio.0040423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besteiro S., Williams R. A., Morrison L. S., Coombs G. H., Mottram J. C.2006Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281, 11 384–11 396 (doi:10.1074/jbc.M512307200) [DOI] [PubMed] [Google Scholar]
- Besteiro S., Williams R. A., Coombs G. H., Mottram J. C.2007Protein turnover and differentiation in Leishmania. Int. J. Parasitol. 37, 1063–1075 (doi:10.1016/j.ijpara.2007.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Garrido J., Carilla-Latorre S., Lázaro-Diéguez F., Egea G., Escalante R.2008Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion, and development. Mol. Biol. Cell 19, 3442–3453 (doi:10.1091/mbc.E08-01-0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E., Reggiori F.2009Regulation of autophagy in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421 (doi:10.1016/j.bbamcr.2009.01.008) [DOI] [PubMed] [Google Scholar]
- Cunha-e-Silva N., Sant'Anna C., Pereira M. G., Porto-Carreiro I., Jeovanio A. L., de Souza W.2006Reservosomes: multipurpose organelles? Parasitol. Res. 99, 325–327 (doi:10.1007/s00436-006-0190-3) [DOI] [PubMed] [Google Scholar]
- De Souza W.2002Basic cell biology of Trypanosoma cruzi. Curr. Pharm. Des. 8, 269–285 [DOI] [PubMed] [Google Scholar]
- Epple U. D., Suriapranata I., Eskelinen E. L., Thumm M.2001Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 183, 5942–5955 (doi:10.1128/JB.183.20.5942-5955.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple U. D., Eskelinen E.-L., Thumm M.2003Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278, 7810–7821 (doi:10.1074/jbc.M209309200) [DOI] [PubMed] [Google Scholar]
- Farré J. C., Vidal J., Subramani S.2007A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy 3, 230–234 [DOI] [PubMed] [Google Scholar]
- Farré J. C., Manjithaya R., Mathewson R. D., Subramani S.2008PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376 (doi:10.1016/j.devcel.2007.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke de Cazzulo B. M., Martínez J., North M. J., Coombs G. H., Cazzulo J. J.1994Effects of proteinase inhibitors on the growth and differentiation of Trypanosoma cruzi. FEMS Microbiol. Lett. 124, 81–86 (doi:10.1111/j.1574-6968.1994.tb07265.x) [DOI] [PubMed] [Google Scholar]
- Haanstra J. R., van Tuijl A., Kessler P., Reijnders W., Michels P. A., Westerhoff H. V., Parsons M., Bakker B. M.2008Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl Acad. Sci. USA 105, 17 718–17 723 (doi:10.1073/pnas.0806664105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Baba M., Ohsumi Y.2005Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65 (doi:10.1111/j.1600-0854.2004.00245.x) [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y.2007The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37 298–37 302 (doi:10.1074/jbc.C700195200) [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Hashimoto T., Adachi J., Iwabe N., Miyata T.1993Early branchings in the evolution of eukaryotes: ancient divergence of entamoeba that lacks mitochondria revealed by protein sequence data. J. Mol. Evol. 36, 380–388 [DOI] [PubMed] [Google Scholar]
- Herman M., Gillies S., Michels P. A., Rigden D. J.2006Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy 2, 107–118 [DOI] [PubMed] [Google Scholar]
- Herman M., Pérez-Morga D., Schtickzelle N., Michels P. A.2008Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 4, 294–308 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y.2005Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16, 2544–2553 (doi:10.1091/mbc.E04-08-0669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Klionsky D. J.2008Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32 386–32 393 (doi:10.1074/jbc.M802403200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J.2009Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 (doi:10.1016/j.devcel.2009.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin R. H.2001Dictyostelium: the evolution, cell biology, and development of a social organism. Cambridge, UK: Cambridge University Press [Google Scholar]
- Kiel J. A. K. W., Veenhuis M., van der Klei I. J.2006PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291–1303 (doi:10.1111/j.1600-0854.2006.00479.x) [DOI] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y.2001Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 (doi:10.1083/jcb.152.3.519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Scott S. V., Oda M. N., Klionsky D. J.1997Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137, 609–618 (doi:10.1083/jcb.137.3.609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P. E., Klionsky D. J.2002Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773 (doi:10.1074/jbc.M109134200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J.2005The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18 (doi:10.1242/jcs.01620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., et al. 2003A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 (doi:10.1016/S1534-5807(03)00296-X) [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cuervo A. M., Dunn W. A., Jr, Levine B., van der Klei I., Seglen P. O.2007How shall I eat thee? Autophagy 3, 413–416 [DOI] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M.2008Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 (doi:10.1038/ncb1723) [DOI] [PubMed] [Google Scholar]
- Kraft C., Reggiori F., Peter M.2009Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793, 1404–1412 (doi:10.1016/j.bbamcr.2009.02.006) [DOI] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E. L., Millen J., Goldfarb D. S., Thumm M.2008Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 19, 4492–4505 (doi:10.1091/mbc.E08-04-0363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J. B., Schwarz H., Mayer A.2004Determination of four sequential stages during microautophagy in vitro. J. Biol. Chem. 279, 9987–9996 (doi:10.1074/jbc.M307905200) [DOI] [PubMed] [Google Scholar]
- Kvam E., Goldfarb D. S.2004Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 117, 4959–4968 (doi:10.1242/jcs.01372) [DOI] [PubMed] [Google Scholar]
- Kvam E., Gable K., Dunn T. M., Goldfarb D. S.2005Targeting of Tsc13p to nucleus–vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol. Biol. Cell 16, 3987–3998 (doi:10.1091/mbc.E05-04-0290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. R.2005The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118, 283–290 (doi:10.1242/jcs.01649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J. A. K. W.2007ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3, 106–116 [DOI] [PubMed] [Google Scholar]
- Michels P. A., Bringaud F., Herman M., Hannaert V.2006Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763, 1463–1477 [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R. J.2006Endoplasmic reticulum and Golgi complex: contributions to, and turnover by, autophagy. Traffic 7, 1590–1595 (doi:10.1111/j.1600-0854.2006.00495.x) [DOI] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A. D., Li Z., Klionsky D. J.2008Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell 19, 1962–1975 (doi:10.1091/mbc.E07-09-0892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H., Baba M., Osumi M., Aoyagi S., Kato N., Ohsumi Y., Sakai Y.2004Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70 (doi:10.1091/mbc.E03-05-0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice D. C., Sato T. K., Stromhaug P. E., Emr S. D., Klionsky D. J.2002Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30 198–30 207 (doi:10.1074/jbc.M204736200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y.2001Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 (doi:10.1038/35056522) [DOI] [PubMed] [Google Scholar]
- Otto G. P., Wu M. Y., Kazgan N., Anderson O. R., Kessin R. H.2003Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278, 17 636–17 645 (doi:10.1074/jbc.M212467200) [DOI] [PubMed] [Google Scholar]
- Otto G. P., Wu M. Y., Kazgan N., Anderson O. R., Kessin R. H.2004Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 279, 15 621–15 629 (doi:10.1074/jbc.M311139200) [DOI] [PubMed] [Google Scholar]
- Petri W. A., Jr2002Pathogenesis of amebiasis. Curr. Opin. Microbiol. 5, 443–447 (doi:10.1016/S1369-5274(02)00335-1) [DOI] [PubMed] [Google Scholar]
- Picazarri K., Nakada-Tsukui K., Nozaki T.2008aAutophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect. Immun. 76, 278–288 (doi:10.1128/IAI.00636-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picazarri K., Nakada-Tsukui K., Sato D., Nozaki T.2008bAnalysis of autophagy in the enteric protozoan parasite Entamoeba. Methods Enzymol. 451, 359–371 (doi:10.1016/S0076-6879(08)03224-2) [DOI] [PubMed] [Google Scholar]
- Priault M., Salin B., Schaeffer J., Vallette F. M., di Rago J. P., Martinou J. C.2005Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12, 1613–1621 (doi:10.1038/sj.cdd.4401697) [DOI] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J.2004The Atg1–Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90 (doi:10.1016/S1534-5807(03)00402-7) [DOI] [PubMed] [Google Scholar]
- Rigden D. J., Michels P. A., Ginger M. L.2009Autophagy in protists: examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 5, 784–794 [DOI] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., Goldfarb D. S.2003Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 (doi:10.1091/mbc.E02-08-0483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropolo A., et al. 2007The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282, 37 124–37 133 (doi:10.1074/jbc.M706956200) [DOI] [PubMed] [Google Scholar]
- Sakai Y., Oku M., van der Klei I. J., Kiel J. A. K. W.2006Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta 1763, 1767–1775 [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Hall M. N.2000TOR, a central controller of cell growth. Cell 103, 253–262 (doi:10.1016/S0092-8674(00)00117-3) [DOI] [PubMed] [Google Scholar]
- Seibenhener M. L., Geetha T., Wooten M. W.2007Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581, 175–179 (doi:10.1016/j.febslet.2006.12.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J.2004Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29 889–29 894 (doi:10.1074/jbc.M404399200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stege J. T., Laub M. T., Loomis W. F.1999tip genes act in parallel pathways of early Dictyostelium development. Dev. Genet. 25, 64–77 (doi:10.1002/(SICI)1520-6408(1999)25:1<64::AID-DVG7>3.0.CO;2-1) [DOI] [PubMed] [Google Scholar]
- Strømhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J.2004Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566 (doi:10.1091/mbc.E04-02-0147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriapranata I., Epple U. D., Bernreuther D., Bredschneider M., Sovarasteanu K., Thumm M.2000The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 113, 4025–4033 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kamada Y., Ohsumi Y.2002Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell 3, 815–824 (doi:10.1016/S1534-5807(02)00359-3) [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y.1992Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 (doi:10.1083/jcb.119.2.301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekinay T., Wu M. Y., Otto G. P., Anderson O. R., Kessin R. H.2006Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot. Cell 5, 1797–1806 (doi:10.1128/EC.00342-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter S. A., Klionsky D. J.2000Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Semin. Cell Dev. Biol. 11, 173–179 [DOI] [PubMed] [Google Scholar]
- Tolkovsky A. M.2009Mitophagy. Biochim. Biophys. Acta 1793, 1508–1515 (doi:10.1016/j.bbamcr.2009.03.002) [DOI] [PubMed] [Google Scholar]
- Tovar J., Fischer A., Clark C. G.1999The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 32, 1013–1021 (doi:10.1046/j.1365-2958.1999.01414.x) [DOI] [PubMed] [Google Scholar]
- Waller R. F., McConville M. J.2002Developmental changes in lysosome morphology and function Leishmania parasites. Int. J. Parasitol. 32, 1435–1445 (doi:10.1016/S0020-7519(02)00140-6) [DOI] [PubMed] [Google Scholar]
- Williams R. A., Tetley L., Mottram J. C., Coombs G. H.2006Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol. Microbiol. 61, 655–674 (doi:10.1111/j.1365-2958.2006.05274.x) [DOI] [PubMed] [Google Scholar]
- Williams R. A., Woods K. L., Juliano L., Mottram J. C., Coombs G. H.2009Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 5, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A. E., Emr S. D.2002Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 158, 761–772 (doi:10.1083/jcb.200112050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D. J.2006Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17, 5094–5104 (doi:10.1091/mbc.E06-06-0479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W. L., Legakis J. E., Nair U., Klionsky D. J.2007Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell 18, 581–593 (doi:10.1091/mbc.E06-07-0612) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J.2005Autophagy: molecular machinery for self-eating. Cell Death Differ. 12, 1542–1552 (doi:10.1038/sj.cdd.4401765) [DOI] [PMC free article] [PubMed] [Google Scholar]