Abstract
Protist mitochondrial genomes show a very wide range of gene content, ranging from three genes for respiratory chain components in Apicomplexa and dinoflagellates to nearly 100 genes in Reclinomonas americana. In many organisms the rRNA genes are fragmented, although still functional. Some protist mitochondria encode a full set of tRNAs, while others rely on imported molecules. There is similarly a wide variation in mitochondrial genome organization, even among closely related groups. Mitochondrial gene expression and control are generally poorly characterized. Transcription probably relies on a ‘viral-type’ RNA polymerase, although a ‘bacterial-type’ enzyme may be involved in some cases. Transcripts are heavily edited in many lineages. The chloroplast genome generally shows less variation in gene content and organization, although greatly reduced genomes are found in dinoflagellate algae and non-photosynthetic organisms. Genes in the former are located on small plasmids in contrast to the larger molecules found elsewhere. Control of gene expression in chloroplasts involves transcriptional and post-transcriptional regulation. Redox poise and the ATP/ADP ratio are likely to be important determinants. Some protists have an additional extranuclear genome, the nucleomorph, which is a remnant nucleus. Nucleomorphs of two separate lineages have a number of features in common.
Keywords: kinetoplast, maxi-circle, mini-circle, plastid, apicoplast, nucleomorph
1. Introduction
The mitochondrion originated from an α-proteobacterial endosymbiont acquired by a host that was unable to carry out aerobic respiration, but whose nature is otherwise controversial (Embley & Martin 2006). The majority of the symbiont's genes were either transferred to the nucleus or lost completely. A number of eukaryotic lineages have secondarily adopted an anaerobic lifestyle, with consequent modification of the mitochondrion to form hydrogenosomes or mitosomes, as discussed elsewhere in this issue. This modification has been accompanied by partial or complete loss of the mitochondrial genome. The chloroplast originated from the acquisition of an oxygenic photosynthetic eubacterial endosymbiont by a non-photosynthetic host, with subsequent reduction of the symbiont genome. Although it is frequently supposed that a single primary endosymbiosis gave rise to all chloroplasts, the evidence for this has been questioned (Larkum et al. 2007), and there is increasing evidence that the photosynthetic chromatophore of the amoeba Paulinella may represent a clearly independent chloroplast origin (Nakayama & Ishida 2009). It is recognized that multiple ‘secondary’ endosymbiotic acquisitions of photosynthetic eukaryotes have occurred. In some secondary endosymbiotic lineages, the nucleus of the intermediate photosynthetic eukaryote persists as a ‘nucleomorph’ between two of the four membranes surrounding the chloroplast.
Some lineages have lost photosynthetic function, with consequent reduction of the chloroplast genome, analogous to the reduction of the mitochondrial genome in anaerobic eukaryotes. It is not clear whether any formerly photosynthetic eukaryotes have completely lost a chloroplast genome, or indeed a chloroplast compartment (Barbrook et al. 2006). There is tremendous variation among mitochondrial genomes, less among chloroplast genomes, which we illustrate in the following discussion. For the purposes of this discussion, we include macroalgal relatives of the microalgae. For simplicity, we use the term chloroplast to cover all photosynthetic organelles, whatever their pigment type, as well as non-photosynthetic organelles derived from them during evolution or development.
2. Gene content and organization of mitochondrial genomes
Eukaryotic microbes exhibit highly divergent mitochondrial genome structure and organization. Variable features include genome size (ranging from nothing in the case of mitosomes, as discussed elsewhere in this volume, to thousands of kilobase pairs in the kinetoplastids), gene complement, the presence of a single mitochondrial DNA (mtDNA) unit or multiple chromosomes, the morphology of the mtDNA (whether existing as discrete units or concatenated in multiple copies) and linear or circular organization (table 1). Mitochondrial genomes of fungi such as Saccharomyces cerevisiae have been well characterized over many years, and will not be discussed here.
Table 1.
Genome size, coding content and topology of protist mitochondrial genomes. n.d., not determined. ORF, open reading frame; Ox. Phos. oxidative phosphorylation.
| organism | size (bp) | % coding | total protein coding genes | Ox. Phos. genes | gene expression genes | other protein coding genes | RNA genes (rRNA/tRNA) | topology |
|---|---|---|---|---|---|---|---|---|
| Spizellomyces punctatus (Chytridiomycota) | Chr 1: 58 830 Chr 2: 1381 Chr 3: 1136 | Chr 1: 39 Chr 2: 16 Chr 3: 0 | Chr 1: 31 Chr 2: 1 Chr 3: 0 | Chr 1: 13 Chr 2: 1 Chr 3: 0 | none | Chr 1: 18 ORFs | Chr 1: 10 | circular |
| Acanthamoeba castellanii (Amoebozoa) | 41 591 | 77 | 40 | 16 | 16 | 8 ORFs | 18 | circular |
| Dictyostelium discoideum (Amoebozoa) | 55 564 | 73 | 42 | 17 | 15 | 9 ORFs plus endonuclease | 21 | circular |
| Chlamydomonas reinhardtii (Chlorophyta) | 15 758 | 58 | 8 | 7 | none | rtl | 17 | linear |
| Chondrus crispus (Rhodophyta) | 25 836 | 71 | 29 | 17 | 5 | 6 ORFs plus ymf16 | 28 | circular |
| Porphyra purpurea (Rhodophyta) | 36 753 | 70 | 31 | 17 | 4 | 7 ORFs plus ymf16, dpo, rtl | 26 | circular |
| Cafeteria roenbergensis (Chromista) | 43 159 | 82 | 34 | 18 | 12 | 4 ORFs | 24 | circular |
| Fucus vesiculosus (Chromista) | 36 392 | 77 | 38a | 17 | 17 | 3 ORFs plus tatC | 28 | circular |
| Rhodomonas salina (Cryptophyta) | 48 063 | 69 | 44 | 22 | 14 | 7 ORFs plus ymf16 | 29 | circular |
| Plasmodium gallinaceum (Alveolata) | 6003 | 55 | 3 | 3 | none | none | fragmented SSU and LSU rRNA | linear |
| Oxyrrhis marina (Alveolata) | n.d. | n.d. | 3 identifiedb | cox1, cob–cox3 | n.d. | n.d. | fragmented LSU rRNA | linear and circular |
| Tetrahymena pyriformis (Alveolata) | 47 296 | 80 | 44 | 15 | 9 | 19 ORFs plus yejR | 14 | linear |
| Reclinomonas americana (Jakobida) | 69 034 | 80 | 67 | 25 | 32 | 10 | 30 | circular |
| Malawimonas pyriformis (Malawimonada) | 47 328 | 67 | 49 | 17 | 22 | 6 ORFs plus yejR, yejU, yejV | 40 | circular |
| Naegleria gruberi (Heterolobosea) | 49 843 | 81 | 46 | 21 | 17 | 4 ORFs, yejR, yejU, ymf16, cox11 | 23 | circular |
aAlso encodes one pseudogene.
bGenes exist in various genetic contexts (fragmented/complete, repeated/alone etc.).
(a). Protein-coding genes
Proteins encoded in mitochondrial genomes are typically involved in translation and oxidative phosphorylation (http://www.ncbi.nlm.nih.gov/Genomes/). A core set of mitochondrial genes, encoding subunits of the ribosome, NADH dehydrogenase, succinate dehydrogenase, cytochrome oxidase and ATP synthase, as well as cytochrome b is present to differing extents in different species (Gray et al. 2004). In some cases, proteins involved in protein translocation and maturation, RNA processing and transcription are also mitochondrially encoded. Gene content and organization have been used to help establish evolutionary relationships among organisms (e.g. Sankoff et al. 1992). The mitochondrial genome of the jakobid protozoan Reclinomonas americana carries 97 genes encoding proteins and stable RNAs. This is the most gene-rich and least derived mtDNA studied to date (Lang et al. 1997). Eighteen mitochondrial genes in R. americana have not been reported from any other mtDNA. Some are involved in mitochondrial electron transport and respiration but there are genes for proteins involved in other functions, such as tufA (a translation factor) and secY (involved in protein translocation). The observation that the R. americana mtDNA carries rpoA–D encoding the α, β, β′ and σ subunits of prokaryotic RNA polymerases is particularly notable, since other mitochondria are believed to use a viral T3/T7-type single subunit RNA polymerase. Consequently, it has been suggested that the R. americana mitochondrial genome is more reminiscent than others of ancestral mitochondria (Lang et al. 1997).
In terms of gene content, the Apicomplexa (including the malaria parasite Plasmodium and the cattle pathogen Theileria) lie at the opposite end of the scale to R. americana. At around 6 kbp, their mtDNAs represent the smallest discovered, carry the fewest genes and lack introns, representing perhaps a minimal gene complement for mitochondrial genomes. They carry three protein coding genes, for subunits I and III of cytochrome c oxidase (cox1 and cox3, respectively) and cytochrome b (cob) (Wilson & Williamson 1997).
The dinoflagellate algae have a particularly unusual mitochondrial genome. Like their sister group, the Apicomplexa, the mitochondrial genome contains cox1, cox3 and cob, but also contains a large number of (presumably) non-functional fragments of them in a wide range of configurations separated by extensive regions of repetitive non-coding DNA (Nash et al. 2008; Kamikawa et al. 2009). In the early-branching dinoflagellate Oxyrrhis marina, cob and cox3 are encoded and expressed as a fusion protein (Slamovits et al. 2007).
(b). RNA genes
Large and small subunit rRNAs (LSU and SSU respectively) are encoded in essentially all mtDNAs studied to date, although in many species the rRNA genes are fragmented. In a few cases (such as SSU rRNA in dinoflagellate mitochondria), it has not been possible to identify rRNA genes conclusively. It may be that they are present, but highly divergent and fragmented (Nash et al. 2008). In some organisms, a gene rrn5 for 5S rRNA is also found. The sporadic distribution of rrn5 suggests it was lost multiple times from different lineages (Gray et al. 2004).
Fragmentation of rRNA genes has occurred in many groups, most markedly in Apicomplexa, dinoflagellates and many green algal (e.g. Chlamydomonas) lineages. In these instances, rRNA segments are encoded separately and the transcripts associate to form functional ribosomes. In some cases, such as Plasmodium falciparum and Chlamydomonas eugametos, the order of rDNA fragments in the genome is different from the order of their homologues in the Escherichia coli 16S and 23S rRNA. In these organisms, LSU and SSU coding fragments are interspersed with each other as well as other coding sequences (Feagin et al. 1992; Denovan-Wright & Lee 1994; Denovan-Wright et al. 1998). The order of rDNA fragments varies between Chlamydomonas species. In Chlamydomonas reinhardtii, SSU fragments are encoded in the same order as in E. coli 16S rRNA, whereas the LSU fragments are scrambled and in a different order from C. eugametos (Denovan-Wright & Lee 1994). Fragmentation of rRNA genes has occurred to a lesser degree in the ciliates, where LSU and SSU rDNA each split into two separate coding regions. However, in Tetrahymena pyriformis, the LSU rRNA gene is represented twice with sequence differences between the two forms (Burger et al. 2000).
The number of tRNAs encoded in protist mitochondrial genomes varies widely. The mitochondrial genome of the ichthyosporean (an independent group located between animals and fungi in the opisthokonts) Amoebidium parasiticum is organized into many 0.3–8.3 kbp linear chromosomes (Burger et al. 2003) and encodes at least 25 tRNAs in multiple copies. Conversely, tRNA genes are absent from kinetoplastid mitochondrial genomes, although they can be thousands of kilobases in size. Studies in trypanosomes and the apicomplexan Toxoplasma gondii demonstrate that tRNAs are imported from the cytosol (Crausaz Esseiva et al. 2004). At least two pathways exist for tRNA uptake, one of which has elements in common with protein import (Alfonzo & Söll 2009). Obtaining a formylmethionyl-tRNA (fMet-tRNA) for translation initiation is potentially problematic. Trypanosome mitochondria are believed to take up and formylate cytosolic elongator Met-tRNA for this purpose (Tan et al. 2002), whereas it has been suggested that in Apicomplexa the initiator fMet-tRNA may be supplied by the apicoplast (the relic secondary chloroplast maintained within the parasite) (Howe & Purton 2007). In Chlamydomonas, the mitochondrial genome also does not encode an initiator fMet-tRNA. A recent study failed to detect chloroplast initiator fMet-tRNAs in the Chlamydomonas mitochondrion and suggested that the initiator fMet-tRNA is derived from cytosolic elongator Met-tRNA, as with trypanosomes (Vinogradova et al. 2009).
(c). Genome organization
The organization, morphology and topology of the mtDNA is highly diverse among protists. Although the majority of mtDNAs are topologically circular, linear monomers and linear tandem arrays are also found. Genome organization can differ widely among related protist species. For example (with the exception of Physarum polycephalum), mtDNA gene content is largely conserved between members of the Amoebozoa (which include the ‘solitary’ amoeba Acanthamoeba castellanii as well as the ‘social’ slime molds Polysphondylium pallidum and Dictyostelium sp.). They share very similar mtDNA sizes, AT richness and gene content, and the genomes are compact (for example, 93.2% of the A. castellanii mtDNA comprises coding sequences) (Burger et al. 1995). They also share a chimeric cox1 and cox2 gene and all genes are transcribed from the same strand (Burger et al. 1995; Heidel & Glöckner 2008). However, there are large variations in gene order. While only a single segmental rearrangement occurs between the sister groups that make up the social amoebae (Dictyostelium discoideum and Dictyostelium citrinum, and P. pallidum and Dictyostelium fasciculatum), conservation of gene order in A. castellanii is limited to pairs of genes (Heidel & Glöckner 2008).
There is a wide variation in mitochondrial genome size and structure among closely related green algal species. For example, C. eugametos and C. reinhardtii have similar gene contents, but the order of genes is entirely different. The C. eugametos mtDNA contains introns and displays a more disperse distribution of rRNA fragments (Denovan-Wright et al. 1998). Furthermore, whereas the mtDNA of C. eugametos is circular mapping with all genes located on the same strand, in C. reinhardtii it exists as linear monomers with genes on both strands. The related green alga Polytomella parva also exhibits similar gene content and rDNA organization to Chlamydomonas, but its genome exists as two linear DNA molecules—one of 13.5 kbp, which contains the majority of coding DNA, and one of 3.5 kbp, which contains only nad6 (Fan & Lee 2002). By contrast, mitochondrial genome size and structure are largely conserved among the red algae (Gray et al. 2004). Within individual phyla of the Chromista, mitochondrial genomes are generally reasonably well conserved. For example, the mitochondrial genomes of Phaeophyceae (brown algae) are similar in gene order and content. Differences are primarily due to variation in the presence and length of intergenic regions and introns (Oudot-Le Secq et al. 2006). However, significant diversity exists between phyla, for example in the presence of repeated sequences and introns (Puerta et al. 2004; Kim et al. 2008).
Mitochondrial DNA structure also varies widely within other protist groups. In the Apicomplexa, for example, Plasmodium mtDNAs are arranged in linear tandem repeats (Wilson & Williamson 1997), whereas the mtDNA in Theileria exists as linear monomers (Kairo et al. 1994). Mitochondrial genome size is largely independent of gene content and organization, and a particularly striking example of this is found in one of the most unusual mitochondrial genomes, in kinetoplastids. All members of the Euglenozoa (i.e. kinetoplastids, diplonemids and euglenids) contain large amounts of mtDNA. In many kinetoplastid flagellates, mitochondrial DNA is present as a compact disc in a region of the mitochondrion known as the kinetoplast (and is often referred to as kinetoplast DNA, or kDNA). Electron microscopic imaging has shown that it exists as a network of large and small circular DNAs known as maxi-circles and mini-circles (Lukeš et al. 2002). Maxi-circles encode proteins for mitochondrial function and exist at around 12 copies per mitochondrion. They are usually 20–40 kbp in size, although coding DNA is always localized within a 17 kbp region known as the ‘conserved region’. The remaining ‘variable’ region contains repeated sequences, some of which are involved in maxi-circle replication (Shapiro & Englund 1995).
Mini-circles are present in thousands of copies per mitochondrion. They encode guide RNAs (gRNAs) that direct the insertion/deletion of Us in RNAs to yield complete coding mRNAs. Some gRNAs are also encoded on maxi-circles. Mini-circles are smaller (0.5–10 kbp) than maxi-circles and contain conserved regions involved in the initiation of their replication and compaction into the kDNA network (Shapiro & Englund 1995). The kDNA is organized differently in different species. In trypanosomatids (which include Trypanosoma and Leishmania), it exists as a catenated network of maxi- and mini-circles found in a distinct periflagellar region of the mitochondrion, while the kDNA of Bodo caudatus and Cryptobia helicis are uncatenated and dispersed through the mitochondrion (Hajduk et al. 1986; Lukeš et al. 1998). Dimastigella mimosa, Dimastigella trypaniformis and Cruzella marina display multiple bundles of typically monomeric mini-circles throughout their mitochondria, while in Trypanoplasma borreli the mini-circles are linked in tandem to form large circular molecules termed mega-circles (for a detailed review, see Lukeš et al. 2002).
The diplonemids have very different mtDNA structure, although they are sister group to the kinetoplastids. Diplonema papillatum appears to have two types of circular mitochondrial chromosomes, one of 6 kbp and one of 7 kbp, both apparently existing in relaxed circular conformation. Fluorescence microscopy studies have shown that the mtDNA is not localized to a distinct region of the mitochondrion as in trypanosomatids, but is distributed throughout the mitochondrion. Furthermore, analysis of cox1 coding sequence showed that the gene appears to exist in approximately 250 bp fragments distributed between the two types of chromosomes (Marande et al. 2005).
3. Mitochondrial gene expression and control
Relatively little is known about mitochondrial gene expression in eukaryotic micro-organisms (other than in some well-studied species such as S. cerevisiae) and even less about its control. In some lineages, such as the Apicomplexa, studies have elucidated the size and processing of mitochondrial transcripts. There has also been considerable focus on editing in a number of lineages, notably the kinetoplastids. However, advances in our knowledge of mitochondrial gene expression in protists have been rather few in recent years.
Expression of mitochondrial genes appears to be generally mediated by a single subunit T3/T7 phage-like RNA polymerase, which was recruited relatively early in the evolution of the organelle (Shutt & Gray 2006a). Sequences encoding such an enzyme have been specifically amplified from many protists (Cermakian et al. 1996; Li et al. 2001) and can typically be found in sequenced protist nuclear genomes. The corresponding activity has been experimentally verified in some lineages (Grams et al. 2002; Miller et al. 2006). However, as noted above, the mitochondrial genomes of R. americana and other jakobids encode a bacterial-type RNA polymerase (Gray et al. 2004). This is presumed to be involved in mitochondrial gene expression, although the presence of a viral-type enzyme in addition cannot be excluded. In the stramenopile Pylaiella littoralis a T7-like polymerase is encoded on the mitochondrial genome, but sequences characteristic of promoters of a bacterial-type polymerase are also present (Oudot-Le Secq et al. 2001). It is therefore possible that in certain protists multiple functional polymerases exist within mitochondria. This would be analogous to the presence of multiple polymerases in the chloroplasts of higher plants (Smith & Purton 2002).
In animals and fungi, mitochondrial factors accessory to the core polymerase have been shown to be important for expression of mitochondrial genes (Asin-Cayuela & Gustafsson 2007). Outside the opisthokonts, putative homologues for such transcription factors have been found in amoebozoans and trypanosomatids (Shutt & Gray 2006b). However, it remains to be established whether the phage-type polymerases acting in the mitochondria of other protists require transcription factors for effective mitochondrial gene expression, perhaps unrelated to those found in animals and yeasts.
The identity of promoter sequences has been suggested for a handful of protists (Wolff & Kück 1996; Richard et al. 1998; Oudot-Le Secq et al. 2001). The viral-type promoters are typically proposed to be nonanucleotide sequences close to the transcription initiation site, with little or no sequence similarity to promoter sequences in yeasts, humans and higher plants. Definitive identification of promoters in protists requires the development of mitochondrial transformation systems. Encouragingly, such a system has been reported for C. reinhardtii (Remacle et al. 2006).
Transcript maps have been generated for various protist mitochondria, including members of the Amoebozoa, Chlorophyta (green algae), Rhodophyta (red algae), Chromista, Apicomplexa and Ciliophora (Gray & Boer 1988; Jones et al. 1990; Wolff & Kück 1996; Richard et al. 1998; Edqvist et al. 2000; Rehkopf et al. 2000). Taken with gene organization data, these maps are consistent with a model of transcription in which a few transcription initiation sites in each genome give rise to large precursor RNAs that are processed to much smaller mono- or polycistronic mRNAs. Such endonucleolytic processing of larger precursors would appear to be highly specific, since mRNAs are frequently generated with very little or no 5′ untranslated region. In some cases, and similarly to humans, the excision of tRNA sequences appears to generate the mature mRNAs (Wolff & Kück 1996). In Chondrus crispus, there is evidence for specific processing within tRNA sequences to generate the mature flanking cistron ends, as well as alternative processing to generate functional tRNAs (Richard et al. 1999).
The model of transcription described above is by no means universal. The kinetoplastids have a similar system of transcription for maxi-circle non-gRNA genes. However, while the gRNAs are transcribed by the same polymerase (Hashimi et al. 2009), there are numerous distinct transcriptional units present on the mini-circles and maxi-circles of the kDNA for their expression (Lukeš et al. 2005). Diplonemid circular chromosomes carrying different gene fragments also require at least one promoter element each, implying the existence of more than 100 transcription units across the set (Marande & Burger 2007). Given the large spacers between genes, and the indication from expressed sequence tags (ESTs) of predominantly monocistronic transcripts, dinoflagellate mitochondria must also employ a large number of separate promoters (Nash et al. 2008). Similar systems are also likely to be used in the less compact and more disperse mitochondrial genomes such as the alga Pseudendoclonium akinetum (Pombert et al. 2004), as with land plants, whose mitochondrial genomes also contain a large number of separate transcriptional units.
A variety of different forms of editing occur in different protist lineages (table 2). Editing has been most closely studied in kinetoplastids (Lukeš et al. 2005). In this system, mRNA editing occurs via the insertion or deletion of U nucleotides. Determination of the positions of editing is carried out via gRNAs that are encoded principally on the kDNA mini-circles. Editing of mitochondrial transcripts also appears to be prevalent within the myxomycetes of the Amoebozoa. The most complex form of mRNA, rRNA and tRNA editing has been elucidated in Physarum polycephalum. In P. polycephalum, a variety of single and dinucleotide insertions take place as well as C to U substitutions (Horton & Landweber 2002). Editing takes place simultaneously with transcription in P. polycephalum and may involve the RNA polymerase itself, whereas kinetoplast editing is post-transcriptional (Byrne & Gott 2004; Miller & Miller 2008). Editing of tRNAs has also been documented for organisms from the Amoebozoa, Chytridiomycota and Jakobida (Gray et al. 2004). Unusually, in trypanosomatids, the anticodon of cytosolic tryptophanyl-tRNA is edited following import into the mitochondrion, to allow recognition of mitochondrial UGA tryptophan codons (Alfonzo et al. 1999). An apparently unrelated mRNA and rRNA substitutional editing system occurs in dinoflagellates (Nash et al. 2008). However, the mechanism is unknown.
Table 2.
Distribution of mitochondrial RNA editing in protist microbes. Arrows indicate the direction of substitutional editing events. Editing has also been characterized in further members of the Dinophyceae and Euglenozoa. However, the editing is of the same RNAs and is essentially the same type in these cases. In many organisms, the full extent of editing is not determined and may extend to further classes of RNA.
| organism | phylum | kingdom | class of RNA edited | types of editing |
|---|---|---|---|---|
| Physarum polycephalum | Mycetozoaa | Amoebozoa | mRNA, rRNA, tRNA | C/U/dinucleotide insertion, C→U |
| Didymium nigripes | Mycetozoaa | Amoebozoa | mRNA, tRNA | C/U/dinucleotide insertion, C→U |
| Stemonitis flavogenita | Mycetozoaa | Amoebozoa | mRNA | C/U/dinucleotide insertion |
| Arcyria cinerea | Mycetozoaa | Amoebozoa | mRNA | U insertion, C→U |
| Clastoderma debaryanum | Mycetozoaa | Amoebozoa | mRNA | U insertion |
| Dictyostelium discoideum | Mycetozoab | Amoebozoa | rRNA | C→U |
| Acanthamoeba castellani | Centramoebida | Amoebozoa | tRNA | U→A, U→G, A→G |
| Spizellomyces punctatus | Chytridiomycota | Opisthokonta | tRNA | A→G, U→G, U→A, C→A |
| Alexandrium catenella | Dinophyceae | Chromalveolata | mRNA, rRNA | A→G, U→C, C→U, G→C, G→A |
| Amphidinium carterae | Dinophyceae | Chromalveolata | mRNA | A→G, U→C, C→U, G→A, A→U, G→U |
| Crypthecodinium cohnii | Dinophyceae | Chromalveolata | mRNA | A→G, U→C, C→U, G→C |
| Karlodinium micrum | Dinophyceae | Chromalveolata | mRNA, rRNA | A→G, U→C, C→U, G→C, G→A, C→G |
| Prorocentrum minimum | Dinophyceae | Chromalveolata | mRNA | A→G, U→C, C→U, G→C, G→A, U→G |
| Pfiesteria piscicida | Dinophyceae | Chromalveolata | mRNA | A→G, U→C, C→U, G→C, G→A, U→A |
| Symbiodinium sp. | Dinophyceae | Chromalveolata | mRNA | A→G, U→C, C→U, G→C, A→C |
| Trypanosoma brucei | Euglenozoac | Excavata | mRNA, tRNA | U insertion/deletion, C→U |
| Bodo caudatus | Euglenozoac | Excavata | mRNA | U insertion/deletion |
| Seculamonas ecuadoriensis | Jakobida | Excavata | tRNA | U→A, A→C, G→U, U→C, A→G |
aPlasmodial slime mould.
bCellular slime mould.
cKinetoplastid.
Other types of post-transcriptional modification also occur. Mitochondrial transcripts have been found to be polyadenylated in Apicomplexa, dinoflagellates, kinetoplastids (maxi-circle mRNAs) and diplonemids (Gray et al. 2004; Lukeš et al. 2005; Marande & Burger 2007; Nash et al. 2008). In Apicomplexa and dinoflagellates, the poly(A) tails are relatively short (less than 22 nucleotides) and of uncertain function. In kinetoplastids, mitochondrial transcripts have either short (approx. 20–25 nucleotides) or long (approx. 120–250 nucleotides) poly(A) tails (Etheridge et al. 2008). The short tails are added first and are required for maintenance of transcripts undergoing editing. The longer tails are added after the completion of editing and contain a significant proportion of uridyl nucleotides. Polyuridylation of transcripts has also been observed for certain transcripts within the myxomycetes, as well as for gRNAs and rRNAs in trypansomes (Adler et al. 1991; Horton & Landweber 2000). Furthermore, the presence of a short 5′ poly(U) cap has been demonstrated for transcripts of O. marina (Slamovits et al. 2007). Trans-splicing to generate mature mRNAs has been discovered in diplonemids and dinoflagellates (Marande & Burger 2007; Nash et al. 2008).
Control of gene expression in protists is probably post-transcriptional in most cases, given the existence of relatively few promoters and the synthesis of large precursor polycistronic transcripts. Results in Trypanosoma brucei support this model (Michelotti et al. 1992; Lukeš et al. 2005).
4. Gene content and organization of chloroplast genomes
Although there is diversity in the content and organization of chloroplast genomes, there is less variation than among mitochondrial genomes. A ‘core’ of genes is present in the chloroplast genome of essentially all photosynthetic organisms, encoding subunits of the two photosystems, the cytochrome b6f complex linking them and the ATP synthase, together with the large subunit of ribulose bis-phosphate carboxylase, components of both ribosomal subunits, a bacterial-type RNA polymerase, rRNAs and tRNAs (table 3).
Table 3.
Core genes (except tRNAs) present in essentially all chloroplast genomes from photosynthetic organisms. Those underlined are present on the mini-circular chloroplast genome of dinoflagellates (adapted from Howe et al. 2008).
| photosystem I | photosystem II | cytochrome b6f complex | ATP synthase | ribosomal protein large subunit | ribosomal protein small subunit | RNA polymerase | hypothetical protein | Rubisco | rRNA |
|---|---|---|---|---|---|---|---|---|---|
| psaA | psbA | petA | atpA | rpl2 | rps2 | rpoA | ycf4 | rbcL | 23S |
| psaB | psbB | petB | atpB | rpl14 | rps3 | rpoB | 16S | ||
| psaC | psbC | petD | atpE | rpl16 | rps4 | rpoC1 | |||
| psaJ | psbD | petG | atpF | rpl20 | rps7 | rpoC2 | |||
| psbE | atpH | rpl36 | rps8 | ||||||
| psbF | rps11 | ||||||||
| psbH | rps12 | ||||||||
| psbI | rps14 | ||||||||
| psbJ | rps18 | ||||||||
| psbK | rps19 | ||||||||
| psbL | |||||||||
| psbN | |||||||||
| psbT | |||||||||
| psbZ |
The genome of green algal chloroplasts typically ranges between 100 and 200 kbp, but with numerous exceptions. Some Acetabularia species are reported to have chloroplast genomes up to 1.5 Mbp in size (Simpson & Stern 2002) while the smallest conventional chloroplast genome of a photosynthetic organism belongs to Ostreococcus tauri, with only 86 genes (excluding duplications) closely packed into 72 kbp (Robbens et al. 2007). Diversity is also observed within genera. For example, Chlamydomonas moewusii (chloroplast genome size 292 kbp) and Chlamydomonas pitchmanii (187 kbp) owe their difference to two intergenic insertions. Chlamydomonas reinhardtii (203 kbp) and Chlamydomonas gelatinosa (285 kbp) differ in intergenic spacers and gene order. The difference in the latter is attributable to several inversions and an expansion/contraction of the inverted repeat (Boudreau & Turmel 1996). Sequenced red algal chloroplast genomes range between 150 and 192 kbp and generally contain more genes, up to 251, than the 103–135 chloroplast genes of green algae (http://www.ncbi.nlm.nih.gov/Genomes/; Hagopian et al. 2004). The glaucophyte Cyanophora paradoxa has a chloroplast of the same pigment type as red algae, but surrounded by a remnant peptidoglycan wall. However, it has a smaller chloroplast genome than red algae, of 136 kbp (Hagopian et al. 2004). Sequenced stramenopile and other chromist chloroplast genomes are typically smaller than red algal ones, at 105–160 kbp, and with a similar gene content to green chloroplasts (http://www.ncbi.nlm.nih.gov/Genomes/).
Chloroplast genomes are typically represented as circular, with an inverted repeat region (containing rRNA and some other genes) dividing the chloroplast genome into large and small single copy regions. Whether the genome is really topologically circular in vivo has been reviewed elsewhere (Bendich 2004). There are some exceptions to this organization. For example, although the red alga Porphyra yezoensis was reported to have rRNA genes organized in inverted repeats (Shivji 1991), sequence analysis indicates that they are in direct configuration (NCBI accession NC_007932). This could represent a strain difference, but the rRNA genes are also in direct configuration in Porphyra purpurea, although they are not repeated in the red algae Gracilaria tenuistipitata and Cyanidium caldarium (Hagopian et al. 2004). In some Euglena gracilis strains, there are three consecutive rRNA repeats and one additional partial one (Hallick et al. 1993).
Some algal chloroplasts have lost photosynthetic capability, with a reduction and compaction of their genome, as demonstrated in the parasitic, non-photosynthetic green alga Helicosporidium. This has a 37.5 kbp chloroplast genome lacking all genes encoding the photosynthetic machinery, with only 5.1 per cent non-coding DNA, small intergenic spaces and encoding only the minimal complement of tRNAs required (de Koning & Keeling 2006). The non-photosynthetic Apicomplexa generally possess a circular genome of approximately 35 kbp that has inverted repeats and is clearly of chloroplast origin (Gardner et al. 1991; Howe 1992) but no longer encodes any of the proteins directly involved in photosynthesis (Wilson & Williamson 1997). A photosynthetic apicomplexan has also been described, but its chloroplast genome has not yet been fully characterized (Moore et al. 2008). The sister group to the Apicomplexa, the dinoflagellates, have a unique chloroplast genome organization where the single large chloroplast genome is replaced by multiple, small plasmid-like DNA molecules termed ‘minicircles’ (Zhang et al. 1999; Barbrook & Howe 2000). They range between 2 and 4 kbp in size and generally carry one gene per minicircle, although minicircles carrying more than one gene have been reported. The minicircles contain a conserved core region, probably responsible for replication and transcription (Howe et al. 2008; Nisbet et al. 2008). They have a much reduced gene content, with most of the conventional core set of chloroplast genes relocated to the nucleus (Howe et al. 2008).
5. Chloroplast gene expression and control
Most of our understanding of gene expression and its control in chloroplasts comes from work on land plants and the unicellular green alga Chlamydomonas. The latter is particularly valuable for such studies as it is (i) readily amenable to genetic analysis and (ii) able to grow heterotrophically on medium containing fixed carbon, so non-photosynthetic mutants remain viable.
Land plant chloroplasts contain a nuclear-encoded, viral-type RNA polymerase (NEP, or nuclear-encoded polymerase), which is used to transcribe ‘genetic system’ genes in the chloroplasts, such as those for the ‘plastid-encoded RNA polymerase’ (PEP), which in turn transcribes genes for proteins involved in photosynthesis. However, algae appear only to have a chloroplast-encoded bacterial-type RNA polymerase (with the possible exception of dinoflagellates), so the possibility of control by modulation of the NEP does not exist (Smith & Purton 2002). The PEP is thought to resemble that of E. coli, where the RNA polymerase is encoded by the rpoA, B and C genes. In algae, the rpoC gene is normally divided into two separate genes rpoC1 and rpoC2. An exception to this is in Chlamydomonas where significant gene rearrangement has occurred and where the location and identity of the rpoA and rpoC1 genes are still unclear (Maul et al. 2002). The PEP presumably requires sigma factors, and there is variation in the number of these. Land plants have several nuclear genes for them, as does the red alga Cyanidioschyzon merolae (Minoda et al. 2005). Many other algae, such as Chlamydomonas reinhardtii, appear to have only a single sigma factor (Carter et al. 2004), and in Guillardia theta, a sigma factor is encoded in the nucleomorph (Smith & Purton 2002).
Early studies on the control of gene expression concentrated on changes in levels of transcripts on illumination, and identified a number of genes whose transcript levels increased. Because chloroplasts were recognized as being of prokaryotic origin, it was assumed that this could be interpreted as an increase in transcription initiation. Subsequent studies determining half-lives of chloroplast transcripts in plants showed that transcript stability and translation efficiency are important determinants of the overall rate of synthesis of many proteins (e.g. Mullet & Klein 1987), although there are nevertheless examples where transcriptional control is important, such as redox regulation of genes for photosystem components (see below). In Chlamydomonas reinhardtii, run-on experiments showed that transcriptional and post-transcriptional processes are both important in determining transcript levels (Lilly et al. 2002). For the red alga Cyanidioschyzon merolae transcript levels of a number of chloroplast genes also increase on illumination, and run-on experiments indicate that transcription is a major determinant of these changes (Minoda et al. 2005).
Pfannschmidt et al. (1999) showed that transcription of genes for photosystem II components increased when plants were transferred to light favouring photosystem I and vice versa. This was shown to depend on the redox state of the plastoquinone pool. Puthiyaveetil et al. (2008) identified a nuclear-encoded prokaryotic-type sensor kinase in chloroplasts that is required for this regulation. Putative chloroplastic sensor kinases also exist in green and other algal lineages, so a similar regulation may operate there (Duplessis et al. 2007; Puthiyaveetil et al. 2008; Puthiyaveetil & Allen 2009).
Sigma factors may also be a means of modulating transcription. Thus the nuclear SIG5 gene of Arabidopsis thaliana encodes a sigma factor that is expressed in blue light and activates the transcription of the chloroplast psbD gene (Tsunoyama et al. 2004). Regulation of one of the sigma factors of C. merolae has been implicated in light-dependent changes in chloroplast transcription (Minoda et al. 2005).
A number of chloroplast transcripts require splicing. For example, the Chlamydomonas chloroplast genome sequence reveals group I and II introns located in the psbA, psaA and 23S rRNA genes. Splicing of the group I introns is through a guanosine-dependent mechanism (Herrin et al. 1990) likely to be similar to that reported for the Tetrahymena rRNA intron. At physiological temperatures, splicing in vitro is much slower than splicing in vivo. Moreover, the splicing of these introns was shown to be highly upregulated by light for photoautotrophically growing Chlamydomonas (Herrin & Nickelsen 2004, and references therein). Editing of chloroplast transcripts is largely restricted to land plants, with the possible exception of some dinoflagellates (Zauner et al. 2004). Chloroplast transcripts also undergo complex cleavage and degradation reactions (for which they may be marked by transient polyadenylation), mediated by nuclear-encoded proteins (Zimmer et al. 2008). Translation is by bacterial-type ribosomes, consistent with the origin of chloroplasts.
Nuclear-encoded factors bind to and determine the stability and translation efficiency of many mRNAs in Chlamydomonas chloroplasts. A well-characterized example is the MCA1 protein, which stabilizes the mRNA for petA (cytochrome f) of the electron transfer chain. In transgenic lines expressing different levels of MCA1, the level of petA mRNA varies accordingly and the level of PetA correlates closely. In wild-type cells, levels of MCA1 vary with parameters such as culture age and environmental conditions, and levels of the petA mRNA vary in the same way. Translation of the petA mRNA depends on a factor, TCA1, levels of which are also modulated according to environmental conditions (Raynaud et al. 2007).
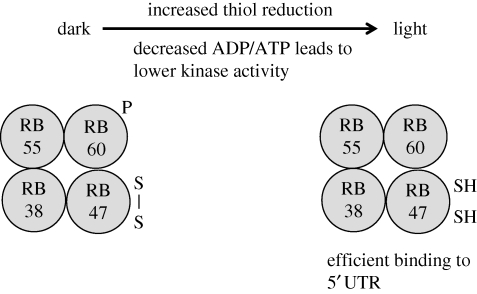
Redox poise and the ATP/ADP balance are also likely to be important determinants of translation, a well-studied example being psbA mRNA in Chlamydomonas (Marin-Navarro et al. 2007) (figure 1). A complex of four proteins, RB60, RB55, RB47 and RB38, binds the 5′ UTR. RB60 regulates the redox state of RB47 (an RNA-binding protein). High light levels lead to reduction of RB60 and increased PsbA synthesis (satisfying the need for increased replacement of PsbA after photodamage), probably through reduction of RB47, which stimulates its binding to the psbA mRNA and translation. In addition, an ADP-dependent kinase inhibits binding of the complex by phosphorylation of RB60. In the light the ATP/ADP ratio is high, so there will be less phosphorylation of RB60 and increased PsbA synthesis.
Figure 1.
Light-dependent regulation of psbA mRNA translation in Chlamydomonas. Increased illumination leads to disulphide reduction on RB47 and decreased phosphorylation of RB60, resulting in increased mRNA-binding activity of the complex and stimulation of translation.
A final determinant of the level of chloroplast-encoded proteins is the fate of protein that has not been assembled into a functional complex (reviewed by Marin-Navarro et al. 2007). For PetA (cytochrome f) in Chlamydomonas, protein that has not been assembled into the cytochrome b6f complex represses its own synthesis. Deletion of a C-terminal region of PetA leads to an increase in synthesis, consistent with this part of the protein being involved in an autoregulatory circuit. In many cases, the inability to assemble a functional complex because of loss of one or more components leads to degradation of the unassembled components. Thus, the final level of control may simply be turnover of protein that is surplus to requirements.
Redox poise therefore seems to be an important determinant of gene expression in plants and green algae. (The requirement for gene expression to respond to redox poise provides an elegant justification for the retention of a chloroplast genome in photosynthetic organisms (Allen 2003), although why non-photosynthetic ones should also retain a genome is less clear (Barbrook et al. 2006).) It seems likely that the regulation of chloroplast gene expression in response to redox poise and the ATP/ADP ratio will be a general feature. It is striking that chloroplast gene expression in Chlamydomonas requires many different nuclear-encoded proteins that bind specific transcripts to determine their stability or their translation. It will be interesting to see to what extent this applies to other unicellular photosynthetic organisms.
6. Nucleomorph
An interesting feature of some complex chloroplasts is the retention of a relic eukaryotic nucleus—the nucleomorph—located between the inner two and outer chloroplast membranes. They have been found only within the chloroplasts of chlorarachniophytes and cryptophytes, which were derived through the secondary endosymbiosis of green and red algae, respectively. Despite their different origins, nucleomorphs of green and red algal ancestry show remarkable similarities. They have both retained a nuclear envelope, which contains pores and encloses the miniature genome. The first complete nucleomorph genome sequence was from the cryptophyte Guillardia theta (Douglas et al. 2001), comprising 551 kbp. Comparison with the smaller (373 kbp) nucleomorph genome of the chlorarachniophyte Bigelowiella natans (Gilson et al. 2006) shows that, despite originating from different algal lineages, the organization of these retained endosymbiont-derived nuclear genomes is similar. All nucleomorph genomes studied so far are very compact, with high gene density, overlapping genes and few repeated genes. On average, the cryptophyte nucleomorph genome is larger than those of the chlorarachniophytes, ranging between 440 and 845 kbp, compared with 330 and 610 kbp, respectively (Lane et al. 2006).
All nucleomorph genomes characterized to date consist of just three chromosomes, which display characteristics of eukaryotic chromosomes such as telomeric repeats. Subtelomeric regions contain rDNA operons encoding 18S/5.8S/28S rRNAs. In G. theta and B. natans, these occur at all six chromosome ends. However, they occur at only three in the cryptophyte Hemiselmis andersenii, at both ends of chromosome 1 and at one end of chromosome 3 (Lane et al. 2007). The 5S rDNA is absent from B. natans, but is present within G. theta's telomeric repeats, and is found separately near the telomere ends in H. andersenii. Centromeres have proved hard to identify, although candidate sequences exist within non-coding regions near the midpoint of chromosomes, and centromere-binding proteins are encoded on the nucleomorph genomes (Gilson et al. 2006).
Consistent with their reduction in size, the gene content of nucleomorph genomes is greatly reduced compared with nuclear genomes. Many nucleomorph-targeted proteins are encoded in the cell's nucleus. Although sequenced nucleomorph genomes have quite dissimilar gene contents, the proteins encoded are primarily involved in the maintenance of the organelle and housekeeping processes such as transcription and translation. The nucleomorph also encodes some chloroplast-targeted proteins. However, of the 17 such proteins encoded on the B. natans nucleomorph genome and the 30 on that of G. theta, only Hsp60, RpoD/Sig2 and isoforms of a Clp protease are common to both (Gilson et al. 2006).
Cryptophyte and chlorarachniophyte nucleomorph genomes differ markedly in the number and sizes of introns present. Cryptophyte nucleomorph genomes appear to have lost the majority of their ancestral introns. The G. theta nucleomorph genome contains only 29 introns, which range in size between 42 and 52 bp. Seventeen of these are within protein coding genes, including 11 in ribosomal protein genes. The remainder are found in tRNA genes (Douglas et al. 2001). The 572 kbp nucleomorph genome of the cryptophyte H. andersenii is slightly larger than that of G. theta (551 kbp) but contains no introns at all, despite having homologues of all but two of G. theta's intron-containing genes (Lane et al. 2007). Unsurprisingly, the H. andersenii nucleomorph genome has lost numerous genes encoding RNA splicing machinery. Only four genes are present that have very weak homology to spliceosome components and rRNA processing machinery (Lane et al. 2007). In contrast to cryptophytes, chlorarachniophyte nucleomorph genomes have retained many introns, although these introns are very small. The B. natans nucleomorph genome contains 852 introns (Gilson et al. 2006), which range in size from 18 to 21 bp. They are AT-rich and have typical eukaryotic GT/AG borders. The position of the majority of these introns is conserved when compared with homologues in Chlamydomonas or Arabidopsis. Concomitantly, a selection of proteins involved in RNA splicing and maturation is also encoded in the nucleomorph. A recent EST study of the nucleomorph genome of the chlorarachniophyte Gymnochlora stellata identified 153 introns (Slamovits & Keeling 2009). They are also reduced in size, ranging between 18 and 27 bp. As for B. natans, analyses revealed GT/AG intron borders and high AT content within the introns, although no branch-point recognition motifs or polypyrimidine tracts were identified (Slamovits & Keeling 2009). This study also revealed a degree of aberrant splicing of G. stellata nucleomorph transcripts—a phenomenon that was not observed in B. natans (Gilson et al. 2006). The efficiency of intron splicing may be linked to intron size. A ‘calliper’ model for splicing has been suggested whereby the splicing machinery recognizes introns by virtue of their strict 18–21 bp size (Gilson et al. 2006). However, the presence of 24 and 27 bp introns in G. stellata contradicts this proposal, although it appears that smaller introns are more efficiently spliced (Slamovits & Keeling 2009).
Little is known about the regulation of nucleomorph gene expression. The presence of genes for histone acetylases and deacetylases on nucleomorph genomes may indicate the involvement of histone modification. Given the small number of non-housekeeping genes, it is possible that sensitive control of gene expression is not necessary within this compartment. However, any control of nucleomorph gene expression may have to respond to both nuclear and chloroplast signals. It will be interesting to see how far eukaryotic mechanisms of gene regulation have been retained in the evolution of this organelle.
7. Conclusions
There is a huge range of mitochondrial genome organization and content among protists. The chloroplast genome is generally more conserved, with a few notable exceptions in isolated groups. Understanding the mechanism of gene expression and control is generally limited to a small number of model organisms. Given the importance of the group as whole, more information will be valuable. However, this will rely on the availability of better genetic tools.
Acknowledgements
Work in the authors’ laboratory is funded by the Leverhulme Trust, the BBSRC and the University of Cambridge Isaac Newton Trust.
Footnotes
One contribution of 12 to a Theme Issue ‘Evolution of organellar metabolism in unicellular eukaryotes’.
References
- Adler B. K., Harris M. E., Bertrand K. I., Hajduk S. L.1991Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell Biol. 11, 5878–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonzo J. D., Söll D.2009Mitochondrial tRNA import—the challenge to understand has just begun. Biol. Chem. 390, 717–722 (doi:10.1515/BC.2009.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonzo J., Blanc V., Estévez A. M., Rubio M. A. T., Simpson L.1999C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 18, 7056–7062 (doi:10.1093/emboj/18.24.7056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. F.2003The function of genomes in bioenergetic organelles. Phil. Trans. R. Soc. Lond. B 358, 19–37 (doi:10.1098/rstb.2002.1191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asin-Cayuela J., Gustafsson C. M.2007Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 32, 111–117 (doi:10.1016/j.tibs.2007.01.003) [DOI] [PubMed] [Google Scholar]
- Barbrook A. C., Howe C. J.2000Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol. Gen. Genet. 263, 152–158 (doi:10.1007/s004380050042) [DOI] [PubMed] [Google Scholar]
- Barbrook A. C., Howe C. J., Purton S.2006Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 11, 101–108 (doi:10.1016/j.tplants.2005.12.004) [DOI] [PubMed] [Google Scholar]
- Bendich A. J.2004Circular chloroplast chromosomes: the grand illusion. Plant Cell 16, 1661–1666 (doi:10.1105/tpc.160771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Turmel M.1996Extensive gene rearrangements in the chloroplast DNAs of Chlamydomonas species featuring multiple dispersed repeats. Mol. Biol. Evol. 13, 233–243 [DOI] [PubMed] [Google Scholar]
- Burger G., Plante I., Lonergan K. M., Gray M. W.1995The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J. Mol. Biol. 245, 522–537 (doi:10.1006/jmbi.1994.0043) [DOI] [PubMed] [Google Scholar]
- Burger G., Littlejohn T. G., Greenwood S. J., Schnare M. N., Lang B. F., Gray M. W.2000Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J. Mol. Biol. 297, 365–380 (doi:10.1006/jmbi.2000.3529) [DOI] [PubMed] [Google Scholar]
- Burger G., Forget L., Zhu Y., Gray M. W., Lang B. F.2003Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl Acad. Sci. USA 100, 892–897 (doi:10.1073/pnas.0336115100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne E. M., Gott J. M.2004Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol. Cell Biol. 24, 7821–7828 (doi:10.1128/MCB.24.18.7821-7828.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. L., Smith A. C., Kobayashi H., Purton S., Herrin D. L.2004Structure, circadian regulation and bioinformatic analysis of the unique sigma factor gene in Chlamydomonas reinhardtii. Phot. Res. 82, 339–349 (doi:10.1007/s11120-004-4213-6) [DOI] [PubMed] [Google Scholar]
- Cermakian N., Ikeda T. M., Cedergren R., Gray M. W.1996Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucl. Acids Res. 24, 648–654 (doi:10.1093/nar/24.4.648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crausaz Esseiva A., Naguleswaran A., Hemphill A., Schneider A.2004Mitochondrial tRNA import in Toxoplasma gondii. J. Biol. Chem. 279, 42 363–42 368 (doi:10.1074/jbc.M404519200) [DOI] [PubMed] [Google Scholar]
- de Koning A. P., Keeling P. J.2006The complete plastid genome of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 4, 12 (doi:10.1186/1741-7007-4-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denovan-Wright E. M., Lee R. W.1994Comparative structure and genomic organization of the discontinuous mitochondrial ribosomal RNA genes of Chlamydomonas eugametos and Chlamydomonas reinhardtii. J. Mol. Biol. 241, 298–311 (doi:10.1006/jmbi.1994.1505) [DOI] [PubMed] [Google Scholar]
- Denovan-Wright E. M., Nedelcu A. M., Lee R. W.1998Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol. Biol. 36, 285–295 (doi:10.1023/A:1005995718091) [DOI] [PubMed] [Google Scholar]
- Douglas S., Fraunholz M., Beaton M., Penny S., Deng L. T., Wu X. N., Reith M., Cavalier-Smith T., Maier U. G.2001The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096 (doi:10.1038/35074092) [DOI] [PubMed] [Google Scholar]
- Duplessis M. R., Karol K. G., Adman E. T., Choi L. Y. S., Jacobs M. A., Cattolico R. A.2007Chloroplast His-to-Asp signal transduction: a potential mechanism for plastid gene regulation in Heterosigma akashiwo (Raphidophyceae). BMC Evol. Biol. 7, 70 (doi:10.1186/1471-2148-7-70) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist J., Burger G., Gray M. W.2000Expression of mitochondrial protein-coding genes in Tetrahymena pyriformis. J. Mol. Biol. 297, 381–393 (doi:10.1006/jmbi.2000.3530) [DOI] [PubMed] [Google Scholar]
- Embley T. M., Martin W.2006Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (doi:10.1038/nature04546) [DOI] [PubMed] [Google Scholar]
- Etheridge R. D., Aphasizheva I., Gershon P. D., Aphasizhev R.20083′ Adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 27, 1596–1608 (doi:10.1038/emboj.2008.87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Lee R. W.2002Mitochondrial genome of the colorless green alga Polytomella parva: two linear DNA molecules with homologous inverted repeat termini. Mol. Biol. Evol. 19, 999–1007 [DOI] [PubMed] [Google Scholar]
- Feagin J. E., Werner E., Gardner M. J., Williamson D. H., Wilson R. J. M.1992Homologies between the contiguous and fragmented rRNAs of the two Plasmodium falciparum extrachromosomal DNAs are limited to core sequences. Nucl. Acids Res. 20, 879–887 (doi:10.1093/nar/20.4.879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. J., Williamson D. H., Wilson R. J. M.1991A circular DNA in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol. Biochem. Parasitol. 44, 115–124 (doi:10.1016/0166-6851(91)90227-W) [DOI] [PubMed] [Google Scholar]
- Gilson P. R., Su V., Slamovits C. H., Reith M. E., Keeling P. J., McFadden G. I.2006Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc. Natl Acad. Sci. USA 103, 9566–9571 (doi:10.1073/pnas.0600707103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams J., Morris J. C., Drew M. E., Wang Z., Englund P. T., Hajduk S. L.2002A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 277, 16 952–16 959 (doi:10.1074/jbc.M200662200) [DOI] [PubMed] [Google Scholar]
- Gray M. W., Boer P. H.1988Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Phil. Trans. R. Soc. Lond. B 319, 135–147 (doi:10.1098/rstb.1988.0038) [DOI] [PubMed] [Google Scholar]
- Gray M. W., Lang B. F., Burger G. E.2004Mitochondria of protists. Ann. Rev. Genet. 38, 477–524 (doi:10.1146/annurev.genet.37.110801.142526) [DOI] [PubMed] [Google Scholar]
- Hagopian J. C., Reis M., Kitajima J. P., Bhattacharya D., de Oliveira M. C.2004Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J. Mol. Evol. 59, 464–477 (doi:10.1007/s00239-004-2638-3) [DOI] [PubMed] [Google Scholar]
- Hajduk S. L., Siqueira A. M., Vickerman K.1986Kinetoplast DNA of Bodo caudatus: a noncatenated structure. Mol. Cell. Biol. 6, 4372–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R. B., Hong L., Drager R. G., Favreau M. R., Monfort A., Orzat B., Spielmann A., Stutz E.1993Complete sequence of Euglena gracilis chloroplast DNA. Nucl. Acids Res. 21, 3537–3544 (doi:10.1093/nar/21.15.3537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H., Čičová Z., Novotná L., Wen Y.-Z., Lukeš J.2009Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15, 588–599 (doi:10.1261/rna.1411809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel A. J., Glöckner G.2008Mitochondrial genome evolution in the social amoebae. Mol. Biol. Evol. 25, 1440–1450 (doi:10.1093/molbev/msn088) [DOI] [PubMed] [Google Scholar]
- Herrin D. L., Nickelsen J.2004Chloroplast RNA processing and stability. Photosynth. Res. 82, 301–314 (doi:10.1007/s11120-004-2741-8) [DOI] [PubMed] [Google Scholar]
- Herrin D. L., Chen Y.-F., Schmidt G. W.1990RNA splicing in Chlamydomonas chloroplasts. J. Biol. Chem. 265, 21 134–21 140 [PubMed] [Google Scholar]
- Horton T. L., Landweber L. F.2000Mitochondrial RNAs of myxomycetes terminate with non-encoded 3′ poly(U) tails. Nucl. Acids Res. 28, 4750–4754 (doi:10.1093/nar/28.23.4750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton T. L., Landweber L. F.2002Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr. Opin. Microbiol. 5, 620–626 (doi:10.1016/S1369-5274(02)00379-X) [DOI] [PubMed] [Google Scholar]
- Howe C. J.1992Plastid origin of an extrachromosomal DNA molecule from Plasmodium, the causative agent of malaria. J. Theor. Biol. 158, 199–205 (doi:10.1016/S0022-5193(05)80718-0) [DOI] [PubMed] [Google Scholar]
- Howe C. J., Purton S.2007The little genome of Apicomplexan plastids: its raison d'etre and a possible explanation for the ‘delayed death’ phenomenon. Protist 158, 121–133 (doi:10.1016/j.protis.2007.02.002) [DOI] [PubMed] [Google Scholar]
- Howe C. J., Nisbet R. E. R., Barbrook A. C.2008The remarkable chloroplast genome of dinoflagellates. J. Exp. Bot. 59, 1035–1045 (doi:10.1093/jxb/erm292) [DOI] [PubMed] [Google Scholar]
- Jones E. P., Mahendran R., Spottswood M. R., Yang Y. C., Miller D. L.1990Mitochondrial DNA of Physarum polycephalum: physical mapping, cloning and transcription mapping. Curr. Genet. 17, 331–337 (doi:10.1007/BF00314881) [DOI] [PubMed] [Google Scholar]
- Kairo A., Fairlamb A. H., Gobright E., Nene V.1994A 7.1 kb linear DNA molecule of Theileria parva has scrambled rDNA sequences and open reading frames for mitochondrially encoded proteins. EMBO J. 13, 898–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R., Nishimura H., Sako Y.2009Analysis of the mitochondrial genome, transcripts, and electron transport activity in the dinoflagellate Alexandrium catenella (Gonyaulacales, Dinophyceae). Phycol. Res. 57, 1–11 (doi:10.1111/j.1440-1835.2008.00511.x) [Google Scholar]
- Kim E., Lane C. E., Curtis B. A., Kozera C., Bowman S., Archibald J. M.2008Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae). BMC Genomics 9, 215 (doi:10.1186/1471-2164-9-215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane C. E., Khan H., MacKinnon M., Fong A., Theophilou S., Archibald J. M.2006Insight into the diversity and evolution of the cryptomonad nucleomorph genome. Mol. Biol. Evol. 23, 856–865 (doi:10.1093/molbev/msj066) [DOI] [PubMed] [Google Scholar]
- Lane C. E., van den Heuvel K., Kozera C., Curtis B. A., Parsons B. J., Bowman S., Archibald J. M.2007Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc. Natl Acad. Sci. USA 104, 19 908–19 913 (doi:10.1073/pnas.0707419104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B. F., Burger G., O'Kelly C. J., Cedergren R., Golding G. B., Lemieux C., Sankoff D., Turmel M., Gray M. W.1997An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387, 493–497 (doi:10.1038/387493a0) [DOI] [PubMed] [Google Scholar]
- Larkum A. W. D., Lockhart P. J., Howe C. J.2007Shopping for plastids. Trends Plant Sci. 12, 189–195 (doi:10.1016/j.tplants.2007.03.011) [DOI] [PubMed] [Google Scholar]
- Li J., Maga J. A., Cermakian N., Cedergren R., Feagin J. E.2001Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol. 113, 261–269 (doi:10.1016/S0166-6851(01)00223-7) [DOI] [PubMed] [Google Scholar]
- Lilly J. W., Maul J. E., Stern D. B.2002The Chlamydomonas reinhardtii organellar genomes respond transcriptionally and post-transcriptionally to abiotic stimuli. Plant Cell 14, 2681–2706 (doi:10.1105/tpc.005595) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lukeš J., Jirkû M., Avliyakulov N., Benada O.1998Pankinetoplast DNA structure in a primitive bodonid flagellate, Cryptobia helicis. EMBO J. 17, 838–846 (doi:10.1093/emboj/17.3.838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J., Lys Guilbride D., Votypka J., Zikova A., Benne R., Englund P. T.2002Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1, 495–502 (doi:10.1128/EC.1.4.495-502.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J., Hashimi H., Zíková A.2005Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48, 277–299 (doi:10.1007/s00294-005-0027-0) [DOI] [PubMed] [Google Scholar]
- Marande W., Burger G.2007Mitochondrial DNA as a genomic jigsaw puzzle. Science 318, 415 (doi:10.1126/science.1148033) [DOI] [PubMed] [Google Scholar]
- Marande W., Lukeš J., Burger G.2005Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot. Cell 4, 1137–1146 (doi:10.1128/EC.4.6.1137-1146.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Navarro J., Manuell A. L., Wu J., Mayfield S. P.2007Chloroplast translation regulation. Phot. Res. 94, 359–374 (doi:10.1007/s11120-007-9183-z) [DOI] [PubMed] [Google Scholar]
- Maul J. E., Lilly J. W., Cui L., dePamphilis C. W., Miller W., Harris E. H., Stern D. B.2002The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14, 2659–2679 (doi:10.1105/tpc.006155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti E. F., Harris M. E., Adler B., Torri A. F., Hajduk S. L.1992Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processing and developmentally regulated expression. Mol. Biochem. Parasitol. 54, 31–41 (doi:10.1016/0166-6851(92)90092-X) [DOI] [PubMed] [Google Scholar]
- Miller M. L., Miller D. L.2008Non-DNA-templated addition of nucleotides to the 3′ end of RNAs by the mitochondrial RNA polymerase of Physarum polycephalum. Mol. Cell. Biol. 28, 5795–5802 (doi:10.1128/MCB.00356-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. L., Antes T. J., Qian F., Miller D. L.2006Identification of a putative mitochondrial RNA polymerase from Physarum polycephalum: characterization, expression, purification, and transcription in vitro. Curr. Genet. 49, 259–271 (doi:10.1007/s00294-005-0053-y) [DOI] [PubMed] [Google Scholar]
- Minoda A., Nagasawa K., Hanaoka M., Horiuchi M., Takahashi T., Tanaka K.2005Microarray profiling of plastid gene expression in a unicellular red alga, Cyanidioschyzon merolae. Plant Mol. Biol. 59, 375–385 (doi:10.1007/s11103-005-0182-1) [DOI] [PubMed] [Google Scholar]
- Moore R. B., et al. 2008A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451, 959–963 (doi:10.1038/nature06635) [DOI] [PubMed] [Google Scholar]
- Mullet J. E., Klein R. R.1987Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 6, 1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Ishida I.2009Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 19, R284–R285 (doi:10.1016/j.cub.2009.02.043) [DOI] [PubMed] [Google Scholar]
- Nash E. A., Nisbet R. E. R., Barbrook A. C., Howe C. J.2008Dinoflagellates: a mitochondrial genome all at sea. Trends Genet. 24, 328–335 (doi:10.1016/j.tig.2008.04.001) [DOI] [PubMed] [Google Scholar]
- Nisbet R. E. R., Hiller R. G., Barry E. R., Skene P., Barbrook A. C., Howe C. J.2008Transcript analysis of dinoflagellate plastid gene minicircles. Protist 159, 31–39 (doi:10.1016/j.protis.2007.07.002) [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M. P., Fontaine J. M., Rousvoal S., Kloareg B., Loiseaux-De Goër S.2001The complete sequence of a brown algal mitochondrial genome, the ectocarpale Pylaiella littoralis (L.) Kjellm. J. Mol. Evol. 53, 80–88 [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M.-P., Goër S., Stam W., Olsen J.2006Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr. Genet. 49, 47–58 (doi:10.1007/s00294-005-0031-4) [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Nilsson A., Allen J. F.1999Photosynthetic control of chloroplast gene expression. Nature 397, 625–628 (doi:10.1038/17624) [Google Scholar]
- Pombert J. F., Otis C., Lemieux C., Turmel M.2004The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol. Biol. Evol. 21, 922–935 (doi:10.1093/molbev/msh099) [DOI] [PubMed] [Google Scholar]
- Puerta M. V. S., Bachvaroff T. R., Delwiche C. F.2004The complete mitochondrial genome sequence of the haptophyte Emiliania huxleyii and its relation to heterokonts. DNA Res. 11, 1–10 (doi:10.1093/dnares/11.1.1) [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S., Allen J. F.2009Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression. Proc. R. Soc. B 276, 2133–2145 (doi:10.1098/rspb.2008.1426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S., et al. 2008The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl Acad. Sci USA 105, 10 061–10 066 (doi:10.1073/pnas.0803928105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollmann F.-A., Choquet Y.2007Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl Acad. Sci USA 104, 9093–9098 (doi:10.1073/pnas.0703162104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehkopf D. H., Gillespie D. E., Harrell M. I., Feagin J. E.2000Transcriptional mapping and RNA processing of the Plasmodium falciparum mitochondrial mRNAs. Mol. Biochem. Parasitol. 105, 91–103 (doi:10.1016/S0166-6851(99)00170-X) [DOI] [PubMed] [Google Scholar]
- Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N.2006High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl Acad. Sci. USA 103, 4771–4776 (doi:10.1073/pnas.0509501103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard O., Bonnard G., Grienenberger J. M., Kloareg B., Boyen C.1998Transcription initiation and RNA processing in the mitochondria of the red alga Chondrus crispus: convergence in the evolution of transcription mechanisms in mitochondria. J. Mol. Biol. 283, 549–557 (doi:10.1006/jmbi.1998.2112) [DOI] [PubMed] [Google Scholar]
- Richard O., Kloareg B., Boyen C.1999mRNA expression in mitochondria of the red alga Chondrus crispus requires a unique RNA-processing mechanism, internal cleavage of upstream tRNAs at pyrimidine 48. J. Mol. Biol. 288, 579–584 (doi:10.1006/jmbi.1999.2725) [DOI] [PubMed] [Google Scholar]
- Robbens S., Derelle E., Ferraz C., Wuyts J., Moreau H., Van de Peer Y.2007The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: organelle genomes of the smallest eukaryote are examples of compaction. Mol. Biol. Evol. 24, 956–968 (doi:10.1093/molbev/msm012) [DOI] [PubMed] [Google Scholar]
- Sankoff D., Leduc G., Antoine N., Paquin B., Lang B. F., Cedergren R.1992Gene order comparisons for phylogenetic inference: evolution of the mitochondrial genome. Proc. Natl Acad. Sci. USA 89, 6575–6579 (doi:10.1073/pnas.89.14.6575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro T. A., Englund P. T.1995The structure and replication of kinetoplast DNA. Ann. Rev. Microbiol. 49, 117–143 (doi:10.1146/annurev.mi.49.100195.001001) [DOI] [PubMed] [Google Scholar]
- Shivji M. S.1991Organization of the chloroplast genome in the red alga Porphyra yezoensis. Curr. Genet. 19, 49–54 (doi:10.1007/BF00362087) [Google Scholar]
- Shutt T. E., Gray M. W.2006aBacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 22, 90–95 (doi:10.1016/j.tig.2005.11.007) [DOI] [PubMed] [Google Scholar]
- Shutt T. E., Gray M. W.2006bHomologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. Mol. Biol. Evol. 23, 1169–1179 (doi:10.1093/molbev/msk001) [DOI] [PubMed] [Google Scholar]
- Simpson C. L., Stern D. B.2002The treasure trove of algal chloroplast genomes. Surprises in architecture and gene content, and their functional implications. Plant Physiol. 129, 957–966 (doi:10.1104/pp.010908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits C. H., Keeling P. J.2009Evolution of ultrasmall spliceosomal introns in highly reduced nuclear genomes. Mol. Biol. Evol. 26, 1699–1705 (doi:10.1093/molbev/msp081) [DOI] [PubMed] [Google Scholar]
- Slamovits C. H., Saldarriaga J. F., Larocque A., Keeling P. J.2007The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 372, 356–368 (doi:10.1016/j.jmb.2007.06.085) [DOI] [PubMed] [Google Scholar]
- Smith A. C., Purton S.2002The transcriptional apparatus of plastids. Eur. J. Phycol. 37, 301–311 (doi:10.1017/S0967026202003694) [Google Scholar]
- Tan T. H. P., Bochud-Allemann N., Schneider A.2002Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc. Natl Acad. Sci. USA 99, 1152–1157 (doi:10.1073/pnas.022522299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama Y., Ishizaki Y., Morikawa K., Kobori M., Nakahira Y., Takeba G., Toyoshima Y., Shiina T.2004Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl Acad. Sci USA 101, 3304–3309 (doi:10.1073/pnas.0308362101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova E., Salinas T., Cognat V., Remacle C., Marechal-Drouard L.2009Steady-state levels of imported tRNAs in Chlamydomonas mitochondria are correlated with both cytosolic and mitochondrial codon usages. Nucl. Acids Res. 37, 1521–1528 (doi:10.1093/nar/gkn1073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. J. M., Williamson D. H.1997Extrachromosomal DNA in the Apicomplexa. Microbiol. Mol. Biol. Rev. 61, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Kück U.1996Transcript mapping and processing of mitochondrial RNA in the chlorophyte alga Prototheca wickerhamii. Plant Mol. Biol. 30, 577–595 (doi:10.1007/BF00049333) [DOI] [PubMed] [Google Scholar]
- Zauner S., Greilinger D., Laatsch T., Kowallik K. V., Maier U. G.2004Substitutional editing of transcripts of genes of cyanobacterial origin in the dinoflagellate Ceratium horridum. FEBS Lett. 577, 535–538 (doi:10.1016/j.febslet.2004.10.060) [DOI] [PubMed] [Google Scholar]
- Zhang Z., Green B. R., Cavalier-Smith T.1999Single gene circles in dinoflagellate chloroplast genomes. Nature 400, 155–159 (doi:10.1038/22099) [DOI] [PubMed] [Google Scholar]
- Zimmer S. L., Fei Z., Stern D. B.2008Genome-based analysis of Chlamydomonas reinhardtii exoribonucleases and poly(A) polymerases predicts unexpected organellar and exosomal features. Genetics 179, 125–136 (doi:10.1534/genetics.107.086223) [DOI] [PMC free article] [PubMed] [Google Scholar]