Abstract
The dramatic expansion of the geographical range of coyotes over the last 90 years is partly explained by changes to the landscape and local extinctions of wolves, but hybridization may also have facilitated their movement. We present mtDNA sequence data from 686 eastern coyotes and measurements of 196 skulls related to their two-front colonization pattern. We find evidence for hybridization with Great Lakes wolves only along the northern front, which is correlated with larger skull size, increased sexual dimorphism and a five times faster colonization rate than the southern front. Northeastern haplotype diversity is low, suggesting that this population was founded by very few females moving across the Saint Lawrence River. This northern front then spread south and west, eventually coming in contact with an expanding front of non-hybrid coyotes in western New York and Pennsylvania. We suggest that hybridization with wolves in Canada introduced adaptive variation that contributed to larger size, which in turn allowed eastern coyotes to better hunt deer, allowing a more rapid colonization of new areas than coyotes without introgressed wolf genes. Thus, hybridization is a conduit by which genetic variation from an extirpated species has been reintroduced into northeastern USA, enabling northeastern coyotes to occupy a portion of the niche left vacant by wolves.
Keywords: hybridization, adaptive introgression, range expansion, genetic variation, morphological variation
1. Introduction
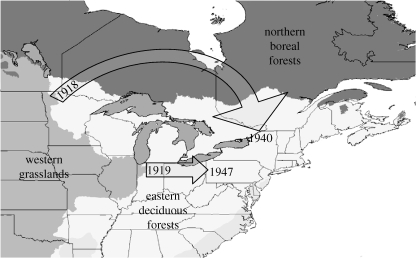
Dramatic expansions in the distribution of a species without being introduced by humans are rare, and are typically explained by habitat change or release from competitors (Sakai et al. 2001). The coyote (Canis latrans) evolved as hunter of small prey in the Great Plains, but has rapidly colonized all of eastern North America in the last 90 years. The spread of agriculture and the extinction of wolves (C. lupus sensu lato) in parts of the region are thought to have facilitated coyote expansion, but genetic interchange with remnant wolf populations may have played a roll. Coyote colonization was fivefold faster via the northern route through Ontario, which exposed them to wolf populations, compared with the southern route through Ohio, where wolves were extirpated prior to coyote expansion (figure 1).
Figure 1.
Colonization routes of coyotes moving from their historic range in the grasslands of western states into eastern deciduous forests (shading shows biomes). Dates are for the first coyote records from Ontario (Young & Jackson 1951), New York (Fener et al. 2005), Ohio (Weeks et al. 1990) and western Pennsylvania (Williams et al. 1985).
The hybridization of colonizing coyotes with wolves (C. lupus lycaon) in Ontario has been demonstrated by recent studies (Leonard & Wayne 2008; Koblmuller et al. 2009; Schwartz & Vucetich 2009; Wheeldon & White 2009; Wilson et al. 2009). Mitochondrial phylogenies reveal three main lineages within Ontario wolves (grey wolf, Great Lakes wolf (GLW) and coyote), suggesting high rates of hybridization in the region. Nuclear loci reveal similar patterns, but suggest that the GLW remains a discrete ecotype despite hybridization.
Less attention has been paid to the effect of this hybridization on eastern coyotes, which are now the largest predator in the region, are abundant in many areas, and are thus thought to play important ecological roles. Although northeastern coyotes are clearly smaller than wolves, they are larger than western coyotes, and have a unique ecology (Lawrence & Bossert 1969; Parker 1995; Kays et al. 2008). Here we examine both genetics and morphology from a large sample of coyotes to evaluate the potential introgression of adaptive variation through hybridization with wolves.
2. Material and methods
Coyote specimens were obtained by New York State Museum specimen salvage efforts, donations by fur trappers, hunters, state and provincial government agencies, scat collection (Kays et al. 2008), and by loans from other museums (see the electronic supplementary material, table S1). Our sample includes three large wolf-like canids from Vermont and New York that were not judged to be escaped pets based on the condition of their claws and teeth, and therefore are presumed to be recent immigrants (USFWS 2002, 2004, 2007). DNA was extracted using FastDNA (MP Biomedicals) or QIAmp Stool (Qiagen) kits. We amplified and sequenced a 369 bp part of the 5′ end of the mitochondrial control region for 687 individuals using primers L15926 5′-TCAAAGCTTACACCAGTCTTG TAAAC-3′ and H16498 5′-CCTGAAGTAGGAACCAGATG-3′ (modified from Vilà et al. 1999). Double-stranded sequences were aligned and edited in Sequencher v. 3.1.1. DnaSP v. 5 (Librado & Rosas 2009) was used to calculate nucleotide diversity (π), haplotypic diversity and the population genetic parameter θ = 2Neμ, all which we use to estimate relative levels of polymorphism. The HKY+Γ model of DNA evolution was selected by the Akaike information criterion implemented in Modeltest 3.6.6 (Posada & Crandall 1998) and used in a heuristic, maximum likelihood tree search conducted in PAUP* v. 4.0b10. Support for nodes was assessed by bootstrapping (100 replicates).
We took 10 cranial and four mandibular measurements on 196 coyote skulls, including 126 animals with corresponding mtDNA data. All measurements were made by one person (A.C.), with an estimated error of 0.15 per cent. All specimens were of known sex and judged to be adults by the fusion of the basioccipital-basisphenoid suture. We used Statistica6 to conduct analysis of variance (ANOVA), grouping specimens into three regions based on colonization history and our genetic results; northeast (n = 154), west (n = 21) and Ohio (n = 21). Western specimens were represented by specimens from Montana (n = 2), Nebraska (n = 8) and Arizona (n = 11).
3. Results
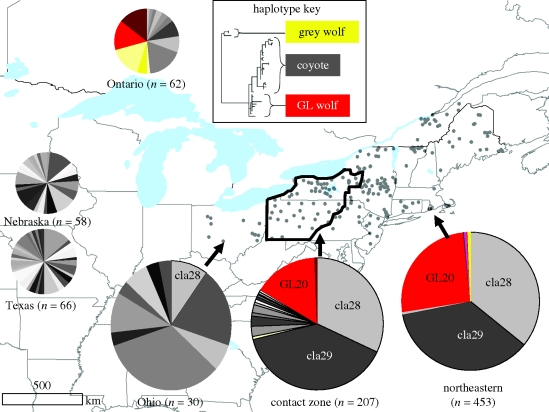
The phylogeny placing our new haplotypes within the context of published haplotypes is consistent with previous studies (Koblmuller et al. 2009) in showing that the eastern coyote population has both western coyote and GLW mtDNA, indicative of a history of hybridization among mitochondrial lineages (inset figure 2; complete tree in the electronic supplementary material, figure S1). From the geographical distribution of haplotypes and historical records of first occurrences in Ontario and eastern states (figure 1), we infer a region in western New York and Pennsylvania representing the contact zone between two advancing fronts of colonizing coyotes (figure 2). Haplotype diversity was highest in the west (Ohio) and lowest in the northeast (table 1). Our subsample east of the contact zone had the lowest value of θ, consistent with a very small founding population. Comparison with the previously published surveys indicates that our sample from Ohio comprises a subset of the diversity found in western coyote populations, whereas nearly all northeastern coyotes (447 of 453 individuals) carry one of three common haplotypes (cla28, cla29, GL20; figure 2). The six remaining individuals in the northeastern sample include three coyotes carrying rare haplotypes (GL21, GL22 and cla37), and three large wolves (two lu32, one GL23, not included in our coyote population genetics calculations). In addition, one partial sequence of a dog-like haplotype was obtained from a coyote in Vermont. The inferred contact zone has an intermediate value of haplotype diversity (table 1), but the highest number of haplotypes, indicating admixture between the diverse western coyote source and the coyote-wolf hybridized source.
Figure 2.
Mitochondrial haplotype frequencies of eastern coyotes and source populations. Sampling localities from this paper are shown as grey dots and summarized as large pie charts showing the proportion of animals with haplotypes categorized according to their position on the phylogeny. Smaller pie charts show haplotype frequencies representative of the two source populations: Ontario wolves and coyotes (Wilson et al. 2000; Leonard & Wayne 2008) and coyotes from western states (Hailer & Leonard 2008).
Table 1.
Summary of genetic diversity observed in the total sample and three geographical subsamples of coyotes (figure 1).
| total | Ohio | contact | northeast | |
|---|---|---|---|---|
| sample size | 687 | 30 | 207 | 450 |
| haplotypes | 22 | 11 | 16 | 6 |
| haplotype diversity | 0.708 | 0.844 | 0.721 | 0.664 |
| nucleotide diversity, π (per site) | 0.0158 | 0.0133 | 0.0152 | 0.0158 |
| θ (per site) | 0.0128 | 0.0179 | 0.0140 | 0.0077 |
| average pairwise number of nucleotide differences, k | 5.57 | 4.69 | 5.36 | 5.56 |
Northeast coyote skulls from both sexes were larger than those from Ohio and western states (table 2), especially in skull width (zygomatic width (ZW), 4–9% larger) and the area of muscle attachment on the mandible (width anterior portion of the ramus (WAR), 8–15% larger). In addition, unlike western coyotes, northeastern coyotes were sexually dimorphic in all measurements (table 2). Within the northeast, we found correlations between genotype and phenotype, but these varied by sex. In males, two measurements were related to mtDNA haplotype (ANOVA, d.f. = 62, p < 0.005 and <0.05, respectively) in that animals with the GL20 (Tukey HSD post hoc tests p < 0.0) and cla29 (p < 0.05) had wider skulls (ZW) than individuals with cla28, and that WAR was larger in males with cla29 than those with cla28 (p < 0.05). In northeastern female coyotes the height from the base of the first upper molar to the orbit was correlated with haplotype (ANOVA, d.f. = 51, p < 0.05), which was primarily driven by larger skulls in animals with GL20 compared with cla28 (Tukey HSD post hoc tests p < 0.05).
Table 2.
Sexual and regional differences in skull measurements for male = M and female = F coyotes. (Mean sexual differences (MSD) are calculated as male/female and the number of specimens measured from each group is in parenthesis. Mean values (mm): length upper carnassial (LP4), width upper second molar (WM2), alveolar length of maxillary toothrow (ALM), maximum (MXP) and minimum (MNP) palate width, greatest length skull (GLS), zygomatic width (ZW), height jugal (HJ), height from the base of the first upper molar to the orbit (M1O), width postorbital processes (WPOP), height coronoid process (HCP), width anterior portion of the ramus (WAR), width mandible at carnassial (WMm1), height mandible at carnassial (HMm1). Significance of an ANOVA using Tukey's HSD post hoc test: *p ≤ 0.05, **p ≤ 0.01, p > 0.05 = n.s.)
| measurement | sexual dimorphism |
regional comparisons |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| northeastern |
Ohio |
western |
northeastern versus Ohio |
northeastern versus western |
|||||||||
| M (88) | F (66) | MSD | M (13) | F (8) | MSD | M (10) | F (11) | MSD | M | F | M | F | |
| LP4 | 20.9 | 19.9 | 1.05** | 20.5 | 19.8 | 1.04 | 20.1 | 19.6 | 1.03 | n.s. | n.s. | n.s. | n.s. |
| WM2 | 12.3 | 11.7 | 1.05** | 11.6 | 12.1 | 0.96 | 11.9 | 11.3 | 1.05 | * | n.s. | n.s. | n.s. |
| ALM | 72.5 | 70.0 | 1.04** | 73.5 | 70.7 | 1.04* | 71.5 | 70.2 | 1.02 | n.s. | n.s. | n.s. | n.s. |
| MXP | 61.3 | 58.1 | 1.06** | 59.8 | 57.6 | 1.04 | 54.5 | 53.6 | 1.02 | n.s. | n.s. | ** | ** |
| MNP | 22.7 | 21.4 | 1.06** | 22.1 | 21.7 | 1.02 | 19.7 | 19.5 | 1.01 | n.s. | n.s. | ** | ** |
| GSL | 201.6 | 192.8 | 1.05** | 201.5 | 195.4 | 1.03* | 194.4 | 189.6 | 1.03 | n.s. | n.s. | * | n.s. |
| ZW | 106.8 | 101.8 | 1.05** | 101.7 | 97.9 | 1.04* | 98.2 | 95.4 | 1.03 | ** | * | ** | ** |
| HJ | 14.1 | 13.2 | 1.07** | 13.6 | 12.9 | 1.05 | 12.1 | 12.4 | 0.98 | n.s. | n.s. | ** | n.s. |
| M1O | 28.2 | 26.8 | 1.05** | 27.4 | 25.8 | 1.06 | 25.8 | 25.2 | 1.02 | n.s. | n.s. | ** | * |
| WPOP | 50.1 | 47.8 | 1.05** | 47.1 | 45.6 | 1.03 | 45.1 | 44.5 | 1.01 | * | n.s. | ** | ** |
| HCP | 55.3 | 52.2 | 1.06** | 53.5 | 52.6 | 1.02 | 50.8 | 49.9 | 1.02 | n.s. | n.s. | ** | * |
| WAR | 9.1 | 8.7 | 1.05** | 8.4 | 8.0 | 1.05 | 7.9 | 8.0 | 0.99 | * | * | ** | * |
| WMm1 | 10.2 | 9.6 | 1.06** | 10.1 | 9.2 | 1.10* | 9.2 | 9.1 | 1.01 | n.s. | n.s. | ** | n.s. |
| HMm1 | 23.1 | 21.8 | 1.06** | 21.9 | 21.5 | 1.02 | 20.7 | 20.4 | 1.01 | * | n.s. | ** | ** |
4. Discussion
The ecological differences between western and northeastern coyotes, on average, are that northeastern animals eat more deer (Odocoileus sp.) but fewer small mammals (Parker 1995), and show no avoidance of forested habitats (Kays et al. 2008). The larger body size of northeastern coyotes is widely accepted as advantageous for hunting large prey, but there has been debate about the origin of this variation through hybridization versus phenotypic plasticity (Lariviere & Crete 1993; Peterson & Thurber 1993). Our results show that northeastern coyote populations are a hybrid swarm resulting from the widespread introgression of GLW genes. This suggests that hybridization introduced genetic variation for the rapid adaptation of more efficient predation on deer, including larger predator body size and skull dimensions. This is further supported by our finding that northeastern coyotes were larger than those from Ohio, which are living in similar eastern forests, but have not hybridized with wolves. Mitochondrial genes are surely not responsible for the large body size, so the observed associations of particular haplotypes with skull morphology suggest that this hybrid swarm is young. That is, the linkage of mtDNA and morphology has not had sufficient time to break down through recombination.
Northeastern coyote skulls are not simply larger versions of their western relatives, but show additional craniodental characteristics similar to wolves, supporting the hypothesis of the introgression of genetic variation; northeastern skulls are proportionally broader, with greater areas of attachment for masticatory musculature. In large-prey hunters, such as wolves, these traits are associated with strong bite forces and resistance to the mechanical stresses imposed by large, struggling prey (Slater et al. 2009). Furthermore, the sexual dimorphism we found in northeastern coyotes is absent in western coyotes, but similar to that reported for wolves (Gittleman & Van Valkenburgh 1997). We suggest that these traits confer similar adaptive advantages in northeastern coyotes and allow them to be more proficient in the capture of deer than western and Ohio coyotes. These adaptations presumably allowed the rapid movement of coyote-wolves through Ontario, in comparison with the slower colonization rate of the smaller non-hybridized coyotes across Ohio.
The geographical pattern of genetic diversity we found in eastern coyotes reflects the reported colonization route (Parker 1995), with a pure coyote front advancing from the west and meeting a coyote-wolf hybrid swarm that colonized earlier from the north. The low number of haplotypes in the northeast is consistent with the idea that the population was founded by a few animals crossing the Saint Lawrence River, with little or no genetic contribution from the scattered introductions of coyotes by hunt clubs (Fener et al. 2005). The presence of C. lupus haplotypes in two large wolf-like animals suggest that there is limited migration of large wolves into the region, but that they are not contributing to the coyote gene pool via hybridization as Canadian wolves are. Likewise, our finding of only one dog-like haplotype suggests that hybridization with dogs has not been significant, at least not between male northeastern coyotes and female dogs.
Our course-grained genetic approach allowed us to show the widespread distribution of introgressed wolf mitochondria across many hundreds of northeastern coyotes but does not provide the detailed ancestry of individuals. Additional analyses with nuclear markers will be needed to provide more resolution on the relative contributions of coyote, GLW, grey wolf and dog to this hybrid swarm, and evaluate the related taxonomic and conservation implications of the dynamic population genetics of the region's Canis.
Acknowledgements
This research was covered by collection permits from the New York Department of Conservation.
Thanks to the hundreds of coyote hunters and state wildlife officials who helped us obtain specimens, and especially to J. Bopp who oversaw their collection and specimen preparation. We thank M. Frye, J. Monzon and J. Murtaugh for laboratory assistance and J. Chupasko, P. Work and D. Lunde for help with museum collections. Funding was provided by the NYSM and NYSDEC.
References
- Fener H. M., Ginsberg J. R., Sanderson E., Gompper M. E.2005Chronology of range expansion of the coyote Canis latrans, in New York. Can. Field Nat. 119, 1–5 [Google Scholar]
- Gittleman J. L., Van Valkenburgh B.1997Sexual dimorphism in the canines and skulls of carnivores: effects of size, phylogeny, and behavioural ecology. J. Zool. Lond. 242, 97–117 (doi:10.1111/j.1469-7998.1997.tb02932.x) [Google Scholar]
- Hailer F., Leonard J. A.2008Hybridization among three native North American canis species in a region of natural sympatry. PLoS ONE 3, e3333 (doi:10.1371/journal.pone.0003333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays R. W., Gompper M. E., Ray J. C.2008Landscape ecology of eastern coyotes based on large-scale estimates of abundance. Ecol. Appl. 18, 1014–1027 (doi:10.1890/07-0298.1) [DOI] [PubMed] [Google Scholar]
- Koblmuller S., Nord M., Wayne R. K., Leonard J. A.2009Origin and status of the Great Lakes wolf. Mol. Ecol. 11, 2313–2326 [DOI] [PubMed] [Google Scholar]
- Lariviere S., Crete M.1993The size of eastern coyotes (Canis latrans): a comment. J. Mammal. 74, 1072–1074 (doi:10.2307/1382446) [Google Scholar]
- Lawrence B., Bossert W. H.1969The cranial evidence for hybridization in New England canis. Breviora 330, 1–13 [Google Scholar]
- Leonard J. A., Wayne R. K.2008Native Great Lakes wolves were not restored. Biol. Lett. 4, 95–98 (doi:10.1098/rsbl.2007.0354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rosas J.2009DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- Parker G.1995Eastern coyote: the story of its success Halifax, Nova Scotia: Nimbus Publishing [Google Scholar]
- Peterson R. O., Thurber J. M.1993The size of eastern coyotes (Canis latrans): a rebuttal. J. Mammal. 74, 1075–1076 (doi:10.2307/1382447) [Google Scholar]
- Posada D., Crandall K. A.1998Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- Sakai A. K., et al. 2001The population biology of invasive species. Ann. Rev. Ecol. Syst. 32, 305 (doi:10.1146/annurev.ecolsys.32.081501.114037) [Google Scholar]
- Schwartz M. K., Vucetich J. A.2009Molecules and beyond: assessing the distinctness of the Great Lakes wolf. Mol. Ecol. 18, 2307–2309 (doi:10.1111/j.1365-294X.2009.04177.x) [DOI] [PubMed] [Google Scholar]
- Slater G. J., Dumont E. R., Van Valkenburgh B.2009Implications of predatory specialization for cranial form and function in canids. J. Zool. Lond 278, 81–188 [Google Scholar]
- USFWS 2002Agency case no. 00FW01519. Report of investigation, US Fish and Wildlife Service, Washington, DC, USA. [Google Scholar]
- USFWS 2004Saratoga county canid. Report of investigation, US Fish and Wildlife Service, Washington, DC, USA. [Google Scholar]
- USFWS 2007Examination report of Vermont Canid, US Fish and Wildlife Service, Washington, DC, USA. [Google Scholar]
- Vilà C., et al. 1999Mitochondrial DNA phylogeny and population history of the grey wolf Canis lupus. Mol. Ecol. 8, 2089–2103 (doi:10.1046/j.1365-294x.1999.00825.x) [DOI] [PubMed] [Google Scholar]
- Weeks J. L., Tori G. M., Shieldcastle M. C.1990Coyotes (Canis latrans) in Ohio. Ohio J. Sci. 90, 142–145 [Google Scholar]
- Wheeldon T., White B. N.2009Genetic analysis of historic western Great Lakes region wolf samples reveals early Canis lupus/lycaon hybridization. Biol. Lett. 5, 101–109 (doi:10.1098/rsbl.2008.0516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. L., McLaren S. B., Burgwin M. A.1985Paleo-archaeological and historical records of selected Pennsylvania mammals. Ann. Carnegie Mus. 54, 77–188 [Google Scholar]
- Wilson P. J., et al. 2000DNA profiles of the eastern Canadian wolf and the red wolf provide evidence for a common evolutionary history independent of the gray wolf. Can. J. Zool. 78, 2156–2166 (doi:10.1139/cjz-78-12-2156) [Google Scholar]
- Wilson P. J., Grewal S. K., Mallory F. F., White B. N.2009Genetic characterization of hybrid wolves across Ontario. J. Hered. 100, S80–S89 (doi:10.1093/jhered/esp034) [Google Scholar]
- Young S. P., Jackson H. H. T.1951The clever coyote Harrisburg, PA: The Stackpole Company [Google Scholar]