Abstract
Microbial systems have become the preferred testing grounds for experimental work on the evolution of traits that benefit other group members. This work, based on conceptual and theoretical models of frequency-dependent selection within populations, has proven fruitful in terms of understanding the dynamics of group beneficial or ‘public goods’ traits within species. Here, we expand the scope of microbial work on the evolution of group-beneficial traits to the case of multi-species communities, particularly those that affect human health. We examined whether β-lactamase-producing Escherichia coli could protect ampicillin-sensitive cohorts of other species, particularly species that could cause human disease. Both β-lactamase-secreting E. coli and, surprisingly, those engineered to retain it, allowed for survival of a large number of ampicillin-sensitive cohorts of Salmonella enterica serovar Typhimurium, including both laboratory and clinical isolates. The Salmonella survivors, however, remained sensitive to ampicillin when re-plated onto solid medium and there was no evidence of gene transfer. Salmonella survival did not even require direct physical contact with the resistant E. coli. The observed phenomenon appears to involve increased release of β-lactamase from the E. coli when present with S. enterica. Significantly, these findings imply that resistant E. coli, that are not themselves pathogenic, may be exploited, even when they are normally selfish with respect to other E. coli. Thus, Salmonella can gain protection against antibiotics from E. coli without gene transfer, a phenomenon not previously known. As a consequence, antibiotic-resistant E. coli can play a decisive role in the survival of a species that causes disease and may thereby interfere with successful treatment.
Keywords: Salmonella enterica serovar Typhimurium, β-lactamase, antibiotic susceptibility
1. Introduction
The emergence of antibiotic-resistant bacteria (e.g. Shigella dysenteriae (Sack et al. 1997); methicillin/oxacillin-resistant Staphylococcus aureus (Merlino et al. 2002); vancomycin-resistant enterococci (Rice 2001); extended-spectrum β-lactamases that confer resistance to cephalosporins and monobactams (Paterson & Bonomo 2005); also Martinez et al. 2007; Torres et al. 2007) has compromised effective treatment of infections. An extensive body of research has examined how new mechanisms of resistance evolve and how resistance, once it emerges, may spread throughout bacterial communities via promiscuous transfer of the genetic bases for such resistance (Hastings et al. 2004; and, for reviews, see Foster 1983; Smith 2001). More recently, we and others have explored an alternative mechanism of resistance, which depends on some members of the bacterial community acting as altruists and ‘sharing’ their resistance without transferring the genes responsible (Lenski & Hattingh 1986; Levin et al. 1997; Dugatkin et al. 2003; Griffin et al. 2004; Dugatkin et al. 2005a,b; Clark et al. 2009). Such shared resistance can be conceptualized as something akin to a ‘public good’ in economics (Samuelson 1954; Brockhurst et al. 2008). One should note, however, that technically shared antibiotic resistance meets the ‘non-excludability’ condition, but probably not the ‘non-rivalness’ condition for a pure public good. We have demonstrated that E. coli that produces β-lactamase available to other bacteria allow sensitive cohorts of E. coli to survive in environments of otherwise lethal concentrations of antibiotics; similar findings of benefit have been observed with antibiotics or potentially toxic substances in biofilms (Karthikeyan et al. 2001; Donlan & Costerton 2002; Lynch & Robertson 2008) and in planktonic cultures (Lenski & Hattingh 1986; Rosensweig et al. 1994; Treves et al. 1998; Travisano 2001).
We have studied antibiotic resistance within populations of E. coli in the context of frequency-dependent selection. This theoretical framework, which has been extensively developed to understand the evolution of group-beneficial, public-good traits, has proven very useful for elucidating the dynamics of group-beneficial traits within microbial species. Our goal here is to expand this work done within species to the case of multi-species microbial communities, with a particular eye for how such studies can help us understand multi-species microbial communities that affect human health. In the long-run, we are interested in such questions as, can the public good provided by individuals from one microbial species benefit individuals from other species, and if so, what implications does this have for understanding the diversity of microbial species?
The group-beneficial trait we examine is antibiotic-resistance. Some bacteria naturally secrete the molecules that provide the mechanism for their resistance to antimicrobials. Gram-positive bacteria that display resistance to β-lactam antibiotics, such as penicillins (e.g. ampicillin; Foster 1983; Livermore 1997), often do so as a result of β-lactamases, typically high-affinity enzymes secreted into their environment. The degree of resistance conferred to cells in the medium (including both producers and others) is frequency-dependent: the more of these bacteria that are present in the growth medium, the greater the available pool of enzyme to destroy the substrate antibiotics and allow the bacteria to survive. In principle, otherwise antibiotic-susceptible cells may survive, and even thrive, in such settings without contributing to the resistance effort and paying a cost. Secretion of β-lactamases thus constitutes a public good (Brockhurst et al. 2008; Sheratt et al. 2009), which can be exploited by those that contribute neither to its production, nor to the benefit of the group as a whole. In contrast to examples of cooperation where production of the public goods increases with increasing resources, and thus, costs less (Brockhurst et al. 2008), here cooperation depends on the amount of β-lactamase needed for cells to withstand the antibiotics present in the microbial environment. We have explored this process by developing theoretical models (Dugatkin et al. 2003, 2005b, 2008), and by experimental work using near-isogenic bacteria, where the laboratory micro-environment can be controlled (Dugatkin et al. 2005a; Clark et al. 2009).
The evolution of resistance by pathogenic bacteria to the antibiotics used to treat serious infections poses a threat to the continued efficacy of such drugs in treatment. In this study, we examined the possibility that ampicillin-resistant E. coli, gram-negative bacteria that were engineered to secrete β-lactamase into their medium (Dugatkin et al. 2005a; Clark et al. 2009), might benefit ampicillin-sensitive cohorts of other species, especially species that could cause human disease. Salmonella enterica serovar Typhimurium is a causative agent of salmonellosis (Arrach et al. 2008). Most persons infected with this organism develop diarrhoea, fever, vomiting and abdominal cramps 12–72 h after infection. Shigella flexneri is another pathogenic enteric bacterium that accounts for about one-third of all cases of shigellosis in the USA. Shigellosis usually involves bloody diarrhoea, and, as with salmonellosis, fever and stomach cramps starting a day or two after they are exposed to the bacterium. People suffering with these infectious diseases may require hospitalization and shigellosis can lead to seizures, especially in young children. Here, we report that S. enterica serovar Typhimurium strains that are otherwise sensitive to ampicillin can survive in co-cultivation with ampicillin-resistant E. coli—even when E. coli cells produce β-lactamase but were not engineered to actively excrete this into the medium and do not permit antibiotic-sensitive E. coli strains to survive.
2. Material and methods
(a). Host bacterial strains and plasmids
For these studies we used strains that could be distinguished visually on agar plates due to colour differences (e.g. in the presence of chromogenic substrates such as X-gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside)). A complete list of strains and plasmids is found in table S1 in the electronic supplementary material. Escherichia coli background strains KL99, 6925, Lu53 and Wyl, S. enterica serovar Typhimurium strain NI-101, and Enterobacter aerogenes ATCC 13048 are all blue on such media, whereas E. coli 5240 and the remaining Salmonella and Shigella strains are all white.
The plasmids used have been described previously (Clark et al. 2009). All plasmids are derived from pCR2.1 TOPO (Invitrogen, Carlsbad, CA, USA), modified so that the only source of ampicillin resistance that can be provided by any plasmid is that originating from a derivative of the TEM-1 β-lactamase gene cloned into the multiple cloning site and placed under the control of the lac promoter of the vector. In the Alt strains, β-lactamase was engineered to be localized to the outer surface of the outer membrane where the effects of its action might be shared with other cells; the Alt strains contain the pSAR1 plasmid (Dugatkin et al. 2005a). In the Self strains, encoded on plasmid pSLAR1 (Dugatkin et al. 2005a), the β-lactamase was specifically engineered to be ‘locked’ in the periplasm. Hereafter in the text, strains bearing the pSAR1 and pSLAR1 plasmids will be referred to as Alt or Self, respectively. Plasmid pBR322 (Bolivar et al. 1977) is a standard ColE1-derivative bearing both a wild-type copy of the TEM-1 bla gene and a tetracycline-resistance gene.
(b). Measurement of β-lactamase activity, β-galacosidase activity and ampicillin concentrations
Beta-lactamase activity was measured using a spectrophotometric assay (Hedberg et al. 1995) with nitrocefin (Glaxo Research, Oxoid, UK) as the chromogenic cephalosporin substrate. The colour change from yellow to red when nitrocefin was cleaved by β-lactamase was monitored over a 30 min period at 495 nm. Activity was measured from whole cells grown in liquid culture, supernatants of such cultures after centrifugation to collect cells, and sonic extracts of pelleted cells. In the latter two cases, activities were normalized to amount of protein, as measured by Bradford assay (Bradford 1976; Bio-Rad, Hercules, CA, USA). Ampicillin concentrations remaining in culture media were measured by the Folin-Ciocalteu method (Ahmad et al. 2004), using ampicillin standards in Davis Minimal Medium (DMM; Lenski et al. 1994) for the spectrophotometric assay. Activity for β-galactosidase was assayed according to the procedure recommended by Sambrook & Russell (2001).
(c). Conditions for growth and competitions
A dual-flask system (Bellco BIOTECHNOLOGY, Vineland, NJ, USA) was used in these experiments (Clark et al. 2009). The filter used between the two flasks was a Miracloth—0.2 µm Supor-200 membrane (PALL) sandwich. This set-up allowed for dilution of waste products and maintenance of proper nutrient and drug concentrations. In competitions where physical contact among cells was permitted, both strains were diluted to the desired ratio (50∶50) and inoculated on the ‘Cell’ side into DMM, supplemented with dextrose (0.006%), MgSO4 (1 mM), thiamine (0.0005%), Casamino acids (0.004%) and IPTG (isopropylthio-β-d-galactoside, 0.16 mM), with or without ampicillin. All competitions done with both strains on the same side were run for at least 120 h, were done at least in duplicate, and were repeated with each genetic background and plasmid combination. Flasks were re-fed on the ‘Nutrient’ side (i.e. with no bacteria) every 12 h with fresh media. Alternatively, for competitions in which direct physical contact was removed, one strain was inoculated on one side of the apparatus, and the other on the side normally used to replenish nutrients. In these cases, media was not exchanged during the course of the experiment, which normally was conducted for up to 96 h. In both set-ups, samples were taken every 12 h and analysed by measuring the absorbance at 600 nm (A600) and by dilution plating, by spotting 40 µl of 105 and 106 dilutions onto XI agar (containing X-gal (80 µg ml−1) + IPTG (0.16 mM)), AXI agar (XI + 100 µg ampicillin per millilitre) and, for control experiments, KXI agar (XI + 50 µg ml−1 kanamycin). Three spots were counted for each dilution, comparing numbers of white versus blue colonies. From these measurements, colony forming units per millilitre were calculated for each strain for each growth culture.
(d). Statistical analyses
One-factor and univariate two-factor analysis of variance (ANOVA) were used to analyse initial growth and control experiments, as well as survival in competitions. As determined by Levene's test either Tukey HSD or Games-Howell post hoc multiple comparisons were performed. For time-course experiments, repeated-measures ANOVA was used for multiple comparisons with Sidak correction to reduce the possibility of Type I errors. Reject/fail-to-reject decisions for all null hypotheses were based on α = 0.05. All routines were performed with PASW Statistics 17.2 (SPSS Inc., Chicago, IL, USA).
3. Results
(a). In the absence of antibiotic, Salmonella strains outgrow E. coli strains
In nutrient-limiting liquid DMM (Lenski et al. 1994; Dugatkin et al. 2005a), the Shigella and Salmonella strains were compared for growth rates with those for the plasmid-bearing and plasmid-free background E. coli strains. In each case, Salmonella strains had more rapid rates of growth than E. coli or Shigella, either alone or in the same flasks, when in direct competitions with their E. coli counterparts (figure S1a in the electronic supplementary material). This was also true if the E. coli and either Shigella or Salmonella were inoculated into opposite sides of a dual-flask set-up used in competition experiments (Clark et al. 2009). On the other hand, all the Shigella and Salmonella strains in this study were unable to grow in ampicillin concentrations at or above 100 µg ml−1, either in liquid DMM (figure S1b in the electronic supplementary material) or on solid DMM agar or rich Luria–Bertani (LB) agar. To further assess these differences in growth, the relative susceptibilities (minimum inhibitory concentrations (MICs)) to ampicillin of S. flexneri ATCC 12022, S. enterica serovar Typhimurium ATCC strain 14028, and the background E. coli strains 5240 and 6925 were determined after growth in DMM broth with subsequent spotting onto LB agar plates containing different concentrations of ampicillin. From these assessments (see table S2 in the electronic supplementary material), we found that the S. flexneri ATCC 12022 was by far the most susceptible to ampicillin (MIC 10 µg ml−1), whereas the plasmid-free E. coli strains displayed intermediate susceptibility (MIC 25–50 µg ml−1). Interestingly, the S. enterica serovar Typhimurium ATCC strains (14028, NI-1), while susceptible at 50 µg ampicillin per millilitre, persisted at low levels (about 1% of that seen, for example, on 25 µg ampicillin per millilitre) even on agar containing 100 µg ampicillin per millilitre. All plasmid-bearing E. coli in this study had MICs greater than 400 µg ampicillin per millilitre.
(b). Otherwise antibiotic-sensitive Salmonella cells survive when grown together with E. coli engineered to secrete β-lactamase
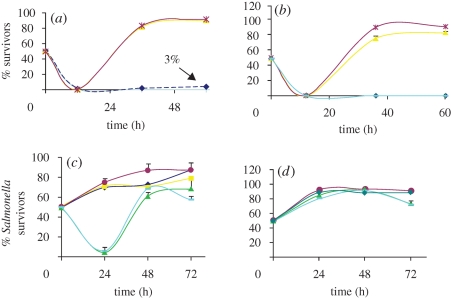
We have previously shown that ampicillin-sensitive E. coli, grown with near-isogenic ampicillin-resistant altruist E. coli bearing the pSAR1 plasmid, were able to survive normally lethal concentrations of the drug (Dugatkin et al. 2005a; Clark et al. 2009). This was again observed here, where sensitive E. coli 5240 survived at approximately 3 per cent when present with the E. coli KL99Alt (Dugatkin et al. 2005a; Clark et al. 2009; figure 1a) or 6925Alt (figure 1a). No such survivors were observed in identical competitions between 5240 and KL99Self (figure 1b). To explore the possibility that such E. coli cells bearing pSAR1 could similarly protect sensitive cohorts of different species, S. flexneri ATCC 12022 and S. enterica serovar Typhimurium strains were each competed with KL99Alt. No S. flexneri ATCC 12022 survivors were observed in such competitions, at either 50 or 100 µg ampicillin per millilitre. As seen in figure 1a, for competitions in 100 µg ampicillin per millilitre, at 12 h sampling, the colony forming units per millilitre of S. enterica serovar Typhimurium ATCC strain 14028 dropped below 2.5 × 106. However, survivors of this strain were detected after 24–36 h of growth in the competition flasks at either concentration of ampicillin; their numbers increased throughout the course of competitions, until they represented roughly 80 per cent of survivors on LB agar containing X-gal and IPTG (XI) plates. However, when bacteria from these same competitions were plated directly onto XI agar plates containing 100 µg ampicillin per millilitre (AXI agar plates), no S. enterica serovar Typhimurium colonies grew. In addition, all S. enterica serovar Typhimurium survivors that appeared on XI agar plates were ampicillin-sensitive (i.e. they failed to grow when re-streaked onto AXI agar plates). As controls, competitions were also conducted between ampicillin-sensitive E. coli plasmid-free background strains KL99 or 6925, and these similarly failed to yield either S. enterica serovar Typhimurium or S. flexneri ATCC 12022 survivors in the presence of 100 µg ampicillin per millilitre of DMM broth (not shown; statistically significant differences in per cent survivors, representing p-values <0.01, from Games-Howell post hoc analyses). Moreover, S. enterica serovar Typhimurium 14028 had significantly greater fitness than S. flexneri strain 12022 or plasmid-free E. coli strains competed with the ampicillin-resistant E. coli.
Figure 1.
Otherwise selfish ampicillin-resistant E. coli support the survival of Salmonella. Salmonella enterica serovar Typhimurium 14028 or S. flexneri strain 12022 were competed against (a) Altruist E. coli strains or (b) Selfish E. coli strains in 100 µg ampicillin per millilitre DMM. Additional competitions with Salmonella were conducted (c) at 50∶50 initial starting proportions of strains in 100 µg ampicillin per millilitre DMM liquid medium; or (d) in DMM liquid medium without ampicillin. Presented are the results of three independent replicates, with the mean per cent ampicillin-sensitive survivors on XI shown, along with standard error bars. Those samples whose cfu ml−1 were less than 2.5 × 106 are represented as 0 in this graph. In each case, the plasmid-free ampicillin-sensitive strain is indicated and, in parentheses, its plasmid-bearing ampicillin-resistant competitor (as, e.g. vKL99Alt). 5240 Free, 5240 without plasmid; Salmonella, S. enterica serovar Typhimurium ATCC 14028; Shigella, S. flexneri strain 12022; Shigella(vKL99Self50), the S. flexneri strain 12022 was competed with E. coli KL99Self at 50 µg ampicillin per millilitre DMM. (a) Dark blue, 5240Free(vKL99Alt); yellow, Salmonella(vKL99Alt); purple, Salmonella(v6925Alt); light blue, Shigella(vKL99Alt). (b) Dark blue, 5240Free(vKL99Self); yellow, Salmonella(vKL99Self); purple, Salmonella(v6925Self); green, Shigella(vKL99Self); light blue, Shigella(vKL99Self50). (c) Dark blue, KL99Alt; yellow, KL99Self; purple, 6925Self; green, Lu53pBR322; light blue, Wyl. (d) Purple, 6925Self; green, Lu53pBR322; light blue, Wyl; dark green, 6925Free.
(c). Other ampicillin-resistant E. coli also allow sensitive Salmonella cells to survive
The E. coli strain KL99Self (Dugatkin et al. 2005a; Clark et al. 2009) is resistant to ampicillin, but does not share resistance with sensitive E. coli (Clark et al. 2009). The survival of sensitive E. coli in ampicillin when competed with KL99Alt was approximately 3 per cent (figure 1a), compared with no observed survivors in competitions of the sensitive E. coli with the Selfish E. coli (figure 1b). This difference was nearly statistically significant (p = 0.093).
To our surprise, this Selfish strain also allowed otherwise ampicillin-sensitive S. enterica serovar Typhimurium to grow (figure 1b), at a rate comparable to that provided by the KL99Alt and this benefit was observed even when competitions were in the presence of starting concentrations of 200 µg ampicillin per millilitre of DMM. At 200 µg ampicillin per millilitre DMM broth, at 36 h, the S. enterica-sensitive cells were roughly 60 per cent of the survivors on XI agar, and this increased to 68.6 per cent by 60 h (not shown). When the starting ratios of E. coli to S. enterica serovar Typhimurium were 75∶25, the proportion of S. enterica serovar Typhimurium became 64.4 per cent by 36 h, and this proportion continued to rise until the S. enterica serovar Typhimurium was 75.8 per cent of the survivors plated onto XI, a level comparable to what was observed with starting ratios of 50∶50 E. coli to S. enterica serovar Typhimurium (not shown). With starting ratios of 10∶90, although at 24 h S. enterica serovar Typhimurium-sensitive cells yielded less than 2.5 × 106 cfu ml−1, by 48 h, they had become 75 per cent of the survivors on XI agar plates (not shown).
Since E. coli KL99 is an Hfr strain, we tested the possibility that survival of S. enterica serovar Typhimurium survivors depended on the transfer (either permanent or transient) of the bla gene from the E. coli strain. As mentioned earlier, all S. enterica serovar Typhimurium survivors obtained from XI plates remained sensitive to ampicillin, and none examined contained plasmids. However, to further rule out gene transfer from the KL99 background as explanation for our findings, we repeated the competition experiments using strains that lack conjugative transfer ability (i.e. 6925Self and Lu53(pBR322); the latter strain bears the wild-type TEM-1 bla gene on the classic plasmid vector, pBR322). Although at 24 h, co-cultivation with the latter strain yielded only about 5 per cent of the otherwise sensitive S. enterica serovar Typhimurium cells observed with 6925Self; by 48 h, the proportions of S. enterica cheaters produced by both E. coli strains (figure 1c) were comparable, although Lu53(pBR322) still yielded a significantly lower proportion. Moreover, similar numbers were obtained when samples were plated onto XI or DMM + XI, or onto AXI versus DMM + AXI. In these experiments, an equilibrium was established, whereby the sensitive S. enterica failed to exclude their ampicillin-resistant E. coli benefactors. Similar results were obtained at both 100 (figure 1c) and 200 µg ampicillin per millilitre of DMM (not shown). On the other hand, when competitions were conducted in LB broth (i.e. rich medium) containing 100 µg ampicillin per millilitre, cultures grew to much higher densities and at 48 h essentially 100 per cent of survivors observed by plating 105 dilutions onto XI agar were S. enterica serovar Typhimurium (not shown).
(d). Survival of otherwise ampicillin-sensitive cells is not limited to one Salmonella strain and does not require plasmid-borne bla
In order to rule out the possibility that the laboratory strain of S. enterica serovar Typhimurium used might be a unique variant or mutant, the above experiments were repeated with several additional isolates, including another source of ATCC 14028, and two S. enterica serovar Typhimurium clinical isolates, NI-1 and NI-101. Competitions between each of these isolates and a Selfish E. coli (6925Self or 5240Self) yielded results comparable to those already described with our original laboratory source of S. enterica serovar Typhimurium ATCC 14028 (figure S2 in the electronic supplementary material).
From the control competitions with plasmid-free, ampicillin-sensitive E. coli, it was clear that survival of otherwise sensitive S. enterica serovar Typhimurium cells required the presence of a β-lactamase-producing E. coli strain. Neither sensitive E. coli, nor a β-lactamase-negative, ampicillin-resistant E. aerogenes strain (figure 2a) yielded S. enterica serovar Typhimurium survivors, which were otherwise sensitive to ampicillin in competition experiments. To examine whether such cell production was dependent on the genetic/physical location of the bla gene in the resistant E. coli, competitions were conducted between an E. coli strain, Wyl, in which the bla gene was integrated via recombination into the chromosome (Chait et al. 2007). As seen in figure 1, Salmonella cells that were otherwise sensitive to ampicillin also survived when co-cultivated with this strain, albeit at significantly less proportions (p < 0.01) than with the plasmid-borne bla in KL99Alt, KL99Self or 6925Self. Moreover, in each case, when no ampicillin was present, the S. enterica ATCC 14028 strain grew to over 80 per cent of the mixed population within 24 h (figure 1d).
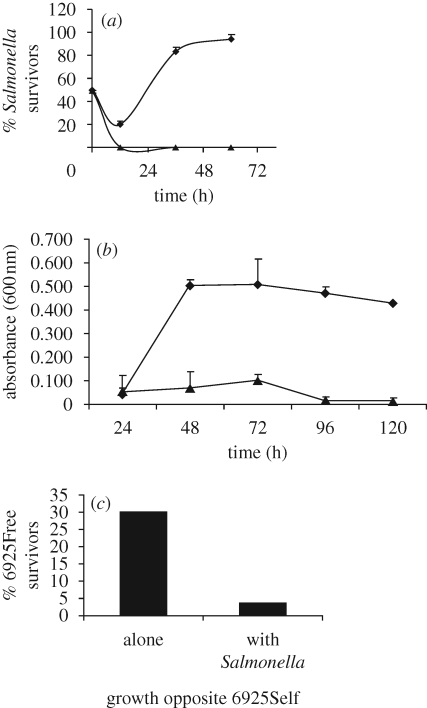
Figure 2.
No physical contact is required for the survival of Salmonella when co-cultivated with ampicillin-resistant E. coli. Salmonella enterica serovar Typhimurium ATCC 14028 was grown in competition with 6925Self or with E. aerogenes ATCC 13048 in the presence of 100 µg ampicillin per millilitre DMM. Presented are the results of three independent replicates with (a) the mean per cent of Salmonella colony forming units per millilitre on XI plates shown or (b) the absorbance at 600 nm of the Salmonella side of dual-flasks. Those samples whose cfu ml−1 were less than 2.5 × 106 are represented as 0 in this graph. Standard error bars are shown. (a) Strains were inoculated into the same side of a dual-flask apparatus; v6925Self, competition of Salmonella against E. coli 6925Self; vEntero, competition of Salmonella against E. aerogenes ATCC 13048. Diamonds, Salmonella v6925Self; triangles, Salmonella vEntero. (b) Strains were inoculated into opposite sides of a dual-flask apparatus. vEntero, competition with one flask containing E. aerogenes and the other side of dual-flask apparatus containing S. enterica serovar Typhimurium ATCC 14028. Diamonds, Salmonella v6925Self; triangles, Salmonella vEntero. (c) 6925Free (plasmid-free, ampicillin-sensitive E. coli) was inoculated into the opposite side of a dual-flask apparatus either: alone, opposite 6925Self and S. enterica serovar Typhimurium ATCC 14028; or, together ‘with Salmonella’ strain 14028, while Selfish E. coli 6925Self was inoculated into the opposite side of the dual-flask set-up.
(e). Physical contact between ampicillin-resistant E. coli and susceptible Salmonella is not required for protection
As a preliminary assessment of additional requirements for S. enterica serovar Typhimurium survival, competitions between ampicillin-resistant E. coli and susceptible S. enterica serovar Typhimurium isolates were conducted, with the competitors inoculated into opposite sides of a dual-flask competition apparatus that allowed exchange of low-molecular weight compounds, including proteins and enzymes, but prevented physical contact of cells on the opposite sides of a membrane (Clark et al. 2009). We determined that neither bacterial strain was able to cross the membrane, and most common coliphages also fail to cross such membranes (Rapp et al. 1992). Although we could not rule out smaller phages crossing the membrane, in preliminary phage titre assays, we did not observe evidence that lytic phage capable of producing plaques were present in any of the cultures (data not shown).
When S. enterica serovar Typhimurium strains were competed without contact against KL99Self or 6925Self in the presence of ampicillin, S. enterica serovar Typhimurium cells grew at proportions comparable to what was observed when the respective strains were competed together in the same flask (and fresh media was exchanged at 12 h intervals). Thus, S. enterica serovar Typhimurium growth does not depend on direct contact with the resistant E. coli (figure 2b).
We additionally sought to determine whether the interaction between resistant E. coli strains and ampicillin-susceptible S. enterica serovar Typhimurium isolates could benefit sensitive E. coli too, resulting in survival of E. coli cells that were otherwise sensitive to ampicillin. In ampicillin, when E. coli 6925Self was grown on the same side of the dual-flask set-up with S. enterica serovar Typhimurium 14028, plasmid-free E. coli 6925 cells survived when inoculated into the opposite side of the set-up, containing the same concentration of antibiotic (figure 2c). After 48 h, E. coli 6925 cells grew when plated onto XI agar (at frequencies about 30% of those seen for S. enterica serovar Typhimurium 14028 (figure 2c); no E. coli 6925 colonies appeared on AXI). When the plasmid-free E. coli 6925 and S. enterica serovar Typhimurium 14028 were inoculated onto the same side of the dual-flask apparatus and 6925Self was inoculated alone into the opposite side (figure 2c), the number of plasmid-free E. coli 6925 survivors only reached about one-seventh the level observed in the previous experiment (i.e. when the plasmid-free E. coli 6925 was in the opposite side of the set-up and E. coli 6925Self grew together with S. enterica serovar Typhimurium 14028).
(f). Co-cultivation of ampicillin-resistant E. coli with susceptible Salmonella leads to free β-lactamase in the medium
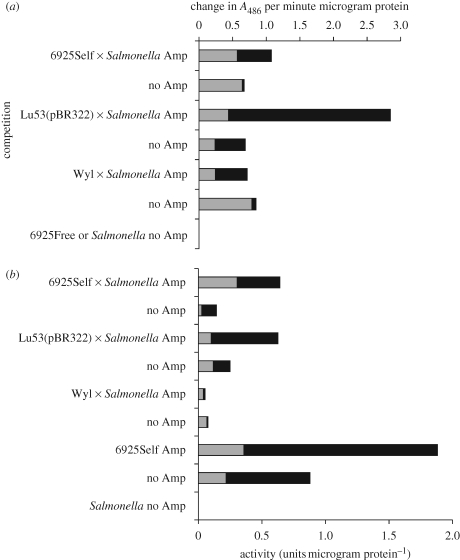
Levels of β-lactamase were measured in medium collected from E. coli and S. enterica serovar Typhimurium cells grown in competitions. Levels were determined for the spent media and for sonic extracts of collected cells, for strains grown individually, strains grown in the same flasks for competitions and strains grown on opposite sides of flasks in the dual-flask set-up. β-Lactamase activity was found in supernatants of ampicillin-resistant E. coli grown with S. enterica. Such activity was also observed on the S. enterica side of the dual-flask apparatus. No β-lactamase activity was detected in supernatants where ampicillin-sensitive E. coli or S. enterica serovar Typhimurium had grown alone. In the absence of ampicillin, for the most part, a larger proportion of β-lactamase activity was found in the supernatant fraction of cultures. On the other hand, in most cases the total levels of β-lactamase activity were higher when ampicillin-resistant E. coli were cultured together with S. enterica in the presence of ampicillin (versus the same comparison in the absence of ampicillin; figure 3). At the same time, the larger proportion of total activity was predominantly in sonic extracts of cells. This may, in part, reflect selection for higher proportions of β-lactamase-producing E. coli in such cultures (due to the presence of ampicillin). As an indicator of cell lysis, we also assayed for activity of a large cytoplasmic enzyme, β-galactosidase. As seen in figure 3b, although the bulk of the enzyme activity remained within the cells, in some cases this enzyme was released at substantial proportions, indicating cell lysis. In the case of the Wyl strain in competitions with S. enterica, the majority of the β-galactosidase was found in the supernatant.
Figure 3.
β-Lactamase present in competitions is found in both pelleted cells and in the collected growth medium. Competitions (50∶50 initial starting proportions of strains or with each strain alone) were conducted at 100 µg ampicillin per millilitre DMM liquid medium or without antibiotic. The values presented here represent samples collected after 48 h growth. (a) β-Lactamase activity expressed as absorbance at 486 nm (A486) per minute per microgram of protein. (b) Activity of β-galactosidase. Salmonella enterica serovar Typhimurium ATCC 14028 is unable to produce this enzyme, hence the absence of activity from it and the reduction in specific activity when this organism dominates the cultures. Dark bars—supernatant, filter-sterilized supernatant from competition; light bars—sonic extract, sonic extract of collected cell pellet from competition.
(g). Media recovered from competitions does not inhibit the growth of newly inoculated strains
Growth media from competitions and from strains grown alone were recovered by centrifugation and subsequent filtration through 0.2 µm filters for sterilization. In this way we sought to characterize properties of the respective media vis-à-vis effects on growth of strains newly inoculated therein. Little or no growth was observed for the E. coli strains 6925 and 6925Self or S. enterica serovar Typhimurium ATCC 14028 when inoculated into any of these media. However, when additional nutrients (final concentrations of dextrose (0.006%), MgSO4 (1 mM), thiamine (0.0005%), Casamino acids (0.004%) and IPTG (0.16 mM)) were added, each of these strains grew in the media (see figure S3 and table S3 in the electronic supplementary material). If, in addition to nutrients, ampicillin was added to a predicted final concentration (ignoring any residual ampicillin already present in the supernatants) of 100 µg ml−1, the growth rates and final cell densities of each culture depended on the source of the supernatant. If obtained from cultures originally containing ampicillin-resistant E. coli, growth was similar to what was observed without the addition of drug, not only for the ampicillin-resistant E. coli and sensitive S. enterica serovar Typhimurium ATCC 14028, but even for the ampicillin-sensitive strains such as E. coli 6925 and S. flexneri ATCC 12022 (figure S3 in the electronic supplementary material). If the supernatant was originally obtained from cultures of S. enterica serovar Typhimurium ATCC 14028 grown alone, then addition of nutrients and ampicillin only allowed the growth of the ampicillin-resistant 6925Self, while the other ampicillin-sensitive strains (plasmid-free 6925 and S. enterica serovar Typhimurium ATCC 14028, or S. flexneri ATCC 12022) failed to grow (figure S3, top panel in the electronic supplementary material).
4. Discussion
In this report, we have extended the previous studies that used a bacterial model for the production of public goods, leading to survival of otherwise antibiotic-susceptible cohorts (Dugatkin et al. 2005a; Clark et al. 2009). Here, we sought to explore the possibility that E. coli engineered to release β-lactamase could lead to the survival of otherwise ampicillin-sensitive cells in a setting that more realistically models a natural setting, where they normally have to compete not only with other E. coli, but with a wide variety of gut flora. Among these are organisms that cause gastrointestinal disease. Salmonella enterica serovar Typhimurium is known to cause gastrointestinal disease and this requires that cells of this species establish themselves in an environment already colonized with other bacteria. Salmonella enterica serovar Typhimurium are apparently highly effective at such colonization (Esteves et al. 2005). What is more, the would-be pathogens are more effective in binding to epithelial cells, such as those that line the gut and they can displace the E. coli that have already attached (Esteves et al. 2005). Thus, when introduced into the gut ecosystem, S. enterica serovar Typhimurium would appear to be somewhat at an advantage in competitions with E. coli. On the other hand, antibiotic-susceptible S. enterica serovar Typhimurium (and other species) should be selected against and eliminated in the presence of sufficiently high levels of drug, such as ampicillin, while ampicillin-resistant cells should survive and have increased fitness.
In the current experiments, ampicillin-resistant E. coli survived, but so did otherwise ampicillin-susceptible S. enterica serovar Typhimurium. These latter cells did not acquire genetically based ampicillin-resistance but rather, remaining sensitive, they survived the conditions of growth. Not only did these cells survive, but they actually thrived under these conditions. When ampicillin was present, an equilibrium developed, whereby the sensitive S. enterica grew to large proportions of the culture, but failed to exclude their ampicillin-resistant E. coli benefactors. This equilibrium took time to establish itself. The S. enterica serovar Typhimurium survived when grown with E. coli that had been engineered to export β-lactamase to the outer membrane; in contrast, ampicillin-sensitive S. flexneri were unable to survive under similar conditions. This is probably due to a much greater intrinsic susceptibility to ampicillin of the S. flexneri strains used in this study compared with either the S. enterica serovar Typhimurium or the ampicillin-sensitive E. coli (table S2 in the electronic supplementary material). On the other hand, apparently the sole requirement for S. enterica serovar Typhimurium survivors was that their E. coli benefactor must themselves produce β-lactamase, which gets released to higher levels when E. coli grow in the presence of the S. enterica serovar Typhimurium. This is clearly an example where an extracellular product or public good can fortuitously benefit others (Sheratt et al. 2009). The results of the present study are of a kind, up to a point, with a variety of reports where resistance to antibiotics is observed but is not heritable (for an overview, see Levin 2004; also, see Barclay et al. 1992; Gradelski et al. 2001). However, a difference here is the added interaction between antibiotic-resistant cells of one species and the sensitive beneficiaries of another. When not constrained by ampicillin, the aggressive growth of the S. enterica serovar Typhimurium ATCC 14028 was compatible with the findings of Esteves et al. (2005), who found that S. enterica had a higher fitness (as measured by overall cell densities achieved) than E. coli when these species were grown together. However, the findings of Esteves et al. (2005) do not provide the mechanism exploited by S. enterica serovar Typhimurium ATCC 14028 to survive when inoculated into lethal ampicillin concentrations.
One possible explanation for the survival of S. enterica serovar Typhimurium ATCC 14028 in the presence of ampicillin in our study is the transient expression in this organism of an otherwise cryptic ampicillin-resistance gene. However, if this is the case, it is only effective when ampicillin-resistant E. coli are also present.
It is also possible that S. enterica serovar Typhimurium produces a substance or entity that increases leakiness of E. coli. The media spent from cultures that contained S. enterica serovar Typhimurium did not seem to interfere with the growth of E. coli. Thus, at present, we have no data that indicate either the presence of a phage or other products released by the S. enterica serovar Typhimurium, which inhibit the growth of E. coli or likely increase its lysis.
For competitions in ampicillin-containing media, the observation that within the first 12–24 h, S. enterica numbers drop from 50 per cent to nearly 0 per cent (data from figures S1 and S2 in the electronic supplementary material) and then rebound to levels of 60–90%, is significant. It suggests that after 12–24 h of competition with bla-containing E. coli, there has been a change in the environment experienced by the competing bacteria that allows cells to survive and grow, even as ampicillin is replenished. Further, the growth experiments with media recovered from competitions suggest that, by 24 h, survival of cells in ampicillin is no longer dependent on the frequency of β-lactamase producers. Thus, a simpler explanation for the observed survival of S. enterica serovar Typhimurium may be related to the release and/or production of β-lactamase by the resistant E. coli.
We observed that β-lactamase-producing E. coli had greater net amounts of enzyme in ampicillin-containing media and greater total enzyme activity was retained within the cells (figure 3). This observation may be due to increased selection for cells that produce the enzyme in sufficiently high amounts to survive under these conditions. Still, large quantities of β-lactamase were also detected in the supernatants from competitions of such strains with S. enterica. The detection of activity for a large cytoplasmic enzyme (i.e. β-galactosidase) in such supernatants suggests that some cell lysis has occurred under these conditions. At the same time, co-culturing with S. enterica serovar Typhimurium was associated with increased β-lactamase levels and/or leakiness of the ampicillin-resistant E. coli cells in the presence of ampicillin. It is known that a variety of growing Gram-negative bacteria release membrane blebs (Katsui et al. 1982; Kadurugamuwa & Beveridge 1997; Li et al. 1998; Beveridge 1999; Post et al. 2005). Such blebs are associated with abnormal in-growth of the outer membrane, and they typically appear around the septa during cell division. They often contain phospholipids and a range of periplasmic enzymes, including β-lactamase (Ciofu et al. 2000), which are eventually further released into the medium (Kadurugamuwa & Beveridge 1997; Li et al. 1998). Moreover, release of blebs increases under conditions of stress, such as antibiotic exposure, serum containing complement or nutrient deprivation (Knox et al. 1966; MacDonald & Beveridge 2002). Perhaps, the individual and combined stresses of nutrient deprivation, antibiotic exposure and competition with a robust and vigorously growing competitor (i.e. S. enterica serovar Typhimurium) increasingly lead β-lactamase-producing E. coli to substantially release their enzymes into the media, to the benefit of ampicillin-sensitive cells.
It has recently been shown that antibiotic treatment has dramatic effects on the composition and diversity of the gut microbiota (Dethlefsen et al. 2008). Moreover, despite the apparent resilience of the gut community, long-lasting changes in composition often do occur (Dethlefsen et al. 2008). The results presented here suggest that the production of substances that break down the antibiotics in one species of gut microbiota (e.g. E. coli) has implications not just for that species, but other species in the community in which it resides. The studies described here are significant, for they show that the β-lactamase produced by strains of E. coli may be associated with maintenance of diversity of bacteria microbiota communities, the evolution of antibiotic resistance, and survival of potential pathogens in otherwise lethal concentrations of drug. It has already been demonstrated that ‘cheats’ may exploit situations where wild-type cells cooperate to enhance virulence in clinical settings (Köhler et al. 2009). Our results demonstrate clearly that non-pathogenic antibiotic-resistant bacteria may protect otherwise susceptible pathogens by a mechanism that does not involve gene transfer. Evolution of such transient survival mechanisms may thereby hinder therapeutic use of antibiotics, and should be considered in devising effective treatment strategies.
Acknowledgements
The authors are deeply indebted to Susanna Remold for extensive review of the manuscript and suggestions for improved presentation. We thank Micah Worley for helpful comments, and Stan Malloy for suggestions regarding possible mechanisms to account for our observations. We also thank the Associate Editor, Dr Lars Raberg, and two anonymous reviewers supplied by the journal, whose comments and suggestions led to an improved version of the manuscript. The assistance of Jessica Stivers and Destinee Woodcox in conducting the competitions is greatly appreciated. We wish to thank the E. coli Stock Center at Yale University for providing the background strains used in this study, Roy Kishony at Harvard University Medical School for E. coli strain Wyl, and the laboratory of Micah Worley at the University of Louisville for providing S. enterica serovar Typhimurium strain 14028, and clinical isolates NI-1 and NI101. This work was supported, in part, by AREA grant 1 R15 AI060667-01A1 from the National Institutes of Health and by URG and URS grants from the Office of the Vice President for Research at the University of Louisville.
M.H.P., L.A.D., R.M.A., D.R.C. and C.P. designed the research; D.R.C., C.M., H.P., N.J., C.K., C.P., A.B. and M.H.P. performed the research; M.H.P., D.R.C., R.M.A., D.A.M. and L.A.D. analysed the data; and M.H.P, L.A.D. and R.M.A. wrote the paper. The authors declare no conflict of interest.
References
- Ahmad A. S., Rahman N., Islam F.2004Spectrophotometric determination of ampicillin, amoxycillin, and carbenicillin using Folin-Ciocalteu phenol reagent. J. Anal. Chem. 59, 119–123 (doi:10.1023/B:JANC.0000014736.59554.5c) [Google Scholar]
- Arrach N., Porwollik S., Cheng P., Cho A., Long F., Choi S. H., McClelland M.2008Salmonella serovar identification using PCR-based detection of gene presence and absence. J. Clin. Microbiol. 46, 2581–2589 (doi:10.1128/JCM.02147-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay M. L., Begg E. J., Chambers S. T.1992Adaptive resistance following single doses of gentimicin in a dynamic in vitro model. Antimicrob. Agents Chemother. 36, 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge T. J.1999Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heynecker H. L., Boyer H. W.1977Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2, 95–113 [PubMed] [Google Scholar]
- Bradford M. M.1976A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (doi:10.1016/0003-2697(76)90527-3) [DOI] [PubMed] [Google Scholar]
- Brockhurst M. A., Buckling A., Racey D., Gardner A.2008Resource supply and the evolution of public-goods cooperation in bacteria. BMC Biol. 14, 20 (doi:10.1186/1741-7007-6-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chait R., Craney A., Kishony R.2007Antibiotic interactions that select against resistance. Nature 446, 668–671 (doi:10.1038/nature05685) [DOI] [PubMed] [Google Scholar]
- Ciofu O., Beveridge T. J., Kadurugamuwa J., Walther-Rasmussen J., Hoiby N.2000Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45, 9–13 (doi:10.1093/jac/45.1.9) [DOI] [PubMed] [Google Scholar]
- Clark D. R., Alton T. M., Bajorek A., Holden P., Dugatkin L. A., Atlas R. M., Perlin M. H.2009Evolution of altruists and cheaters in near-isogenic populations of Escherichia coli. Front. BioSci. 14, 4815–4824 [DOI] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M., Relman D. A.2008The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (doi:10.1371/journal.pbio.0060280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W.2002Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 67–193 (doi:10.1128/CMR.15.2.167-193.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugatkin L., Perlin M., Atlas R. M.2003The evolution of group beneficial traits in the absence of between-group selection: a model. J. Theoret. Biol. 220, 67–74 (doi:10.1006/jtbi.2003.3149) [DOI] [PubMed] [Google Scholar]
- Dugatkin L., Perlin M., Lucas J. S., Atlas R. M.2005aGroup-beneficial traits and genotypic diversity: an antibiotic resistance paradigm. Proc. R. Soc. B 272, 79–83 (doi:10.1098/rspb.2004.2916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugatkin L., Perlin M., Atlas R. M.2005bAntibiotic resistance and the evolution of group-beneficial traits. II: A metapopulation model. J. Theoret. Biol. 236, 392–396 (doi:10.1016/j.jtbi.2005.03.021) [DOI] [PubMed] [Google Scholar]
- Dugatkin L. A., Dugatkin A. D., Atlas R. M., Perlin M. H.2008Cheating on the edge. PLoS One 3, e2763 (doi:10.1371/journal.pone.0002763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves C. L. C., Jones B. D., Clegg S.2005Biofilm formation by Salmonella enterica serovar Typhimurium and Escherichia coli on epithelial cells following mixed inoculations. Infect. Immun. 7, 5198–5203 (doi:10.1128/IAI.73.8.5198-5203.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J.1983Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 47, 361–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradelski E., Valera L., Kolek B., Bonner D., Fung-Tomc J.2001Comparative killing kinetics of the novel des-fluoro(6) quinolone BMS-284756, fluoroquinolones, vancomycin and beta-lactams. Int. J. Antimicrob. Agents 18, 43–48 (doi:10.1016/S0924-8579(01)00343-0) [DOI] [PubMed] [Google Scholar]
- Griffin A. S., West S. A., Buckling A.2004Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 (doi:10.1038/nature02744) [DOI] [PubMed] [Google Scholar]
- Hastings P. J., Rosenberg S. M., Slack A.2004Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12, 401–404 (doi:10.1016/j.tim.2004.07.003) [DOI] [PubMed] [Google Scholar]
- Hedberg M., Lindqvist L., Bergman T., Nord C. E.1995Purification and characterization of a new beta-lactamase from Bacteroides uniformis. Antimicrob. Agents Chemother. 39, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa J. L., Beveridge T. J.1997Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40, 615–621 (doi:10.1093/jac/40.5.615) [DOI] [PubMed] [Google Scholar]
- Karthikeyan S., Korber D. R., Wolfaardt G. M., Caldwell D. E.2001Adaptation of bacterial communities to environmental transitions from labile to refractory substrates. Int. Microbiol. 4, 73–80 [DOI] [PubMed] [Google Scholar]
- Katsui N., Tsuchido T., Hiramatsu R., Fujikawa S., Takano M., Shibasaki I.1982Heat-induced blebbing and vesiculation of the outer membrane of Escherichia coli. J. Bacteriol. 151, 1523–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K. W., Vesk M., Work E.1966Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92, 1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Buckling A., van Delden C.2009Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl Acad. Aci USA 106, 6339–6344 (doi:10.1073/pnas.0811741106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski L. E., Hattingh S. E.1986Coexistence of two competitors on one resource and one inhibitor: a chemostat model based on bacteria and antibiotics. J. Theoret. Biol. 122, 83–93 (doi:10.1016/S0022-5193(86)80226-0) [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Simpson S. C., Nguyen T. T.1994Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176, 3140–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B.2004Noninherited resistance to antibiotics. Science 305, 1578–1579 (doi:10.1126/science.1103077) [DOI] [PubMed] [Google Scholar]
- Levin B. R., Lipsitch M., Perrot V., Schrag S., Antia R., Simonsen L., Walker N. M., Stewart F. M.1997The population genetics of antibiotic resistance. Clin. Infect. Dis. 24(Suppl. 1), S9–S16 [DOI] [PubMed] [Google Scholar]
- Li Z., Clarke A. J., Beveridge T. J.1998Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180, 5478–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M.1997β-Lactamases: quantity and resistance. Clin. Microbiol. Infect. 4(Suppl.), S10–S19 [PubMed] [Google Scholar]
- Lynch A. S., Robertson G. T.2008Bacterial and fungal biofilm infections. Annu. Rev. Med. 59, 415–428 (doi:10.1146/annurev.med.59.110106.132000) [DOI] [PubMed] [Google Scholar]
- MacDonald K. L., Beveridge T. J.2002Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can. J. Microbiol. 48, 810–820 (doi:10.1139/w02-077) [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Baquero F., Andersson D. I.2007Predicting antibiotic resistance. Nat. Rev. Microbiol. 5, 958–965 (doi:10.1038/nrmicro1796) [DOI] [PubMed] [Google Scholar]
- Merlino J., Watson J., Rose B., Beard-Pegler M., Gottlieb T., Bradbury R., Harbour C.2002Detection and expression of methicillin/oxacillin resistance in multidrug-resistant and non-multidrug-resistant Staphylococcus aureus in Central Sydney, Australia. J. Antimicrob. Chemother. 49, 793–801 (doi:10.1093/jac/dkf021) [DOI] [PubMed] [Google Scholar]
- Paterson D. L., Bonomo R. A.2005Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686 (doi:10.1128/CMR.18.4.657-686.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post D. M. B., Zhang D., Eastvold J. S., Teghanemt A., Gibson B. W., Weiss J. P.2005Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J. Biol. Chem. 280, 38383–38394 (doi:10.1074/jbc.M508063200) [DOI] [PubMed] [Google Scholar]
- Rapp M. L., Thiel T., Arrowsmith R. J.1992Model system using coliphage φx174 for testing virus removal by air filters. Appl. Environ. Microbiol. 58, 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. B.2001Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7, 183–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosensweig R. F., Sharp R. R., Treves D. S., Adams J.1994Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137, 903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack R. B., Khan E. H.1997Antimicrobial resistance in organisms causing diarrheal disease. Clin. Infect. Dis. 24(Suppl. 1), S102–S105 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.2001Molecular cloning: a laboratory manual, pp. 2344, 3rd edn Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Samuelson P. A.1954The pure theory of public expenditure. Rev. Econ. Stat. 36, 387–389 (doi:10.2307/1925895) [Google Scholar]
- Sheratt T. N., Roberts G., Kassen R.2009Evolutionary stable investment in products that confer both and individual benefit and public good. Front. BioSci. 14, 340–347 [DOI] [PubMed] [Google Scholar]
- Smith J.2001The social evolution of bacterial pathogenesis. Proc. R. Soc. Lond. B 268, 61–69 (doi:10.1098/rspb.2000.1330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J. A., Villegas M. V., Quinn J. P.2007Current concepts in antibiotic-resistant gram-negative bacteria. Expert Rev. Anti-Infect. Ther. 5, 833–843 (doi:10.1586/14787210.5.5.833) [DOI] [PubMed] [Google Scholar]
- Travisano M.2001Experimental evolution studies yield insights into bacterial diversity. ASM News 67, 403–409 [Google Scholar]
- Treves D. S., Manning S., Adams J.1998Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 15, 789–797 [DOI] [PubMed] [Google Scholar]