Abstract
It has often been suggested that the genome sizes of birds are constrained relative to other tetrapods owing to the high metabolic demands of powered flight and the link between nuclear DNA content and red blood cell size. This hypothesis predicts that hummingbirds, which engage in energy-intensive hovering flight, will display especially constrained genomes even relative to other birds. We report genome size measurements for 37 species of hummingbirds that confirm this prediction. Our results suggest that genome size was reduced before the divergence of extant hummingbird lineages, and that only minimal additional reduction occurred during hummingbird diversification. Unlike in some other avian taxa, the small amount of variation observed within hummingbirds is not explained by variation in respiratory and flight-related parameters. Unexpectedly, genome size appears to have increased in four unrelated hummingbird species whose distributions are centred on humid forests of the upper-tropical elevational zone on the eastern slope of the Andes. This suggests that the secondary expansion of the genome may have been mediated by biogeographical and demographic effects.
Keywords: Apodiformes, cell size, C-value, genome size, Trochilidae
1. Introduction
It has long been recognized that genome size, nucleus size and red blood cell size are positively correlated among vertebrates (Gregory 2001). Given that larger cells exhibit lower surface area to volume ratios and are therefore less efficient for gas exchange, it has been argued that groups with high metabolic demands are constrained to small cells, and therefore possess small genomes (e.g. Szarski 1983). Not surprisingly, vertebrates exhibiting metabolically intense powered flight (i.e. birds and bats) have been hypothesized to be particularly constrained in this regard (Hughes & Hughes 1995). Thus, birds possess smaller average genomes than any other tetrapod group (Andrews et al. 2009), and bats display small genomes relative to most other mammals (Smith & Gregory 2009).
Broad comparisons have revealed inverse correlations between genome size and resting metabolic rate in both birds and mammals (Vinogradov 1995; Gregory 2002), and more recently genome size has been found to correlate negatively with heart index in birds generally (Vinogradov & Anatskaya 2006) and positively with wing loading within the avian order Passeriformes (Andrews et al. 2009). Importantly, it has recently been shown that saurischian dinosaurs (from which birds are derived) had already undergone an initial reduction in genome size prior to the evolution of flight (Organ et al. 2007), though their estimated genome sizes were larger than those of most modern birds. Moreover, genome sizes have been suggested to assort according to flight ability in extant birds, with strong fliers possessing the smallest genomes and flightless birds some of the largest (Hughes 1999; Gregory 2005). In other words, genome reductions appear to have both preceded the origin of flight and become more pronounced in groups that evolved strong flight.
Hummingbirds (order Apodiformes, family Trochilidae) are widely recognized as exhibiting mass-specific metabolic rates of aerobic metabolism that approach theoretical limits for vertebrates (approx. 40 ml O2 g−1 h−1 during stationary hovering flight) (Bartholomew & Lighton 1986). Hummingbirds correspondingly possess the highest relative heart volumes, lung volumes, mitochondrial volume densities, mitochondrial respiration rates and capillary volume densities compared with any vertebrates (Suarez et al. 1991; Suarez 1992).
The challenge of maintaining metabolically intense hovering flight is especially acute for hummingbirds living at high elevations (up to 5000 m) owing to the reduced oxygen availability and increased difficulty of generating lift in air of lower density. However, it has been shown that oxygen delivery in particular, rather than air density, is the primary limiting factor in hummingbird flight at high elevations (Altshuler & Dudley 2006). Thus, it is not surprising that these birds possess very small erythrocytes that can both pass through small, numerous capillaries and exchange respiratory gases efficiently owing to a high surface area to volume ratio (Hartman & Lessler 1963; Opazo et al. 2005).
Flight metabolism, body size and red blood cell size considerations suggest that hummingbirds can be expected to have particularly small genomes. However, despite their obvious relevance to understanding avian genome size evolution, no hummingbird genome size estimates are currently available. Here we rectify this major gap in the bird dataset by providing genome size estimates for 37 species representing eight of the nine major clades of extant hummingbirds (McGuire et al. 2009), along with original data on nucleus size, cell size, cardiac mass, haematocrit (Hct), haemoglobin concentration, body mass, wing loading and elevation, all analysed within a phylogenetic context.
2. Material and methods
(a). Haematology and genome size
Blood was drawn from the brachial vein into heparinized capillary tubes for preparation of blood smears and centrifugation for measurement of Hct. Haemoglobin concentration was measured using the Haemocue system with a correction for avian blood (Simmons & Lill 2006). A 200 : 1 dilution of whole blood in saline was prepared for the quantification of red blood cell concentration (RBC) using a haemocytometer. Mean corpuscular volume (MCV) was calculated as Hct/RBC. Dry nuclear and erythrocyte areas were measured by computer image analysis following Wright staining. Genome size and haematological data are provided in electronic supplementary material, appendix 1.
Genome size was estimated by Feulgen image analysis densitometry using the protocol described in detail by Hardie et al. (2002), using chicken (Gallus gallus, 1.25 pg) as the standard. Air-dried blood smears were post-fixed overnight in 85 methanol : 10 formalin : 5 glacial acetic acid, rinsed in tepid tap water and hydrolyzed for 120 min in 5 N HCl at room temperature before being stained for 120 min in freshly prepared Schiff reagent and passed through a series of bisulphite and distilled water rinses. Integrated optical densities were measured for at least 200 nuclei per individual using the Bioquant Life Science software package and an Optronics DEI-750 CE three-chip CCD camera mounted on a Leica DM LS microscope with a 100× lens.
(b). Morphology and distribution
Birds were weighed in the field with a digital scale. Wing area was quantified using digital photographs of the wings spread on a graph paper, with the leading edge of the wings held parallel. Wing loading was calculated as body mass (g) over wing area (mm2). Morphological data are provided in electronic supplementary material, appendix 1. Voucher specimens were prepared and deposited along with associated frozen tissues at the Museum of Southwestern Biology (University of New Mexico, USA) and Centro de Ornitología y Biodiversidad (Lima, Peru). All specimen data are available online at http://arctos.database.museum/SpecimenSearch.cfm, and are searchable using the catalogue numbers listed in electronic supplementary material, appendix 2. Distribution size data for each species were quantified using species range polygons developed by Ridgely et al. (2003).
(c). Phylogenetic and comparative analyses
The phylogeny with branch lengths for the 37 hummingbird species and one outgroup is a majority-rule consensus tree from a partitioned Bayesian analysis of two mitochondrial and two nuclear genes (electronic supplementary material, appendix 3). Details of laboratory methods, four loci, nine process partitions and Bayesian phylogenetic methods are contained in McGuire et al. (2007). Tissue samples and GenBank accession numbers for all 38 taxa are listed (electronic supplementary material, appendix 2). Non-synonymous to synonymous ratios (ω) were calculated for each branch of the tree using the branch model of the codeml module of PAML (Yang 2007). Genome sizes for each species were analysed against ω for the corresponding terminal branch.
For comparisons of genome size and other species characteristics, Felsenstein's (1985) phylogenetically independent contrasts (PICs) were calculated using log-transformed data in the PDAP module of Mesquite v. 2.5 (Midford et al. 2003; Maddison & Maddison 2008). One degree of freedom was subtracted for each branch in the single polytomy (Purvis & Garland 1993; Garland & Díaz-Uriarte 1999). Mass-corrected PICs were conducted in Mesquite by first performing independent contrast correlations between parameter 1 (e.g. wing loading) versus body mass and saving the residuals, repeating for parameter 2 (e.g. genome size) versus body mass, and then analysing these residuals together by Pearson correlations forced through the origin. Results of correlation analyses are provided in electronic supplementary material, appendix 4. Squared change parsimony was used to reconstruct ancestral genome sizes.
3. Genome sizes of hummingbirds
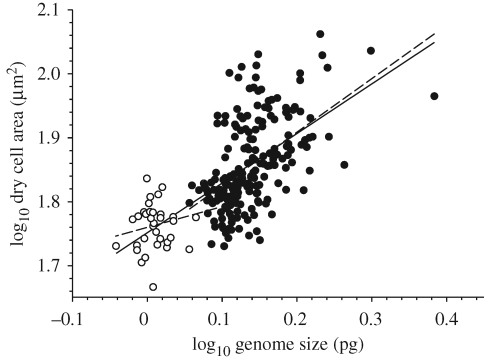
Our results indicate that the genome sizes of hummingbirds are, as predicted, constrained even relative to other amniotes (electronic supplementary material, appendix 1). In the present dataset, these ranged from 0.91 pg in the black-chinned hummingbird (Archilochus alexandri) to 1.29 pg in the wedge-billed hummingbird (Schistes geoffroyi) and averaged only 1.03 pg ± 0.01 s.e. (note that 1 pg = 978 Mb). By contrast, the average for other birds as a whole is 1.42 pg ± 0.01 s.e.; whereas for non-avian reptiles it is 2.24 pg ± 0.04 s.e. and for mammals it is 3.37 pg ± 0.04 s.e. (Gregory 2009). Despite the very limited diversity in genome size among hummingbirds, and following correction for phylogenetic non-independence of species data, genome size was positively correlated with dry nucleus area (r = 0.60, p = 0.0001), dry erythrocyte area (r = 0.33, p < 0.05) and MCV (r = 0.55, p = 0.0004) in this group. The relative sizes of hummingbird genomes and cells as compared with other birds are shown in figure 1.
Figure 1.
The relationship between genome size and red blood cell size (dry area) in hummingbirds (open circles) and other birds (closed circles). Hummingbirds display not only the smallest genomes among birds, but also the smallest erythrocytes. The relationship between these parameters is significant across all birds (solid line), among non-hummingbirds (short-dashed line) and within hummingbirds (long-dashed line).
Neither genome size nor MCV was significantly correlated with body size, Hct or haemoglobin concentration (all p > 0.17). MCV was unrelated to either absolute or relative heart mass (all p > 0.18), whereas genome size was positively correlated with absolute (r = 0.41, p < 0.02), but not relative (p > 0.2), heart mass. Unlike in perching birds (Andrews et al. 2009), genome size was not significantly related to wing loading in hummingbirds (p > 0.44). Hummingbirds are of particular interest in this regard because wing loading is also unrelated to body mass in this group (p > 0.99); this is explained by an unusually large increase in wing area with increasing body mass (r = 0.92, p < 0.0001). Hummingbird wing loading varies in accordance with altitude and in terms of a trade-off between flight efficiency and aerial maneuverability (Altshuler & Dudley 2002). As such, wing loading may not reflect the underlying interspecific variation in metabolic activity in hovering birds as it does in birds with other styles of flight.
It is clear that hummingbirds possess the smallest and least variable genome sizes of any bird family studied thus far. However, two important questions remain regarding the limited amount of diversity that does exist within this group: (i) Are the small genome sizes of hummingbirds derived or ancestral features? (ii) Why do a few hummingbirds have larger genomes than others?
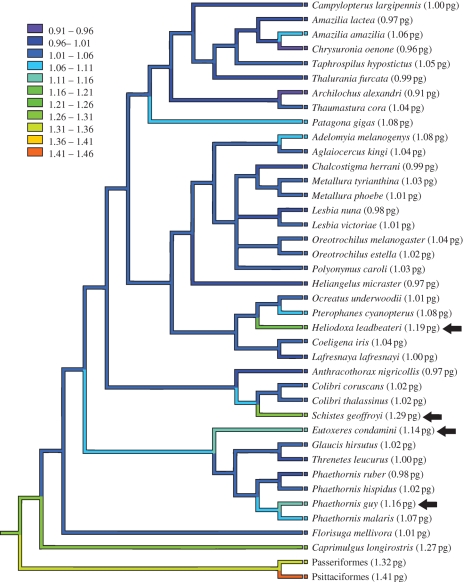
The lineage leading to hummingbirds diverged successively from the nightjars (Caprimulgidae), the owlet-nightjars (Aegothelidae) and the swifts (Apodidae and Hemiprocnidae) between the Late Cretaceous and Eocene, although precise divergence date estimates remain contentious (Ericson et al. 2006; Brown et al. 2007, 2008; Hackett et al. 2008). We estimated the genome size of the band-winged nightjar (Caprimulgidae: Caprimulgus longirostris) to be 1.27 pg, which is larger than all but one of the hummingbird genome sizes assessed. Overall, it is clear that a marked reduction in hummingbird genome size occurred after the divergence of hummingbirds and nightjars, but before the most recent common ancestor of extant hummingbirds (figure 2). We hypothesize that hovering flight and reduced genome size evolved in concert early in the hummingbird lineage. Further genome size estimates for owlet-nightjars and swifts and cell size estimates for fossil Apodiformes will bear on this hypothesis (Mayr 2004; Organ et al. 2007; Organ & Shedlock 2009), but remain unavailable at present.
Figure 2.
Phylogeny of the 37 hummingbirds studied here, showing the distribution (and general lack of diversity) in genome size among species. The band-winged nightjar (Caprimulgidae: C. longirostris) is included as a close outgroup of the hummingbirds, and the clade consisting of perching birds (Passeriformes) and parrots (Psittaciformes) provides a more distant outgroup, with average genome size estimates based on the same methods used in the present analysis (Andrews & Gregory 2009; Andrews et al. 2009). Branch colours indicate the results of a squared change parsimony reconstruction of ancestral genome sizes. Species with a centre of abundance at the humid forests of the upper tropical zone are indicated by arrows. These appear to have independently undergone an increase in genome size; another species sometimes found in the same region (C. thalassinus) displays a broader habitat tolerance and has not undergone an increase.
Interestingly, the species with the largest genomes (and the only ones that exceed the mean by more than 10%)—the buff-tailed sicklebill (Eutoxeres condamini; 1.14 pg), the violet-fronted brilliant (Heliodoxa leadbeateri; 1.19 pg), the green hermit (Phaethornis guy; 1.16 pg) and the wedge-billed hummingbird (S. geoffroyi; 1.29 pg)—all have a centre of abundance in the upper tropical zone, 900–1600 m elevation, where they are restricted to humid, evergreen forests (Parker et al. 1996). As these species are not close relatives (figure 2), they appear to represent four independent increases in genome size associated with movement into the upper tropical zone. One other species in our dataset, the green violet-ear (Colibri thalassinus), shares this centre of abundance but has a typically diminutive genome of 1.02 pg. Unlike the other four species from this zone, the green violet-ear has a broader habitat tolerance that includes secondary forest and scrub as high as 3150 m elevation. One possible explanation for this pattern is that the long, narrow distributions of these four species along the flanks of the Andes support small effective populations, such that selection against mildly deleterious DNA insertions is less effective and genome sizes increase secondarily (Lynch & Conery 2003). In addition, climatic fluctuations may have caused population bottlenecks that were more severe or frequent for species in this ecoclimatic zone than for species occurring at higher or lower elevations.
We found a phylogenetically independent, inverse correlation between geographical range size and genome size (r = −0.50, p = 0.0016) that is consistent with the population size hypothesis. A further test is provided by the ratio of non-synonymous to synonymous substitutions in mitochondrial protein-coding genes, which may tend to be higher in smaller populations owing to genetic drift of slightly deleterious mutations (Popadin et al. 2007). However, non-synonymous to synonymous ratios for mitochondrial ND2 and ND4 genes were not elevated in hummingbirds with enlarged genomes (r = 0.11, p = 0.52).
4. Concluding remarks
Relative to other birds (and indeed, all tetrapods), hummingbirds exhibit small and minimally variable genome sizes. This observation is consistent with the hypothesis that the metabolic demands of powered flight, which are extreme in these hovering birds, have played a role in the evolution of reduced genome sizes of birds, probably through the intermediate of cell size (Hughes & Hughes 1995; Organ et al. 2007; Andrews et al. 2009). However, although genome size and cell size are positively correlated within hummingbirds as they are across birds (and vertebrates generally), metabolic and flight-related factors do not appear to account for the limited genome size variation that does exist in this group. Instead, the clearest pattern relates to habitat, with species that have independently specialized on upper-tropical zone humid forests exhibiting larger genomes. This could reflect demographic impacts on genome evolution, but this hypothesis needs to be tested with additional taxon sampling. In any case, the results of the present study reinforce the growing recognition of an important evolutionary interplay between features at the genomic, cellular, organismal and ecological levels.
Acknowledgements
Hummingbirds were captured in mistnets in Peru and New Mexico during 2007 and 2008 under permits issued by the management authorities of Peru, the USA and New Mexico, and according to University of New Mexico and University of California-Berkeley animal utilization protocols.
We thank Fernando Angulo, Emil Bautista O., Daniel Blanco, Miguel Campos Díaz, Jessica A. Castillo, Ben Cook, Olivio Díaz Idrogo, Robert Dudley, Daniel Echecopar, Zachary R. Hanna, Andrew B. Johnson, Michael Lelevier, Carrie McAtee, Christopher Merkord, Jano Nuñez, José Antonio Otero, Redinson Rodríguez Salazar, Dora Susanibar, Abraham Urbay, Thomas Valqui, and Geneva Williams for assistance with fieldwork. We also thank the personnel of the organizations Peru Verde and CORBIDI, and the communities of Sianbal, Incahuasi and San Pedro de Casta for their support and the Louisiana State University Museum of Natural Science and the University of Copenhagen Museum of Zoology for providing tissues used in phylogenetic analyses. Funding was provided in part by NSF (DEB-0543556 and DEB-0330750) and NSERC. In addition, we thank INRENA for providing permits to conduct fieldwork in Peru (76-2006-INRENA-IFFS-DCB and 087-2007-INRENA-IFFS-DCB).
References
- Altshuler D. L., Dudley R.2002The ecological and evolutionary interface of hummingbird flight physiology. J. Exp. Biol. 205, 2325–2336 [DOI] [PubMed] [Google Scholar]
- Altshuler D. L., Dudley R.2006The physiology and biomechanics of avian flight at high altitude. Integr. Comp. Biol. 46, 62–71 (doi:10.1093/icb/icj008) [DOI] [PubMed] [Google Scholar]
- Andrews C. B., Gregory T. R.2009Genome size is inversely correlated with relative brain size in parrots and cockatoos. Genome 52, 261–267 (doi:10.1139/G09-003) [DOI] [PubMed] [Google Scholar]
- Andrews C. B., Mackenzie S. A., Gregory T. R.2009Genome size and wing parameters in passerine birds. Proc. R. Soc. B 276, 55–61 (doi:10.1098/rspb.2008.1012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew G. A., Lighton J. R. B.1986Oxygen consumption during hover-feeding in free-ranging Anna hummingbirds. J. Exp. Biol. 123, 191–199 [DOI] [PubMed] [Google Scholar]
- Brown J. W., Payne R. B., Mindell D. P.2007Nuclear DNA does not reconcile ‘rocks’ and ‘clocks’ in Neoaves: a comment on Ericson et al. Biol. Lett. 3, 257–259 (doi:10.1098/rsbl.2006.0611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Rest J. S., Garcia-Moreno J., Sorenson M. D., Mindell D. P.2008Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biol. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson P. G. P., et al. 2006Diversification of Neoaves: integration of molecular sequence data and fossils. Biol. Lett. 2, 543–547 (doi:10.1098/rsbl.2006.0523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.1985Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- Garland T., Díaz-Uriarte R.1999Polytomies and phylogenetically independent contrasts: examination of the bounded degrees of freedom approach. Syst. Biol. 48, 547–558 (doi:10.1080/106351599260139) [DOI] [PubMed] [Google Scholar]
- Gregory T. R.2001Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. 76, 65–101 (doi:10.1017/S1464793100005595) [DOI] [PubMed] [Google Scholar]
- Gregory T. R.2002A bird's-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class Aves. Evolution 56, 121–130 [DOI] [PubMed] [Google Scholar]
- Gregory T. R.2005Genome size evolution in animals. In The evolution of the genome (ed. Gregory T. R.), pp. 3–87 San Diego, CA: Elsevier [Google Scholar]
- Gregory T. R.2009Animal genome size database. See http://www.genomesize.com/
- Hackett S. J., et al. 2008A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1768 (doi:10.1126/science.1157704) [DOI] [PubMed] [Google Scholar]
- Hardie D. C., Gregory T. R., Hebert P. D. N.2002From pixels to picograms: a beginners’ guide to genome quantification by Feulgen image analysis densitometry. J. Histochem. Cytochem. 50, 735–749 [DOI] [PubMed] [Google Scholar]
- Hartman F. A., Lessler M. A.1963Erythrocyte measurements in birds. Auk 80, 467–473 [Google Scholar]
- Hughes A. L.1999Adaptive evolution of genes and genomes Oxford, UK: Oxford University Press [Google Scholar]
- Hughes A. L., Hughes M. K.1995Small genomes for better flyers. Nature 377, 391 (doi:10.1038/377391a0) [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S.2003The origins of genome complexity. Science 302, 1401–1404 (doi:10.1126/science.1089370) [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R.2008Mesquite: a modular system for evolutionary analysis. Version 2.5. See http://mesquiteproject.org
- Mayr G.2004Old World fossil record of modern-type hummingbirds. Science 304, 861–864 (doi:10.1126/science.1096856) [DOI] [PubMed] [Google Scholar]
- McGuire J. A., Witt C. C., Altshuler D. L., Remsen J. V.2007Phylogenetic systematics and biogeography of hummingbirds: Bayesian and maximum likelihood analyses of partitioned data and selection of an appropriate partitioning strategy. Syst. Biol. 56, 837–856 (doi:10.1080/10635150701656360) [DOI] [PubMed] [Google Scholar]
- McGuire J. A., Witt C. C., Remsen J. V., Dudley R., Altshuler D. L.2009A higher-level taxonomy for hummingbirds. J. Ornithol. 150, 155–165 (doi:10.1007/s10336-008-0330-x) [Google Scholar]
- Midford P. E., Garland T., Maddison W. P.2003PDAP:PDTREE package for Mesquite. Version 1.12. See http://mesquiteproject.org/pdap_mesquite/
- Opazo J. C., Soto-Gamboa M., Fernández M. J.2005Cell size and basal metabolic rate in hummingbirds. Rev. Chil. Hist. Nat. 78, 261–265 [Google Scholar]
- Organ C. L., Shedlock A. M.2009Palaeogenomics of pterosaurs and the evolution of small genome size in flying vertebrates. Biol. Lett. 5, 47–50 (doi:10.1098/rsbl.2008.0491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ C. L., Shedlock A. M., Meade A., Pagel M., Edwards S. V.2007Origin of avian genome size and structure in non-avian dinosaurs. Nature 446, 180–184 (doi:10.1038/nature05621) [DOI] [PubMed] [Google Scholar]
- Parker T. A., Stotz D. F., Fitzpatrick J. W.1996Ecological and distributional databases. In Neotropical birds: ecology and conservation (eds Stotz D. F., Fitzpatrick J. W., Parker T. A., Moskovits D. K.), pp. 113–460 Chicago, IL: University of Chicago Press [Google Scholar]
- Popadin K., Polishchuk L. V., Mamirova L., Knorre D., Gunbin K.2007Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc. Natl Acad. Sci. USA 104, 13 390–13 395 (doi:10.1073/pnas.0701256104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A., Garland T.1993Polytomies in comparative analyses of continuous characters. Syst. Biol. 42, 569–575 [Google Scholar]
- Ridgely R. S., Allnutt T. F., Brooks T., McNicol D. K., Mehlman D. W., Young B. E., Zook J. R.2003Digital distribution maps of the birds of the Western Hemisphere, Version 1.0 Arlington, VA: NatureServe [Google Scholar]
- Simmons P., Lill A.2006Development of parameters influencing blood oxygen carrying capacity in the welcome swallow and fairy martin. Comp. Biochem. Physiol. 143A, 459–468 [DOI] [PubMed] [Google Scholar]
- Smith J. D. L., Gregory T. R.2009The genome sizes of megabats (Chiroptera: Pteropodidae) are remarkably constrained. Biol. Lett. 5, 347–351 (doi:10.1098/rsbl.2009.0016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez R. K.1992Hummingbird flight: sustaining the highest mass-specific metabolic rates among vertebrates. Experientia 48, 565–570 (doi:10.1007/BF01920240) [DOI] [PubMed] [Google Scholar]
- Suarez R. K., Lighton J. R., Brown G. S., Mathieu-Costello O.1991Mitochondrial respiration in hummingbird flight muscles. Proc. Natl Acad. Sci. USA 88, 4870–4873 (doi:10.1073/pnas.88.11.4870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H.1983Cell size and the concept of wasteful and frugal evolutionary strategies. J. Theor. Biol. 105, 201–209 (doi:10.1016/S0022-5193(83)80002-2) [DOI] [PubMed] [Google Scholar]
- Vinogradov A. E.1995Nucleotypic effect in homeotherms: body mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49, 1249–1259 (doi:10.2307/2410449) [DOI] [PubMed] [Google Scholar]
- Vinogradov A. E., Anatskaya O. V.2006Genome size and metabolic intensity in tetrapods: a tale of two lines. Proc. R. Soc. B 273, 27–32 (doi:10.1098/rspb.2005.3266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.2007PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]