Abstract
The question as to whether cultures evolve in a manner analogous to that of genetic evolution can be addressed by attempting to reconstruct population histories using cultural data. As others have argued, this can only succeed if cultures are isolated enough to maintain and pass on a central core of traditions that can be modified over time. In this study we used a set of cultural data (canoe design traits from Polynesia) to look for the kinds of patterns and relationships normally found in population genetic studies. After developing new techniques to accommodate the peculiarities of cultural data, we were able to infer an ancestral region (Fiji) and a sequence of cultural origins for these Polynesian societies. In addition, we found evidence of cultural exchange, migration and a serial founder effect. Results were stronger when analyses were based on functional traits (presumably subject to natural selection and convergence) rather than symbolic or stylistic traits (probably subject to cultural selection for rapid divergence). These patterns strongly suggest that cultural evolution, while clearly affected by cultural exchange, is also subject to some of the same processes and constraints as genetic evolution.
Keywords: cultural evolution, founder effect, population genetic, stylistic traits, Polynesian societies, canoe design
1. Introduction
Distance measures and phylogenetic techniques applied to population genetic data have proved to be powerful tools for investigating historical relationships between human populations around the globe. After decades of refining both data and methods, such studies have provided insights into questions of human demography, migration, ancestry, adaptation and disease predispositions (Cavalli-Sforza et al. 1994; Rosenberg et al. 2002, 2005; van de Vijver et al. 2002; Ramachandran et al. 2005; Tishkoff et al. 2007; Li et al. 2008). We can also infer the differing roles of males and females in the past from patterns in mitochondrial DNA (mtDNA) and non-recombining Y (NRY) portions of the genome, and the X chromosome (Seielstad et al. 1998; Ramachandran et al. 2004; Wilkins & Marlowe 2006; Segurel et al. 2008; Keinan et al. 2009). In Polynesia, for example, differences in the patterns of mtDNA and NRY between populations suggests ancient matrilineal marriages in which women remained within a community of related families, while men moved around and married into these families (Kayser et al. 2006).
Inferences about the histories, adaptations and relationships of human populations are strongest when multiple lines of evidence are brought to bear (Cavalli-Sforza et al. 1988, 1994; Hurles et al. 2003; Gray et al. 2007). In one of the first papers to bring together genetic, archaeological and linguistic data for human populations around the world, Cavalli-Sforza et al. (1988) showed that every linguistic phylum, with a few easily explained exceptions, corresponded to just one of six major genetic clusters, while the ratio of genetic distances (FST) to time since observed archaeological divergence seemed to be fairly constant. More recently, Hurles et al. (2003) compared archaeological dates, linguistic relationships and genetic data to untangle the prehistory of Oceanic settlement, finding converging lines of evidence for the dispersal of Lapita peoples into Polynesia.
Can data on population-level variation in cultural traits be used to reconstruct population histories? Boyd et al. (1997) concluded that the answer hinges on whether cultures are isolated enough, through geographic or other barriers, such that they can maintain a coherent central core of traditions that is not swamped by cultural exchange with neighbouring groups. Terrell et al. (1997) argued that the level of cultural exchange among Pacific island groups was sufficient to obscure any phylogenetic relationships. Hewlett and others tackled this question empirically for populations in Africa, finding that cultural traits related to kinship, family and community or political stratification were transmitted conservatively within families and groups (Hewlett et al. 2002; see also Hewlett & Cavalli-Sforza 1986; Guglielmino et al. 1995). Vansina (1990) came to a similar conclusion after finding numerous cultural similarities conserved throughout the 4000-year Bantu expansion in equatorial Africa. Ammerman & Cavalli-Sforza (1984) used demographic models and archaeological dates to show that the spread of agriculture in Eurasia was almost certainly due to demic diffusion (expansion of populations through movement of people) rather than horizontal transmission of the technology from group to group; this finding was subsequently confirmed with genetic data (Chikhi et al. 2002).
Gray et al. (2007) acknowledge that both phylogenetic signal and horizontal exchange will always exist in cultural data. They propose techniques for distinguishing the two processes and conclude that, if used cautiously, cultural data can help answer questions about the location of ancestral homelands, the order of cultural divergence, factors affecting cultural rates of change, ancestral states of traits and adaptive cultural responses. Taking the discussion a step further, Greenhill et al. (2009) simulate language evolution with realistic levels of borrowing, and then use phylogenetic techniques to see if they can reconstruct the history despite the presence of horizontal exchange; the answer is yes. Meanwhile, archaeologists are already using population genetic and phylogenetic approaches to learn much about the evolution of material culture and of the populations associated with these artefacts (Shennan & Wilkinson 2001; O'Brien & Lyman 2003; Bentley et al. 2004; Buchanan & Collard 2007; Lycett & von Cramon-Taubadel 2008; Mesoudi & O'Brien 2008; O'Brien 2008; Hamilton & Buchanan 2009; Lycett 2009).
Oceania has been the focus of much effort to answer these kinds of questions. It is often cited as a good test system because of its relatively recent occupation and easily identified boundaries (Kirch & Green 2001; Hurles et al. 2003; Rogers & Ehrlich 2008). Most studies have focused on the early stages of expansion into Oceania, between 5000 and 2500 YBP (years before present; Diamond 1988; Lum et al. 1998; Oppenheimer & Richards 2001; Hurles et al. 2002; Friedlaender et al. 2005, 2008; Kayser et al. 2006, 2008). For this time period, inferences from archaeological, linguistic and genetic data generally support some version of the ‘express train’ model: a rapid expansion of Lapita culture out of Taiwan, through coastal Melanesia and islands near Oceania, and thence into remote Oceania (Hurles et al. 2003).
Genetic, linguistic and archaeological data, however, have not provided clear answers concerning the settlement sequence and phylogenetic relationships for the various island populations within Polynesia. Most studies agree that Fiji was the jumping-off point for the colonization of Polynesia, with the western Polynesian archipelagos of Tonga and Samoa settled earliest in Polynesia proper (Kirch & Green 2001). There also seems to be general agreement that Aotearoa (New Zealand) was settled most recently, although by whom is under dispute (Sutton 1994; Underhill et al. 2001; Hurles et al. 2003). For most of the rest of Polynesia there is no consensus, because dates for archaeological sites are still under revision, linguistic and anthropological data show much inter-island borrowing, and few islands have sufficient genetic data to draw conclusions. Almost completely missing is the incorporation of quantitative analysis of non-linguistic cultural data.
In this study, we analyse a set of material cultural data from Polynesia and ask whether these data can be used to infer population history. Using presence/absence data on canoe design traits for ten Polynesian cultures and Fiji, we investigate the extent to which a phylogenetic signal remains in core cultural traits, and whether horizontal cultural exchange obscures the historical signal. We look for correlations of cultural differences with geographic distance, ask where the ancestral homelands might lie, and attempt to infer cultural origins for the island groups of Polynesia. We conclude by assessing whether some important population genetic findings such as loss of variability through a serial founder effect (Flint et al. 1989; Murray-McIntosh et al. 1998; Lum et al. 2002; Ramachandran et al. 2005; Manica et al. 2007; Lycett & von Cramon-Taubadel 2008; Deshpande et al. 2009) are also evident in our cultural data. Throughout, we are interested in such general methodological questions as whether and how cultural data might be used to draw these types of inferences, as well as specific questions about the sequence of cultural divergence in Polynesia.
2. Material and methods
Characteristics of traditional (pre-contact) Polynesian canoe design and construction were obtained from Haddon & Hornell (1936–1938), presenting descriptions of traditional canoe design for cultures from each island group in Oceania (Rogers & Ehrlich 2008). A data matrix consisting of presence/absence data for these canoe design features was created for 11 island groups (ten Polynesian island groups plus Fiji; see fig. S1 and table S1 in the electronic supplementary material). Each description provided by Haddon and Hornell was coded into the appropriate island group and canoe trait cell for two main canoe types: outrigger and double hull. For pairs of traits that had a Pearson product–moment correlation coefficient greater than 0.7 and that arguably provided redundant information, only one of the pair was used (see table S2 in the electronic supplementary material). The final data matrix had presence/absence data for 134 design traits (96 functional and 38 symbolic; see tables S3 and S4 in the electronic supplementary material). See the electronic supplementary material for details on methods.
Island-by-island cultural distance matrices were created using a Jaccard distance measure between sets of presence/absence records for pairs of islands (Rogers & Ehrlich 2008). Three such distance matrices—based on functional traits only, symbolic traits only and the combined set of traits—were generated (see table S5 in the electronic supplementary material). Three rooted consensus trees (based on functional, symbolic and combined traits) were created using the neighbour-joining algorithm with bootstrapped datasets. Fiji was specified as the out-group because it is widely thought to be the region from which the Polynesian islands were originally colonized (Kirch & Green 2001). The relationships between the various island cultures were explored further by carrying out a principal-components analysis using the three cultural Jaccard distance matrices.
In order to assess the extent to which island-to-island cultural distances might reflect cultural exchange between neighbours due to geographic proximity, two geographic distance matrices were developed: one based on distance (in kilometres) between the centres of each island group (table S6 in the electronic supplementary material) and one based on the number of archipelago-to-archipelago ‘steps’ (counted on a map) between each pair of island groups (table S7 in the electronic supplementary material). Correlations between cultural distances and these geographic and ‘stepwise’ distances were assessed using Mantel tests. To see if ancestral origin could be inferred, we considered each of the 11 island groups as a potential starting point. Cultural distances from that island were then regressed on geographic distances (in kilometres) and on the number of steps from that island (Ramachandran et al. 2005; Lycett & von Cramon-Taubadel 2008).
A new technique was developed to infer cultural origins for related populations in the presence of cultural exchange (see the electronic supplementary material for a detailed description of the method). Potential origins for each of the 11 island groups in our study were identified using the anthropological literature (table S8 in the electronic supplementary material). All plausible cultural origin sequences were then generated by iteratively choosing one of these plausible cultural origins for each of the 11 island groups. For each of these sequences, Fiji was identified as the ancestral island group. Nonlinear sequences (in which one origin led to multiple endpoints) were allowed, but circular sequences (in which an island group could be traced back to itself) were not allowed; thus 38 879 such ‘plausible’ sequences were generated using this algorithm.
For each pair of islands in each of the generated sequences, the most recent common origin (island) was determined. Pairwise island-to-island geographic distances were then calculated through these common origins for each pair of islands. The corresponding pairwise cultural distances (Jaccard distances for combined traits) were then regressed on these geographic distances. The 30 cultural origin sequences that gave the highest r2 values were identified and assessed for common elements by counting how many times each specific two-island pair (or three-island triplet) appeared in these 30 top sequences. Probabilities were then assigned to these counts, assuming that the number of times each origin appears for a given endpoint should have a binomial distribution (see the electronic supplementary material for complete details).
This method suggests particular cultural origins (which have been defined as plausible a priori), but it needed to be validated before relying on the results. This was done by generating a second set of 10 million random cultural origin sequences, not limited to plausible origins. Cultural distances were regressed on geographic distances calculated through the sequence, as before, and the 100 sequences resulting in the highest r2 values were identified. These sequences were then analysed to determine if the method succeeded in identifying ‘plausible’ origins (i.e. those identified in the literature, and including reversed-order pairs) more often than would be expected by chance, as follows.
We tabulated the number of plausible two-island origin–endpoint pairs observed in these sequences, and then calculated the probability of this number of successes (or a more extreme value) in the 100 sequences using a binomial probability distribution. We observed 808 plausible two-island pairs out of the 1100 total pairs. The binomial probability for 808 out of 1100, each with a probability of 0.5372 (65 plausible/121 possible), is 2.3247 × 10−41. Likewise, we observed 571 plausible triplets out of the 1100 total three-island triplets. Again, assuming a binomial distribution for plausible triplets, each with a probability of 0.2007 (223 plausible/1111 possible triplets), this observation was extremely unlikely (probability of 3.7052 × 10−121). This indicates that our method succeeds in identifying plausible origins significantly more often than could be accounted for by chance, and suggests that our approach to finding likely two- and three-island sequences is valid.
The 100 randomly generated sequences with highest r2 values were then assessed for common elements by counting how many times each specific two-island pair appeared, and probabilities were assigned to these results assuming they followed a binomial distribution. Significance levels were corrected by dividing by the number of comparisons made; that is, the total number of pairs that could have appeared (11 possible origins × 11 endpoints = 121). The 100 sequences were also assessed for three-island sequences (triplets), and significance levels corrected by dividing by the number of comparisons made; that is, the total number of triplets that could have appeared (1111).
This entire ‘cultural origins’ technique was based on cultural distances calculated for combined (functional plus symbolic) traits. We also carried out the same analysis using cultural distances based solely on symbolic traits (see §4 for rationale). For the latter analysis, our technique identified 30 top plausible sequences and 150 top randomly generated sequences. We observed 1155 plausible two-island pairs out of the 1650 total pairs generated, each with a probability of 0.5372, giving a binomial probability of 1.101 × 10−41. Likewise, we observed 535 plausible triplets out of the total 1650 triplets generated, each with a probability of 0.2007, giving a binomial probability of 9.283 × 10−31. This indicates that our method applied to symbolic traits also succeeds in identifying plausible origins more often than could be accounted for by chance. We then assessed all top plausible and randomly generated sequences for common elements, and assigned probabilities to these results as before.
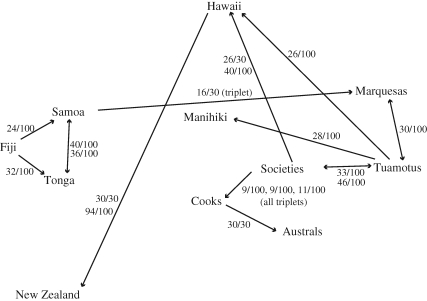
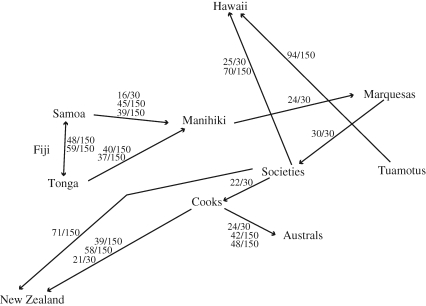
All two-island pairs and three-island triplets that appeared with a corrected significance level of less than 0.001 are presented in table S9 of the electronic supplementary material. These were then used to develop figures 1 and 2, which summarize inferred cultural origins for the 11 island groups based on combined traits and symbolic traits, respectively. See the electronic supplementary material for a detailed description of how figures 1 and 2 were developed. Two matrices of geographic distances were then developed for the 11 island groups, with distances (in km) constrained to pass through the two inferred cultural origin sequences—combined (table S10 in the electronic supplementary material) and symbolic (table S11 in the electronic supplementary material)—and the Mantel correlation test was used to see whether the Jaccard cultural distances were correlated with these geographic distances.
Figure 1.
Cultural origin sequence supported by best regressions of cultural distance (combined traits) on geographic distance constrained through ordered islands. See tables S9, S12 and S13 in the electronic supplementary material for supporting results. Numerators over denominators of 30 or 100 show the number of times this two-island origin–endpoint pair appeared (with p < 0.001) in the two-island analyses conducted on the top 30 plausible sequences or the top 100 random permutations sequences. Fractions labelled ‘triplet’ show the number of times this three-island sequence appeared (with p < 0.001) in the comparable three-island analyses; see §2 and the electronic supplementary material for an explanation.
Figure 2.
Cultural origin sequence supported by best regressions of cultural distance (symbolic traits) on geographic distance constrained through ordered islands. See tables S9, S12 and S13 in the electronic supplementary material for supporting results. Numerators over denominators of 30 or 150 show the number of times this two-island origin–endpoint sequence appeared (with p < 0.001) in the two-island analyses conducted on the top 30 plausible sequences or the top 150 random permutations sequences; see §2 and the electronic supplementary material for an explanation.
Finally, for each island group we counted the total number of canoe design traits (functional, symbolic and combined), and the number of canoe design traits shared in common with Fiji. We then regressed these sets of trait numbers on the direct geographic distance from Fiji and on distance calculated through the two inferred cultural origin sequences identified in our study.
3. Results
Figures 1 and 2 (and tables S9, S12 and S13 in the electronic supplementary material) summarize the results of the cultural origin sequence analysis based on regressions of cultural distances on geographic distances as calculated through various sequences. Note that the analysis does not result in a linear settlement sequence, but instead infers one or more primary cultural origins for each island group.
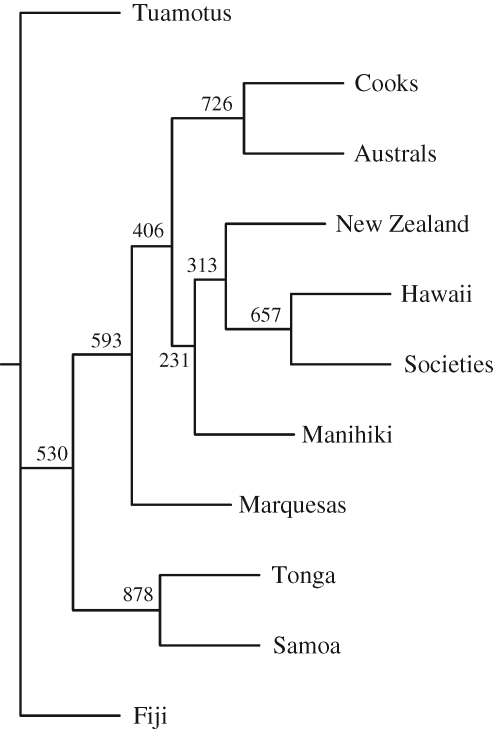
Neighbour-joining consensus trees for combined and for functional canoe design traits (figure 3; see also fig. S2 in the electronic supplementary material) show Tonga and Samoa splitting off soon after Fiji, with Tuamotus also diverging early on; later clusters include Societies and Hawaii with New Zealand, and Australs with Cooks. The consensus trees based on symbolic traits only (fig. S3 in the electronic supplementary material) shows Manihikis splitting off with Tonga and Samoa, but Tuamotus clustering later with Hawaii and the Societies. Principal components analyses (figs S4, S5 and S6 in the electronic supplementary material) show essentially the same groupings as the phylogenetic trees and do not reveal any obvious relationships with geographic or environmental factors.
Figure 3.
Consensus tree using Jaccard distances for combined traits. Numbers indicate branch support from bootstrapped dataset with 1000 iterations.
Bootstrap support for branches on the consensus trees is not high; it ranges from 23–88 per cent for the combined traits tree. This raises the question of the extent to which these cultural data are providing a treelike (sequentially bifurcating) phylogenetic signal or reflect relationships due to horizontal cultural exchange. The treeness indices we used suggest that the phylogenetic signal is not strong. Rambaut's Treeness Index (Phillips & Penny 2003; Rambaut & Drummond 2007) gives a relatively low value for proportion of total tree length taken up by internal branches (0.23–0.28), while the ensemble Consistency Index (0.42–0.65) and Retention Index (0.33–0.5; Kitching et al. 1998; Hammer et al. 2001) were both relatively low as well, indicating that many characters were not in accordance with a strictly bifurcating tree (table S14 in the electronic supplementary material).
Neither the direct geographic distance matrix nor the ‘stepwise’ distance matrix exhibited significant correlations with cultural distances based on canoe traits (table S15 in the electronic supplementary material), although there may be a decline in symbolic traits with stepwise distances (r = 0.27, p = 0.081). Likewise, none of the regressions of cultural distance on geographic distance from a hypothetical ancestral island group were significant (table S16 in the electronic supplementary material). Notably, there was no significant relationship when Fiji was used as the starting point, although archaeologists are relatively certain that Fiji was the point from which settlement originated. Nor was there a decline in trait diversity with direct geographic distance from Fiji (table S17 in the electronic supplementary material).
There is, however, a strong significant correlation between the cultural distances and geographic distances when constrained to pass through the inferred cultural origin sequence based on combined traits (r > 0.7, p < 0.001; table S18 in the electronic supplementary material). The results are not as strong when the cultural origin sequence is based on symbolic traits only (r > 0.48, p = 0.006). There is also an apparent decline in total number of functional canoe design traits (r2 > 0.41, p < 0.099), and a modestly significant decline in number of functional traits shared with Fiji (r2 = 0.48, p < 0.055), as geographic distance from Fiji increases through the inferred cultural origin sequence based on combined traits (figs S7 and S8 and table S17 in the electronic supplementary material). This suggests a possible serial founder effect for Polynesian cultures, as has been proposed on the basis of genetic data (see §1). Again, results for the cultural origin sequence based on symbolic traits alone are not as strong.
4. Discussion
We have proposed a method to identify a sequence of cultural origins for a group of related populations in the presence of cultural exchange. The validity of this method is indicated by the fact that plausible origins (previously identified in the anthropological literature) occur in such sequences far more frequently than is likely to have happened by chance. Using this method, support for Fiji as the ancestral homeland can be inferred, cultural distances can be seen to increase as geographic distance increases through the sequence and a serial founder effect of declining cultural trait diversity with sequential distance from Fiji is suggested. These are the kinds of results that have been observed in genetic data (Ramachandran et al. 2005). Finding them with cultural data indicates that, despite the presence of horizontal exchange, the transmission, modification and drift of cultural traits may generate patterns similar to those seen for genetic traits. This was predictable, as both genetic and phenotypic relationships between populations are also affected by migration and horizontal exchange (Manica et al. 2007; von Cramon-Taubadel & Lycett 2008).
Our cultural origin sequence based on symbolic traits alone differs in some important ways from that based on combined traits. The latter connects all the island groups to one another through a sequence of inferred cultural origins, resulting in strong correlations between cultural and geographic distances, and showing modestly significant regressions for a number of combined traits from Fiji, on distance from Fiji. The sequence based on symbolic traits alone does not connect all the island groups to one another through a sequence of inferred cultural origins; correlations between cultural and geographic distance are not as strong, and the regression for the number of symbolic traits from Fiji on distance from Fiji, while negative, is not significant (r2 < 0.166, p > 0.6).
Based on these considerations, we might conclude that the use of distances based on combined cultural traits is more appropriate for this technique. There are two arguments to the contrary. First, older work within evolutionary archaeology concluded that symbolic (stylistic) traits should be used to infer historical relationships, because functional traits may be subject to selection and thus reflect convergence due to similarities in the social or physical environment (Dunnell 1978). However, symbolic traits may well be under cultural selection; for example, as identity markers that are chosen to differentiate a society from its neighbours (Barth 1969; Nagel 1994; Harrison 1999, 2002). If this were the case, historical relationships could be obscured by intentional cultural choices. More recent approaches to the evolution of material culture recognize that it is the presence of selection, whether natural or cultural, that leads to lower goodness-of-fit of cultural data to the serial founder effect model (Shennan & Wilkinson 2001; Lycett & von Cramon-Taubadel 2008; Hamilton & Buchanan 2009). Second, the fact that the functional trait data may be less treelike than the symbolic trait data (table S14 in the electronic supplementary material) could be a concern. However, our technique to infer cultural origins was developed specifically in order to incorporate cultural exchange by allowing multiple connections or origins in the analysis.
Perhaps the strongest objection to using the combined traits for this analysis is one conclusion it yields: that Hawaii was the primary cultural origin for New Zealand. Sutton (1994) and Kirch (2000) indicate that the current consensus is that New Zealand was colonized from central Eastern Polynesia (most probably the region of the Cooks or Societies). Davidson says that we do not have sufficient archaeological evidence to be more precise in identifying the source of migration to New Zealand than ‘somewhere in Eastern Polynesia’ (Davidson 1994, p. 216). Older ethnographic treatments assert that there was a pronounced cultural connection between Hawaii and New Zealand, including strong similarities in mythology (Dixon 1916, p. 93), and shared elements in oral histories and genealogies (Smith 1904). Davidson (1994) notes the appearance of two-piece fishhooks solely in New Zealand, Hawaii and Easter Island, although she suggests convergent evolution as a possible cause of this shared technology. The clustering of Hawaii with New Zealand (Maori) on Gray's recent linguistic phylogeny provides additional support for a close connection between Hawaii and New Zealand (Gray et al. 2009, p. 51 in electronic supplementary material). In light of questions on the use of symbolic or functional traits, the lack of definitive genetic evidence for Maori origins, the incomplete archaeological record and our cultural origin sequence findings, the early cultural origins of New Zealand are still to be resolved.
The cultural origin sequence based on combined traits (and, to a lesser extent, the symbolic traits sequence) also lends support to the possibility that the Marquesas and Tuamotus were settled early, directly from the region of Samoa and Tonga, with more westerly parts of Eastern Polynesia settled later. This idea has been proposed in the past (Sinoto 1968), but has fallen out of favour more recently because of dates assigned to archaeological sites (Anderson & Sinoto 2002). Whether or not these island groups were settled early, the canoe-building culture of the Tuamotus was quite different from that of nearby island groups, perhaps because this archipelago of low coral atolls needed to import resources from high volcanic islands (Finney 1994).
Although a cladistic (parsimony) approach to phylogenetic inference may have resulted in a different tree, it is clear from the low bootstrap branch support values and treeness indices that our cultural data do not simply reflect a pattern of vertical (i.e. intra-group) trait transmission with sequential bifurcation over time. This means that it is not advisable to take at face value any given phylogenetic tree drawn from these data, regardless of approach. Comparisons of our cultural consensus tree with other published linguistic and genetic data or trees for the same populations agree (in almost every case) on the older, deeper divergence between the Fiji–Tonga–Samoa region and the rest of Polynesia; however, the more recent branches vary widely from tree to tree (table S19 in the electronic supplementary material). This pattern of similar deep branches, but widely varying recent branches, almost always appears when other trees for Polynesian populations (based on various types of data) are compared with one another (O'Shaughnessy et al. 1990; Cavalli-Sforza et al. 1994; Pietrusewsky, 1996; Gray & Jordan 2000; Marck 2000; Capelli et al. 2001; Fischer 2001; Hurles et al. 2003; Gray et al. 2009; p. 41 in electronic supplementary material; but not for Lum & Cann 2000). This clear divergence between the Fiji–Tonga–Samoa region and the rest of Polynesia is consistent with the archaeological record, and with the settlement pulse–pause hypothesis advanced by Gray et al. (2009) from their analysis of some 400 Austronesian languages.
There are several possible reasons why such cultural data may not be strictly treelike. The most obvious possibility is that horizontal cultural exchange between many of these island societies has obscured the phylogenetic divergences that may have developed. There is good evidence for trade networks and other ongoing cultural relationships throughout Polynesia (Rolett 1996, 2002; Petersen 2000; Barnes & Hunt 2005). For example, stone adzes found on Mangaia Island in the southern Cooks were shown to be made of basalt imported from a quarry on Tutuila Island in American Samoa, 1600 km away (Weisler & Kirch 1996); imported basalt adzes and pounders were also found on the Tuamotuan coral atolls (Emory 1975). It has also been suggested that canoes from the relatively large, resource-rich islands of the Societies may have been exported to nearby archipelagos affected by timber depletion (Rolett 2002).
A related reason for the weakness of the treelike signal in these cultural data may be the time scale or geographic scale for settlement of the Polynesian societies. Phylogenies are typically based on genetic or linguistic data for a group of populations that have had sufficient time and geographic isolation from one another to develop and maintain differences. Our phylogenetic tree showed the same split between Eastern and Western Polynesia as those of other biological or linguistic phylogenies, yet the more recent splits and clusters show no consistent pattern. It may be that the more recently settled groups simply have not had enough time or isolation to diverge in the classical phylogenetic manner.
It is also possible, as mentioned earlier, that selection—natural or cultural—has acted on certain traits in such a way as to obscure the phylogenetic signal. Most of our analyses were based on the functional (or combined) canoe design traits. We felt this was appropriate because the functional traits have been shown to change significantly more slowly, making them more likely to retain any phylogenetic signal, and also because the symbolic (or stylistic) traits appeared to be under cultural selection to diversify rapidly, perhaps as identity markers (see Rogers & Ehrlich 2008). There is a precedent within biology for developing phylogenetic trees based on functional traits thought to be under purifying selection: 16S rRNA has been conserved across many life forms, and is thought to provide one of the best sources of data for inferring species phylogenies (Boyer et al. 2001).
Other reasons for the lack of treelike signal might include inadequacies of the data (i.e. missing, inconsistent or incorrect data) or confounding factors such as environmental variability, resource depletion or demographic histories. Notably, these factors and the others mentioned above—including selection, migration, horizontal exchange, and time or geographic scale of divergence—all affect the retention of phylogenetic signal in genetic data as well.
Analyses of genetic distance between populations show that increased geographic distance is correlated with increased genetic distance, presumably through migration (Manica et al. 2005; Ramachandran et al. 2005); similar trends have been observed for phenotypic data (Manica et al. 2007) and cultural data (Lycett & von Cramon-Taubadel 2008). The lack of significant correlation of direct geographic distances with distances based on our cultural data suggests that cultural exchange is mediated by particular cultural–historical relationships between groups, rather than by the convenience of geographic proximity. This is supported by the observation that correlations with cultural distances are greatly strengthened when the geographic distances are constrained to go through our proposed cultural origin sequence. Once the sequence of cultural origins is incorporated, the expected findings—cultural distance increasing with geographic distance and declining trait diversity through serial founder effect—become apparent.
Data on variation in cultural traits can give important insight into the historical and cultural relationships between societies, provided cultural selection and specific relationships of cultural exchange are taken into account. Further confirmation of these findings requires acquisition of additional cultural datasets, development of origin sequences using other cultural, linguistic and genetic data, and quantitative exploration of the effects of horizontal cultural exchange on estimates of cultural distances, rates of change and phylogenetic relationships.
Acknowledgment
Special thanks are extended to D. Akinniyi, B. Beheim, R. Gray and S. Thomas for significant help with data, analysis and interpretation. For helpful suggestions throughout the project, we thank A. Anderson, R. Blust, R. Cann, E. Cochrane, D. DeGusta, J. Diamond, J. Felsenstein, F. Jordan, M. Lubell, R. Mace, J. Marck, R. McElreath, S. Ramachandran, P. Richerson, R. Sapolsky, S. Shennan, J. Van Cleve and D. Weissman. This work was supported by an NSF Graduate Research Fellowship (D.S.R.), the Morrison Institute for Population and Resource Studies (D.S.R.), John P. Gifford Fund (P.R.E.), Mertz Gilmore Foundation (P.R.E.), Peter and Helen Bing (P.R.E.) and NIH grant GM 28016 (M.W.F.).
References
- Ammerman A. J., Cavalli-Sforza L. L.1984The Neolithic transition and the genetics of populations in Europe. Princeton, NJ: Princeton University Press [Google Scholar]
- Anderson A., Sinoto Y.2002New radiocarbon ages of colonization sites in east Polynesia. Asian Perspectives 41, 242–257 (doi:10.1353/asi.2003.0002) [Google Scholar]
- Barnes S. S., Hunt T. L.2005Samoa's pre-contact connections in West Polynesia and beyond. J. Polynesian Soc. 114, 227–266 [Google Scholar]
- Barth F. (ed.) 1969Ethnic groups and boundaries Boston, MA: Little Brown & Company [Google Scholar]
- Bentley R. A., Hahn M. W., Shennan S. J.2004Random drift and culture change. Proc. R. Soc. Lond. B 271, 1443–1450 (doi:10.1098/rspb.2004.2746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd R., Mulder M. B., Durham W., Richerson P. J.1997Are cultural phylogenies possible? In Human by nature: between biology and the social sciences (eds Weingart P., Mitchel S. D., Richerson P. J., Maasen S.), pp. 355–386 Mahwah, NJ: Lawrence Erlbaum Associates [Google Scholar]
- Boyer S. L., Flechtner V. R., Johansen J. R.2001Is the 16S–23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 18, 1057–1069 [DOI] [PubMed] [Google Scholar]
- Buchanan B., Collard M.2007Investigating the peopling of North America through cladistic analyses of Early Paleoindian projectile points. J. Anthropol. Archaeol. 26, 366–393 (doi:10.1016/j.jaa.2007.02.005) [Google Scholar]
- Capelli C., et al. 2001A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am. J. Human Genet. 68, 432–443 (doi:10.1086/318205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L., Piazza A., Menozzi P., Mountain J.1988Reconstruction of human evolution: bringing together genetic, archaeological and linguistic data. Proc. Natl Acad. Sci. USA 85, 6002–6006 (doi:10.1073/pnas.85.16.6002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Menozzi P., Piazza A.1994The history and geography of human genes. Princeton, NJ: Princeton University Press [Google Scholar]
- Chikhi L., Nichols R. A., Barbujani G., Beaumont M. A.2002Y genetic data support the Neolithic demic diffusion model. Proc. Natl Acad. Sci. USA 99, 11 008–11 013 (doi:10.1073/pnas.162158799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J.1994The eastern Polynesian origins of the New Zealand archaic. In The origins of the first New Zealanders (ed. Sutton D. G.), pp. 208–219 Auckland, New Zealand: Auckland University Press [Google Scholar]
- Deshpande O., Batzoglou S., Feldman M. W., Cavalli-Sforza L. L.2009A serial founder effect model for human settlement out of Africa. Proc. R. Soc. B 276, 291–300 (doi:10.1098/rspb.2008.0750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J. M.1988Archaeology—express train to Polynesia. Nature 336, 307–308 (doi:10.1038/336307a0) [Google Scholar]
- Dixon R. B.1916Oceanic mythology. In The mythology of all races Boston, MA: Marshall Jones Company [Google Scholar]
- Dunnell R. C.1978Style and function: a fundamental dichotomy. Am. Antiquity 43, 192–202 (doi:10.2307/279244) [Google Scholar]
- Emory K. P.1975Material culture of the Tuamotu archipelago. In Pacific anthropological records Honolulu, HA: Bernice P; Bishop Museum. [Google Scholar]
- Finney B.1994Voyage of rediscovery: A cultural odyssey through Polynesia Berkeley, CA: University of California Press [Google Scholar]
- Fischer S. R.2001Mangarevan doublets: preliminary evidence for Proto-Southeastern Polynesian. Oceanic Linguistics 40, 112–124 (doi:10.1353/ol.2001.0005) [Google Scholar]
- Flint J., Boyce A. J., Martinson J. J., Clegg J. B.1989Population bottlenecks in Polynesia revealed by minisatellites. Human Genet. 83, 257–263 (doi:10.1007/BF00285167) [DOI] [PubMed] [Google Scholar]
- Friedlaender J., et al. 2005Expanding southwest pacific mitochondrial haplogroups P and Q. Mol. Biol. Evol. 22, 1506, 2005, 2313–2313 [DOI] [PubMed] [Google Scholar]
- Friedlaender J. S., et al. 2008The genetic structure of Pacific islanders. PLoS Genet. 4, 173–190 (doi:10.1371/journal.pgen.0040019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. D., Jordan F. M.2000Language trees support the express-train sequence of Austronesian expansion. Nature 405, 1052–1055 (doi:10.1038/35016575) [DOI] [PubMed] [Google Scholar]
- Gray R. D., Greenhill S. J., Ross R. M.2007The pleasures and perils of Darwinizing culture (with phylogenies). Biol. Theory 2, 360–375 (doi:10.1162/biot.2007.2.4.360) [Google Scholar]
- Gray R. D., Drummond A. J., Greenhill S. J.2009Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323, 479–483 (doi:10.1126/science.1166858) [DOI] [PubMed] [Google Scholar]
- Greenhill S. J., Currie T. E., Gray R. D.2009Does horizontal transmission invalidate cultural phylogenies? Proc. R. Soc. B 276, 2299–2306 (doi:10.1098/rspb.2008.1944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmino C. R., Viganotti C., Hewlett B., Cavalli-Sforza L. L.1995Cultural variation in Africa—role of mechanisms of transmission and adaptation. Proc. Natl Acad. Sci. USA 92, 7585–7589 (doi:10.1073/pnas.92.16.7585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon A. C., Hornell J.1936–1938Canoes of Oceania Honolulu, HA: Bishop Museum Press [Google Scholar]
- Hamilton M. J., Buchanan B.2009The accumulation of stochastic copying errors causes drift in culturally transmitted technologies: quantifying Clovis evolutionary dynamics. J. Anthropol. Archaeol. 28, 55–69 (doi:10.1016/j.jaa.2008.10.005) [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D.2001PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, 9. [Google Scholar]
- Harrison S.1999Cultural boundaries. Anthropol. Today 15, 10–13 (doi:10.2307/2678369) [Google Scholar]
- Harrison S.2002The politics of resemblance: ethnicity, trademarks, head-hunting. J. R. Anthropol. Inst. 8, 211–232 (doi:10.1111/1467-9655.00001) [Google Scholar]
- Hewlett B., Cavalli-Sforza L. L.1986Cultural transmission among Aka Pygmies. Am. Anthropol. 88, 922–934 (doi:10.1525/aa.1986.88.4.02a00100) [Google Scholar]
- Hewlett B. S., De Silvestri A., Guglielmino C. R.2002Semes and genes in Africa. Curr. Anthropol. 43, 313–321 (doi:10.1086/339379) [Google Scholar]
- Hurles M. E., Nicholson J., Bosch E., Renfrew C., Sykes B. C., Jobling M. A.2002Y chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics 160, 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M. E., Matisoo-Smith E., Gray R. D., Penny D.2003Untangling Oceanic settlement: the edge of the knowable. Trends Ecol. Evol. 18, 531–540 (doi:10.1016/S0169-5347(03)00245-3) [Google Scholar]
- Kayser M., et al. 2006Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol. Biol. Evol. 23, 2234–2244 (doi:10.1093/molbev/msl093) [DOI] [PubMed] [Google Scholar]
- Kayser M., et al. 2008Genome-wide analysis indicates more Asian than Melanesian ancestry of Polynesians. Am. J. Human Genet. 82, 194–198 (doi:10.1016/j.ajhg.2007.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Mullikin J. C., Patterson N., Reich D.2009Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nature Genet. 41, 66–70 (doi:10.1038/ng.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch P. V.2000On the road of winds: an archaeological history of the Pacific Islands before European contact Berkeley, CA: University of California Press [Google Scholar]
- Kirch P. V., Green R. C.2001Hawaiki, ancestral Polynesia: an essay in historical anthropology Cambridge, UK: Cambridge University Press [Google Scholar]
- Kitching I. J., Forey P. L., Humphries C. J., Williams D. M.1998Cladistics: the theory and practice of parsimony analysis. Oxford, UK: Oxford University Press [Google Scholar]
- Li J. Z., et al. 2008Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- Lum J. K., Cann R. L.2000mtDNA lineage analyses: origins and migrations of Micronesians and Polynesians. Am. J. Phys. Anthropol. 113, 151–168 (doi:10.1002/1096-8644(200010)113:2<151::AID-AJPA2>3.0.CO;2-N) [DOI] [PubMed] [Google Scholar]
- Lum J. K., Cann R. L., Martinson J. J., Jorde L. B.1998Mitochondrial and nuclear genetic relationships among Pacific Island and Asian populations. Am. J. Human Genet. 63, 613–624 (doi:10.1086/301949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J. K., Jorde L. B., Schiefenhovel W.2002Affinities among Melanesians, Micronesians and Polynesians: a neutral, biparental genetic perspective. Human Biol. 74, 413–430 (doi:10.1353/hub.2002.0031) [DOI] [PubMed] [Google Scholar]
- Lycett S. J.2009Are Victoria West cores ‘proto-Levallois’? A phylogenetic assessment. J. Human Evol. 56, 175–191 (doi:10.1016/j.jhevol.2008.10.001) [DOI] [PubMed] [Google Scholar]
- Lycett S. J., von Cramon-Taubadel N.2008Acheulean variability and hominin dispersals: a model-bound approach. J. Archaeol. Sci. 35, 553–562 (doi:10.1016/j.jas.2007.05.003) [Google Scholar]
- Manica A., Prugnolle F., Balloux F.2005Geography is a better determinant of human genetic differentiation than ethnicity. Human Genet. 118, 366–371 (doi:10.1007/s00439-005-0039-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A., Amos W., Balloux F., Hanihara T.2007The effect of ancient population bottlenecks on human phenotypic variation. Nature 448, 346–348 (doi:10.1038/nature05951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck J.2000Topics in Polynesian language and culture history Pacific Linguistics 504.Canberra, Australia: Pacific Linguistics [Google Scholar]
- Mesoudi A., O'Brien M. J.2008The cultural transmission of Great Basin projectile-point technology I: an experimental simulation. Am. Antiquity 73, 3–28 [Google Scholar]
- Murray-McIntosh R. P., Scrimshaw B. J., Hatfield P. J., Penny D.1998Testing migration patterns and estimating founding population size in Polynesia by using human mtDNA sequences. Proc. Natl Acad. Sci. USA 95, 9047–9052 (doi:10.1073/pnas.95.15.9047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel J.1994Constructing ethnicity: creating and recreating ethnic identity and culture. Social Problems 41, 152–176 (doi:10.1525/sp.1994.41.1.03x0430n) [Google Scholar]
- O'Brien M. J. (ed.) 2008Cultural transmission and archaeology Washington, DC: Society for American Archaeology [Google Scholar]
- O'Brien M. J., Lyman R. L.2003Cladistics and archaeology Salt Lake City, UT: University of Utah Press [Google Scholar]
- O'Shaughnessy D. F., Hill A. V. S., Bowden D. K., Weatherall D. J., Clegg J. B.1990Globin genes in Micronesia: origins and affinities of Pacific Island peoples. Am. J. Hum. Genet. 46, 144–155 [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer S., Richards M.2001Fast trains, slow boats and the ancestry of the Polynesian islanders. Sci. Progress 84, 157–182 (doi:10.3184/003685001783238989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G.2000Indigenous island empires: Yap and Tonga considered. J. Pacific History 35, 5–27 (doi:10.1080/00223340050052275) [Google Scholar]
- Phillips M. J., Penny D.2003The root of the mammalian tree inferred from whole mitochondrial genomes. Mol. Phylogenet. Evol. 28, 171–185 (doi:10.1016/S1055-7903(03)00057-5) [DOI] [PubMed] [Google Scholar]
- Pietrusewsky M.1996The physical anthropology of Polynesia: a review of some cranial and skeletal studies. In Oceanic culture history: essays in honour of Roger Green (eds Davidson J. M., Irwin G., Leach B. F., Pawley A., Brown D.), pp. 343–353 Dunedin North, New Zealand: New Zealand Journal of Archaeology [Google Scholar]
- Ramachandran S., Rosenberg N. A., Zhivotovsky L. A., Feldman M.2004Robustness of the inferrence of human population structure: a comparison of X-chromosomal and autosomal microsatellites. Human Genomics 1, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., Cavalli-Sforza L. L.2005Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15 942–15 947 (doi:10.1073/pnas.0507611102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A. J.2007TreeStat tree statistics generator, Version 1.1. 27 June 2007. See http://tree.bio.ed.ac.uk/software/treestat/.
- Rogers D. S., Ehrlich P. R.2008Natural selection and cultural rates of change. Proc. Natl Acad. Sci. USA 105, 3416–3420 (doi:10.1073/pnas.0711802105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolett B. V.1996Colonisation and cultural change in the Marquesas. In Oceanic culture history: essays in honour of Roger Green. (eds Davidson J., Irwin G., Leach F., Pawley A., Brown D.), pp. 531–540 Dunedin North, New Zealand: New Zealand Journal of Archaeology [Google Scholar]
- Rolett B.2002Voyaging and interaction in ancient East Polynesia. Asian Perspectives 41, 182–194 (doi:10.1353/asi.2003.0009) [Google Scholar]
- Rosenberg N. A., et al. 2002Genetic structure of human populations. Science 298, 2381–2385 (doi:10.1126/science.1078311) [DOI] [PubMed] [Google Scholar]
- Rosenberg N. A., Mahajan S., Ramachandran S., Zhao C. F., Pritchard J. K., Feldman M. W.2005Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1, 660–671 (doi:10.1371/journal.pgen.0010070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurel L., et al. 2008Sex-specific genetic structure and social organization in central Asia: insights from a multi-locus study. PLoS Genet. 4, e1000200. (doi:10.1371/journal.pgen.1000200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seielstad M. T., Minch E., Cavalli-Sforza L. L.1998Genetic evidence for a higher female migration rate in humans. Nature Genet. 20, 278–280 (doi:10.1038/3088) [DOI] [PubMed] [Google Scholar]
- Shennan S. J., Wilkinson J. R.2001Ceramic style change and neutral evolution: a case study from Neolithic Europe. Am. Antiquity 66, 577–593 (doi:10.2307/2694174) [Google Scholar]
- Sinoto Y.1968Position of the Marquesas Islands in east Polynesia prehistory. In Prehistoric culture in Oceania: a symposium (eds Yawata I., Sinoto Y.), pp. 111–118 Honolulu, HA: Bishop Museum Press [Google Scholar]
- Smith S. P.1904Hawaiki: the original home of the Maori. Christchurch, New Zealand: Whitcombe and Tombs Ltd [Google Scholar]
- Sutton D. G.1994Conclusion: origins. In The Origins of the first New Zealanders (ed. Sutton D. G.), pp. 243–258 Auckland, New Zealand: Auckland University Press [Google Scholar]
- Terrell J. E., Hunt T. L., Gosden C.1997The dimensions of social life in the Pacific—human diversity and the myth of the primitive isolate. Curr. Anthropol. 38, 155–195 (doi:10.1086/204604) [Google Scholar]
- Tishkoff S. A., et al. 2007Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 39, 31–40 (doi:10.1038/ng1946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill P. A., et al. 2001Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Human Mutation 17, 271–280 (doi:10.1002/humu.23) [DOI] [PubMed] [Google Scholar]
- van de Vijver M. J., et al. 2002A gene-expression signature as a predictor of survival in breast cancer. New Engl. J. Med. 347, 1999–2009 (doi:10.1056/NEJMoa021967) [DOI] [PubMed] [Google Scholar]
- Vansina J.1990Paths in the rainforests: toward a history of political tradition in equatorial Africa. Madison, WI: University of Wisconsin Press [Google Scholar]
- von Cramon-Taubadel N., Lycett S. J.2008Brief communication: human cranial variation fits iterative founder effect model with African origin. Am. J. Phys. Anthropol. 136, 108–113 (doi:10.1002/ajpa.20775) [DOI] [PubMed] [Google Scholar]
- Weisler M. I., Kirch P. V.1996Interisland and interarchipelago transfer of stone tools in prehistoric Polynesia. Proc. Natl Acad. Sci. USA 93, 1381–1385 (doi:10.1073/pnas.93.4.1381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins J. F., Marlowe F. W.2006Sex-biased migration in humans: what should we expect from genetic data? Bioessays 28, 290–300 (doi:10.1002/bies.20378) [DOI] [PubMed] [Google Scholar]