Abstract
Life is based on replication and evolution. But replication cannot be taken for granted. We must ask what there was prior to replication and evolution. How does evolution begin? We have proposed prelife as a generative system that produces information and diversity in the absence of replication. We model prelife as a binary soup of active monomers that form random polymers. ‘Prevolutionary’ dynamics can have mutation and selection prior to replication. Some sequences might have catalytic activity, thereby enhancing the rates of certain prelife reactions. We study the selection criteria for these prelife catalysts. Their catalytic efficiency must be above certain critical values. We find a maintenance threshold and an initiation threshold. The former is a linear function of sequence length, and the latter is an exponential function of sequence length. Therefore, it is extremely hard to select for prelife catalysts that have long sequences. We compare prelife catalysis with a simple model for replication. Assuming fast template-based elongation reactions, we can show that replicators have selection thresholds that are independent of their sequence length. Our calculation demonstrates the efficiency of replication and provides an explanation of why replication was selected over other forms of prelife catalysis.
Keywords: evolutionary dynamics, origin of life, evolution, replication, selection threshold, mathematical biology
1. Introduction
The defining feature of biological systems is evolution. Biological organisms are products of evolutionary processes and are capable of undergoing further evolution. We think of the evolutionary process as modifying the traits of living systems. But how does evolution get started? How can we formulate a dynamical system that leads to the origin of evolution? What is there just before evolution begins? This paper is an extension of earlier work that tries to approach such questions (Nowak & Ohtsuki 2008; Manapat et al. 2009). In these papers, we have defined ‘prelife’ as a chemical system that can lead to information and diversity and that is capable of selection and mutation, but does not yet have replication. We have modelled prelife as a soup of active monomers, which can give rise to polymers. Here, we assume that some polymers have catalytic activity: they increase the rate of certain reactions in prelife. We study the criteria for the selection of prelife catalysts. We compare prelife catalysts with replicators, which have the ability to make copies of themselves.
The origin of life is a transition from chemistry to biology. There have been many theoretical and empirical studies concerning the origin of life (Oparin 1953; Crick 1968; Orgel 1968, 1992; Eigen 1971; Dyson 1982, 1999; Eigen & Schuster 1982; Kuppers 1983; Stein & Anderson 1984; Farmer et al. 1986; Szathmary & Demeter 1987; Sievers & von Kiedrowski 1994; Fontana & Schuster 1998; Luther et al. 1998; Lifson & Lifson 1999; de Duve 2005, 2007). One line of research attempts to understand how chemical processes on early Earth can spontaneously synthesize the basic building blocks of life (Miller 1953; Allen & Ponnamperuma 1967; Miller & Orgel 1974; Hargreaves et al. 1977; Rao et al. 1982; Rushdi & Simoneit 2001; Benner et al. 2002; Ricardo et al. 2004; Benner & Ricardo 2005; Wächtershäuser 2007). RNA has the ability to store genetic information and catalyse chemical reactions. Therefore, the proposal has been made that early life existed in an ‘RNA world’ (Orgel 1986; Joyce 1989, 2002; Ellington & Szostak 1990; Cech 1993; Johnston et al. 2001; Steitz & Moore 2003; Hughes et al. 2004). Bartel & Szostak (1993) discovered an RNA sequence that can catalyse RNA polymerization.
Some critics, however, argue that RNA is too complicated and fragile to arise spontaneously and that the origin of life must have been based on simpler molecules, metabolic networks or compositional genomes (Shapiro 1984, 2006, 2007; Kauffman 1986; Morowitz et al. 1988; Segre et al. 1998, 2000). Sometimes this debate is called ‘RNA first’ versus ‘metabolism first’. Our own position is the following. All currently known biological organisms use RNA or DNA. At some time, such a system must have evolved. Therefore, it is a valid programme to investigate the principles that govern the emergence of a biological polymer that carries information. When this event took place, complicated chemical cycles must have been present, which generate the compounds needed for the biological polymers. In this sense, metabolism first is certainly true, but an RNA-like system is needed for the emergence of genetic evolution.
A crucial step in the origin of life is the formation of the first cell (Szostak et al. 2001; Hanczyc et al. 2003; Chen & Szostak 2004a,b; Chen et al. 2004, 2005; Chen 2006). Fatty acids are simple molecules that can be synthesized under prebiotic conditions. They can self-assemble into bilayer vesicles, which can undergo growth and division. A decisive question is whether cells preceded information carrying polymers or vice versa. In the context of our theory, the ordering of these two events affects the population structure. If polymers came first, then their emergence can be studied in well-mixed populations. If cells came first, then the emergence of polymers should be studied in structured meta-populations containing ensembles of dividing subpopulations. From the perspective of mathematical analysis, the logical first step is to study well-mixed populations (as we will do here) and later move to evolutionary dynamics in structured populations (Nowak & May 1992; Rousset 2004; Traulsen & Nowak 2006; Ohtsuki et al. 2006; Taylor et al. 2007; Tarnita et al. 2009).
Eigen & Schuster (1977, 1979) developed a hugely influential molecular theory of chemical evolution. Their quasi-species theory studies the competition of different replicators (McCaskill 1984; Eigen et al. 1989; Nowak & Schuster 1989; Nowak 1992). Hypercycles are cooperative interactions between two or more replicators. By contrast, our theory of prelife does not begin with the presence of replicators; instead, we study mutation and selection prior to replication (Nowak & Ohtsuki 2008; Manapat et al. 2009). Therefore, we study the origin of evolution and the competition between life (which is based on replication) and prelife (chemistry without replication). Fontana & Buss (1994a,b) use λ calculus to study a generative chemistry with and without replication.
This paper is structured as follows. In §2, we present prelife and fully symmetric prelife. In §3, we discuss partial and perfect prelife catalysts. They give rise to hysteresis (bistability). In §4, we discuss a simple replicator. A brief summary of our findings is given in §5.
2. Prelife
We consider two types of activated monomers: 0* and 1*. They are produced by prebiotic chemistry, and they decay at certain rates. They can also become deactivated to generate inactivated monomers, 0 and 1. Activated monomers participate in co-polymerization reactions. Let i denote a binary string. We consider the following chemical reactions: i + 0* → i0 and i + 1* → i1. These chemical reactions can generate all binary strings. Inactivated monomers cannot be used for the elongation reactions, but they can react with active monomers; for example, 0 + 1* → 01.
The chemical kinetics of prelife are described by the following system of linear differential equations
| 2.1 |
The index i represents all binary strings (or sequences). The abundance of sequence i is denoted by xi. Longer strings are produced from shorter ones by adding either a 0* or a 1* on the right side. Each string, i, has one precursor, denoted by i′, and two followers, denoted by i0 and i1 (figure 1a). For example, 010 is the precursor of 0101. The two followers of 0101 are 01010 and 01011. For the precursors of strings 0 and 1, we set x0′ = x1′ = 1. The rate constants ai denote the rate at which string i is formed from string i′ by addition of an activated monomer (which is either 0* or 1*). Equation (2.1) assumes that the concentration of activated monomers are at constant steady-state levels. This happens, for example, when the decay rate of activated monomers is greater than the rate at which they are used up in prelife reactions. In the following, we think that the steady-state density of activated monomers are already subsumed in rate constants. All strings are removed (decay) at rate d.
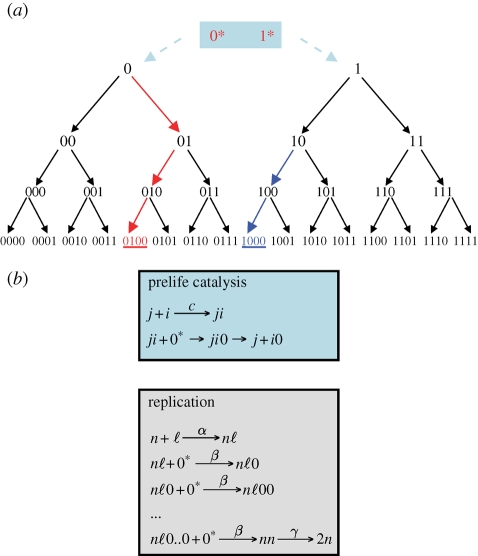
Figure 1.
(a) The tree of prelife. Activated monomers, 0* and 1*, form (random) polymers. Activated monomers can become deactivated, 0* → 0 and 1* → 1. Activated monomers can attach to the end of strings. For simplicity, we assume that all strings grow only on one side. Therefore, each string has one immediate precursor and two immediate followers. Each sequence has exactly one production lineage. The arrows indicate all the chemical reactions of prelife (up to binary strings of length 4). For catalysed prelife, we assume that some strings have the ability to catalyse certain reactions. There can be chemical hysteresis and multiple steady states. The perfect prelife catalyst is a string that enhances the rates of all chemical reactions in its own lineage (as shown in red for the string 0100). Partial catalysis occurs if a string catalyses some reactions in its own lineage (as shown in blue for the string 1000). (b) Reaction mechanisms of prelife catalysis and replication. The prelife catalyst, sequence j, reacts with sequence i to form the complex ji. Then sequence i is extended by addition of an active monomer, 0*. Subsequently, the complex dissociates. For replication, the template n binds to the primer ℓ. Then, the primer is extended by addition of active monomers. The catalytic activity of the template walks along the growing primer. Finally, the completed double strand dissociates. The rate constants of replication are discussed in appendix C.
Prelife dynamics define a tree (more precisely a double tree) with the two roots, 0 and 1. This ‘tree of prelife’ has infinitely many lineages (figure 1a). The half of all lineages starts from 0 and the other half starts from 1. A lineage is a sequence of infinitely many strings that are followers of each other. For example, one such lineage contains all all-0 strings: 0, 00, 000, … . Another lineage contains alternating sequences (that start with 0): 0, 01, 010, 0101, … . We could also consider prelife with more than two types of monomers, but this extension is not necessary for the purpose of this paper.
For fully symmetric prelife, we assume a0 = a1 = λ/2 and ai = a for all other sequences, i. In this case, all sequences of length n have the same equilibrium abundance, [λ/2a][a/(2a + d)]n. The total abundance of all strings is λ/d.
3. Prelife catalysis
Prelife catalysis means that some sequences have the ability to enhance the rates of certain prelife reactions. For example, sequence j might catalyse the reaction i + 0* → i0 at rate c (figure 1b). In this case, the rate of formation of sequence i0 can be written as ai0xi + cxixj. The first term denotes the rate of the uncatalysed reaction. The second term denotes the rate of the catalysed reaction, which is proportional to the abundance of the catalyst, xj. In a subsequent paper, we plan to study sets of prelife catalysts, but here we focus on the dynamics of individual catalysts. We consider a prelife catalyst that enhances some (or all) of its upstream reactions (figure 1a). Our aim is to calculate the equilibrium abundance of such a catalyst. Therefore, we can study the conditions for selection of catalysed over uncatalysed prelife.
Let us consider fully symmetric prelife. Without loss of generality, we assume that the catalyst is the all-0 sequence of length n, which we denote by 0n. There are n − 1 upstream reactions in the lineage, leading from 0 to 0n. Each reaction, 0k + 0* → 0k+1, is enhanced by ck times the abundance of 0n. The parameter ck can be either zero or positive. In order to understand this system, we study the abundances of sequences of the form 0k, where k = 1,2, … . We change our previous notation and let xk denote the abundance of 0k. We have the following system of ordinary differential equations:
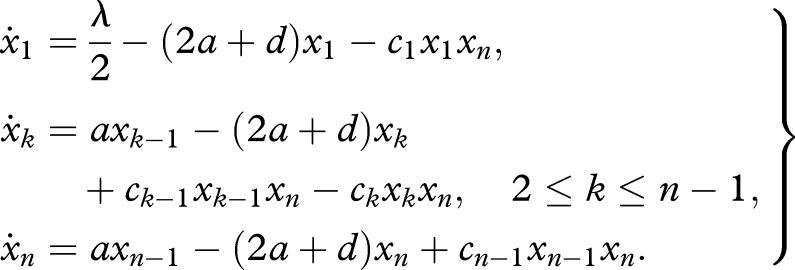 |
3.1 |
We are interested in the equilibrium abundance of the prelife catalyst, which we denote by x̂n. A straightforward calculation shows that it is given as a root of the following polynomial equation:
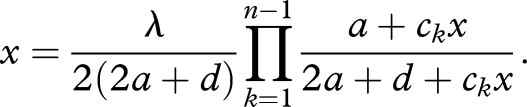 |
3.2 |
(a). Partial catalysis
Imagine a prelife catalyst of length n that catalyses m(1 ≤ m ≤ n − 1) of its n − 1 upstream reactions. For analytical simplicity, we assume that ck is either c or 0. That is, m entries of the vector (c1, …, cn−1) are c and the others are zero. In this case, the equilibrium abundance, x̂n, is given as a root of the equation
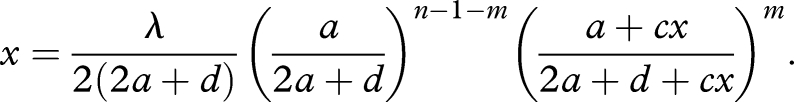 |
3.3 |
Note that equation (3.3) does not depend on which particular m reactions out of the n − 1 upstream reactions are enhanced. For a general c, equation (3.3) cannot be solved explicitly. Nevertheless, we obtain the following result. There exists a critical threshold of m, denoted by mcr. If m ≤ mcr, then the equilibrium abundance, x̂n, is a monotone increasing function of the catalytic activity, c. If m > mcr, then we observe a hysteresis effect: for an interval of intermediate c-values, equation (3.3) has three positive roots: two of them correspond to stable equilibria and one to an unstable equilibrium. Which of the two stable equilibria is reached depends on the initial abundance of the catalyst. For a detailed analysis, see appendix A.
(b). Perfect catalysis
As a special case, let us study a sequence that enhances the rates of all of its upstream reactions. Therefore, we have m = n − 1. The equilibrium abundance, x̂n, is given as a root of the polynomial equation
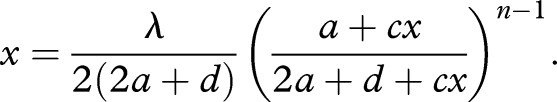 |
3.4 |
For c = ∞, we obtain the maximum abundance, x̂n = λ/2(2a + d)(≡x̂nmax). For a general c, we obtain the following result. There exists a threshold for the length of the catalyst, ncr. When n ≤ ncr (figure 2), the equilibrium abundance x̂n is a monotone increasing function of c. When n > ncr (figure 3), we find the two branches of stable equilibria (the solid lines in figure 3a) and one unstable equilibrium between them (the dotted line in figure 3a). The upper branch exists for c ≥ c1, whereas the lower branch exists for c ≤ c2. For c1 ≤ c ≤ c2, the equilibrium abundance, x̂n, depends on its initial abundance. If the catalyst is initially rare, then it will reach the lower equilibrium (figure 3b). If the catalyst is initially present at high abundance, then it will reach the higher equilibrium (figure 3c).
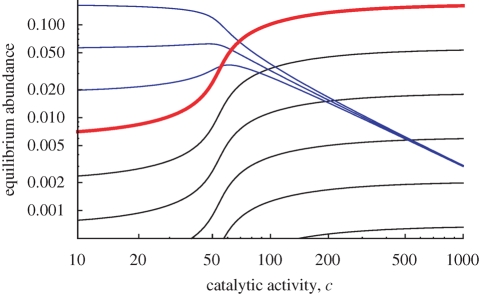
Figure 2.
The equilibrium abundances of the all-0 strings, 01, 02, 03, … , are shown as a function of the catalytic activity, c. The catalyst, 04, is shown in red. Shorter sequences are shown in blue and longer sequences in black. We use a = 1, d = 1 and λ = 1. For these parameters, the critical length of the catalyst is given by ncr = 3 + √3 = 4.732. The length of the catalyst in this example, n = 4, is below this threshold. Therefore, the equilibrium abundance of the catalyst (red curve) increases monotonically with c.
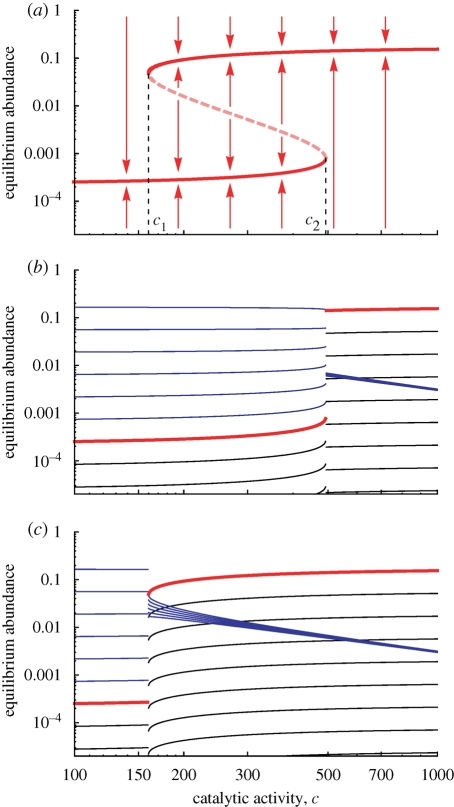
Figure 3.
The equilibrium abundance of the catalyst, x̂n (red), is shown as a function of its catalytic activity, c. The catalyst enhances all of its upstream reactions. We use the same parameter as for figure 2, but the length of the catalyst, n = 7, is above the critical value ncr in this case. (a) The system shows bistability for c1 ≤ c ≤ c2. The solid lines in red represent stable equilibria. The dotted line in red represents unstable equilibria. The red arrows represent the direction of the change from initial abundance to final abundance of the catalyst. (b) When the catalyst is initially rare, we observe a discontinuous jump in the abundance at c = c2. The blue lines represent the abundances of sequences 01 to 06 (upstream). The red line represents the sequence, 07. The black lines represent sequences 08 and longer (downstream). For c < c2, shorter sequences have higher abundance. For c > c2, the catalyst is most abundant. (c) When the catalyst is initially abundant, we observe a discontinuous jump in the abundance at c = c1. For c < c1, shorter sequences have higher abundance. For c > c1, the catalyst is most abundant.
The first threshold, c1, is the critical value of c that is needed to maintain the catalyst at high abundance. The second threshold, c2, is the critical value that is needed to initiate high abundance of the catalyst when it is not common in the beginning. Therefore, we call c1 and c2 ‘maintenance threshold’ and ‘initiation threshold’, respectively. For large n, we obtain
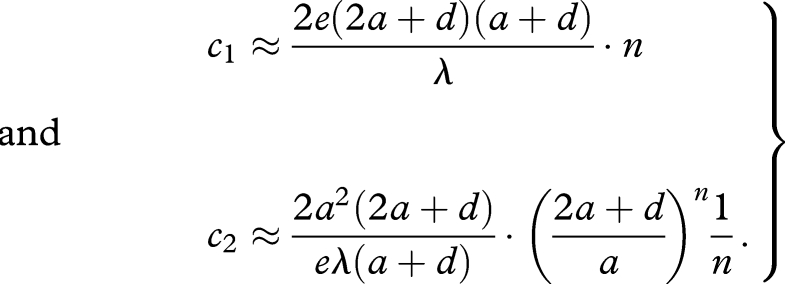 |
3.5 |
Here e = 2.718281… (appendix B). The maintenance threshold, c1, grows as a linear function of the sequence length, n. The initiation threshold, c2, grows (approximately) as an exponential function of the sequence length, n. Therefore, it is extremely difficult to select for a catalyst that has a long sequence. At the same time, it is unlikely that short sequences have good (or any) catalytic activity.
An intuitive biological summary is the following. The system has two equilibria: E1 and E2. At E1, the catalyst has low abundance; all sequences have almost the same abundances as in uncatalysed prelife. At E2, the catalyst has high abundance; it ‘dominates’ the population (figure 3). We say that at equilibrium E2, the catalyst has been selected over uncatalysed prelife. If the catalytic activity, c, is less than the threshold c1, then only E1 is stable. If c is greater than c2, then only E2 is stable. If c is between c1 and c2, then both equilibria are stable. Which one will be chosen depends on the initial condition. Therefore, if the prelife catalyst is already present at high abundance, then it will remain so as long as c is greater than c1. On the other hand, if the catalyst is initially not present at high abundance, then it will gain high abundance only if c is greater than c2. This ‘chemical hysteresis’ is caused by the bistability of our system.
4. Replication
(a). The primer is a monomer
Imagine that a sequence i can make a copy of itself by using activated monomers. For fully symmetric prelife, we can once again assume without loss of generality that the replicator is the all-0 sequence of length n, denoted by 0n. The replication starts from the primer, 0, and incorporates activated monomers 0* for elongation.
The difference between the perfect prelife catalyst and the replicator is the following. The prelife catalyst can attach to a sequence and increase the rate at which the activated monomer is added. Afterwards, the catalyst dissociates from the elongated sequence. By contrast, the replicator attaches to a primer and then holds on to the growing sequence. Therefore, the catalytic activity of the replicator can ‘walk along’ the entire sequence. In both cases, we assume that the catalysed elongation step is not rate limiting. Consequently, for the replicator, a single rate-limiting bimolecular reaction is sufficient (attaching between template and primer). For the perfect prelife catalyst, we need n − 1 rate-limiting bimolecular reactions (see figure 1b).
As before, let xk be the abundance of the sequence in the form of 0k (k = 1, … , n). The consumption of primers is described by the term −rx1xn. If we assume perfect replication, two copies of replicators are produced from one primer and one replicator. Therefore, the production of replicators is described by the term rx1xn. In a general case, we obtain the following system of differential equations:
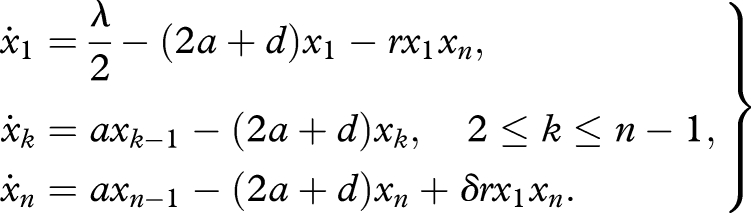 |
4.1 |
Here the parameter δ represents the efficacy of replication. A perfect replication leads to δ = 1. If replication is always unsuccessful, we have δ = −1, because replicators are consumed in vain. In general, δ takes a value between −1 and 1. In appendix C, we provide a derivation of equation (4.1) by examining the detailed mechanism of the replication process. A key assumption there is that the template-based elongation steps are not rate limiting. In the following, we study δ > 0, otherwise replicators are never selected.
From equation (4.1), it is easy to see that the equilibrium abundance of the replicator, x̂n, is given as the positive root of the following quadratic equation:
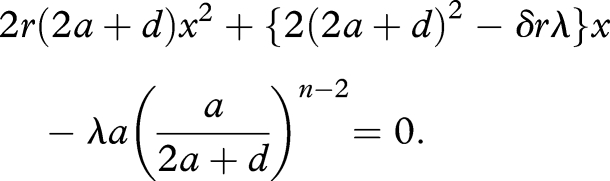 |
4.2 |
For large r, we obtain x̂nmax = δλ/2(2a + d), which agrees with x̂nmax in the case of c = ∞ for prelife catalysts (see §3b), but up to the factor δ. However, the dependence of the equilibrium abundance on r is qualitatively different from that on c in prelife catalysts. It is shown that if the efficacy of replication exceeds δ* = (a/2a + d)n−1, the equilibrium abundance x̂n monotonically increases with r. Bistability is never observed (figure 4). There exists a critical threshold of r given by
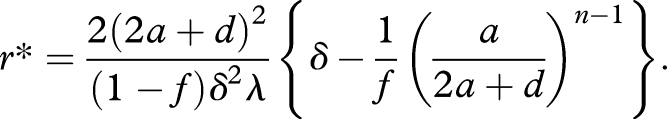 |
4.3 |
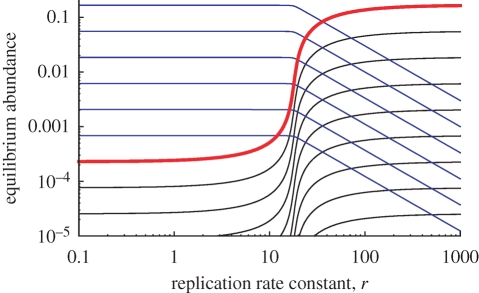
Figure 4.
The equilibrium abundances of the replicator (red), shorter sequences (blue) and longer sequences (black) are shown as functions of the replication rate constant, r. We use a = 1, d = 1, λ = 1, δ = 1 and n = 7. From equation (4.3), the threshold value of r for the equilibrium abundance of replicators to exceed 10 per cent of the theoretical maximum is predicted as r* =19.7256… .
If r > r* holds, the equilibrium abundance of the replicator is more than a fraction f (0 < f < 1) of its theoretical maximum, i.e. x̂n > fx̂nmax. Interestingly, the threshold equation (4.3) converges to a fixed value, 2(2a + d)2/(1 − f)δλ, for large n. In contrast to prelife catalysts, long replicators can be selected over prelife.
(b). The primer is not a monomer
Now we consider a scenario where the primer of replication is not a monomer, but a sequence of length ℓ (>1). As before, suppose that the replicator is 0n. The primer of the replication is given by 0ℓ (1 < ℓ< n). Replication is described by the term rxℓxn. Taking into account the efficacy of replication, we obtain the following system of differential equations:
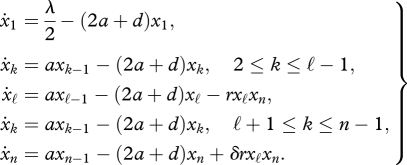 |
4.4 |
A calculation shows that the equilibrium abundance of the replicator, denoted by x̂n, is given by the positive root of the quadratic equation
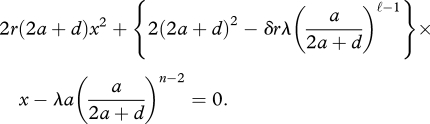 |
4.5 |
The equilibrium abundance of replicators monotonically increases with r if and only if the efficacy exceeds
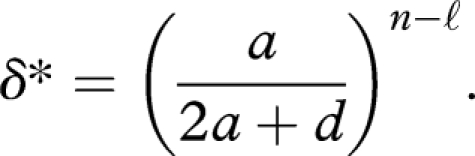 |
4.6 |
Therefore, for a fixed length of the replicator, n, the required efficacy grows exponentially with the length of the primer, ℓ. The replicator that requires a longer primer is less likely to be selected.
Suppose the efficacy exceeds equation (4.6). We obtain x̂n = [δλ/2a]·[a/(2a + d)]ℓ (≡x̂nmax) at r → ∞. The critical threshold of the replication constant, denoted by r*, is given by
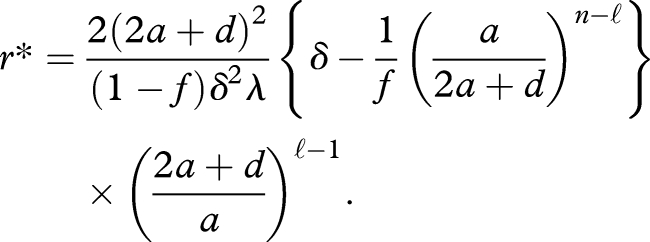 |
4.7 |
This threshold means that if r > r*, then the equilibrium abundance of the replicator exceeds a fraction f (0 < f < 1) of its theoretical maximum, i.e. x̂n > fx̂nmax For a fixed primer length, ℓ, the threshold (4.7) tends to a constant, r* = 2(2a + d)2/[(2a + d)/a]ℓ−1(1 − f)δλ for large n. Thus, the critical threshold (4.7) converges to a fixed value for increasing n, which is consistent with the result found in §4a. The intuitive explanation for this finding is that the catalysed elongation steps of the replication process are not rate limiting. Therefore, the length of the replicator does not affect the rate of replication.
5. Discussion
We have studied the selection criteria for prelife catalysts and replicators. By prelife catalysts, we mean sequences that can enhance certain reactions in prelife. The perfect prelife catalyst is a (hypothetical) sequence that enhances the rates of all reactions in its own production lineage. We show that even for a perfect prelife catalyst, it is very difficult to achieve a high equilibrium abundance, because the catalytic activity has to exceed a threshold value that grows exponentially with the sequence length. By contrast, sequences that can replicate can achieve high equilibrium abundance even if they have considerable length. The critical replication rate is almost independent of the length of the replicator. However, the required efficacy of replication grows with the length of the primer.
Our selection thresholds arise, because there is competition between prelife and catalytic prelife, on the one hand, and between prelife and replication (life), on the other hand. The latter is especially interesting because prelife is needed to build the sequences for replication (the replicator and the primer), but then prelife and life compete for the same resources (activated monomers). This tension between prelife and life leads to the origin of evolution.
Acknowledgements
Support from the John Templeton Foundation, the NSF/NIH joint program in mathematical biology (NIH grant no. R01GM078986), the Bill and Melinda Gates Foundation (Grand Challenges grant 37874), and J. Epstein is gratefully acknowledged.
Appendix A: Thresholds for bistability in prelife catalysts
First, we will study equation (3.4), which is rewritten as
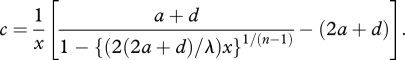 |
A1 |
Therefore, we can regard c as a single-valued function of x. Let c(x) be the right-hand side of equation (A 1). Its derivative with respect to x is
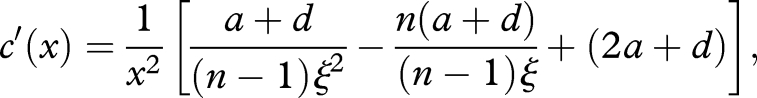 |
A2 |
where ξ ≡ 1 − [2(2a + d)x/λ][1/(n−1)]. As c is non-negative, from equation (A 1), we need 0 < ξ ≤ (a + d)/(2a + d). Solving c′(x) = 0 leads to
| A3 |
Let D be the discriminant of the quadratic equation of ξ, equation (A 3). D vanishes at
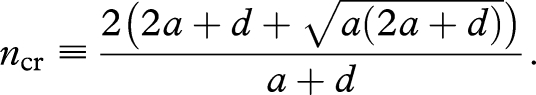 |
A4 |
Also, D is strictly negative at n = 2. Thus, if 2 ≤ n ≤ ncr, then D ≤ 0, which means that c′(x) is always non-negative. Therefore, c = c(x) is a monotone increasing function of x, so is its inverse function x = x(c). If n> ncr, then D > 0, which means that equation (A 3) has two distinct roots. We can prove that these two roots satisfy 0 < ξ< (a + d)/(2a + d). Therefore, c = c(x) has one local maximum and one local minimum, leading to the S-shaped curve in figure 3a.
Next we study equation (3.3), which can be rewritten in the same form as equation (3.4) by setting n′ ≡ m + 1 and λ′ ≡ λ[a/(2a + d)]n−1−m. Therefore, similar conclusions can be drawn. If n′ > ncr, or equivalently, if
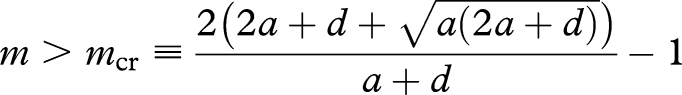 |
A5 |
holds, then the system shows bistability. There are the maintenance threshold, c1, and the initiation threshold, c2.
Appendix B: Asymptotic values of c1 and c2
First, we study a perfect catalyst that catalyses all of its upstream reactions. When n > ncr, solving equation (A 3) yields
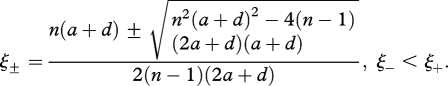 |
B1 |
Remember that we have defined ξ as ξ = 1 − [2(2a + d)x/λ][1/(n−1)], so x± = [λ/2(2a + d)](1 − ξ±)n−1. Note that x+ < x−. The function c = c(x) has its local maximum at x = x+ and its local minimum at x = x−. We obtain c1 = c(x−) and c2 = c(x+) (figure 3a). A direct calculation shows the asymptotic estimates of these values shown in the main text. We use (1 + 1/n)n ≈ e = 2.718281… . for large n.
Similarly, for a catalyst that catalyses a fraction θ(=m/(n − 1)) of its upstream reactions with the catalytic activity c, we obtain the following asymptotic estimates of c1 and c2 for large n:
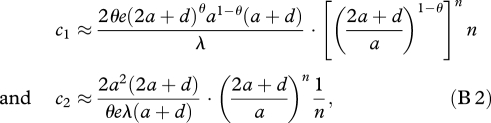 |
where 0 < θ ≤ 1. Therefore, the two thresholds grow (approximately) exponentially with n when the catalyst enhances some of its upstream reactions (0 < θ< 1). Only when the catalyst enhances all of its upstream reactions (θ = 1) does the maintenance threshold, c1, grow linearly with n.
Appendix C: A detailed derivation of equation (4.1) in the main text
Here we explain the underlying mechanics of replication and provide a detailed derivation of equation (4.1). Let 0n denote the replicator. As in the main text, we denote the abundance of sequence 0k by xk (k = 1, … , n). We assume direct as opposed to complementary replication. The replication process starts when a (inactivated) monomer 0, which is a primer, attaches to a replicator, which is a template. This reaction is described by the term αx1xn. The resulting complex between the template and the primer grows in length by incorporating activated monomers 0* one by one until it becomes the full double strand of 0n. We call these steps elongation reactions. Let yk denote the abundance of the complex between the template (of length n) and the growing sequence that has reached length k. The abundance of the full double strand is given by yn. For simplicity, we assume that the reaction rate of each elongation step is constant and given by β. The full double strand separates at rate γ (for example, via temperature oscillations). All sequences and complexes decay at rate d. We obtain the following system of differential equations:
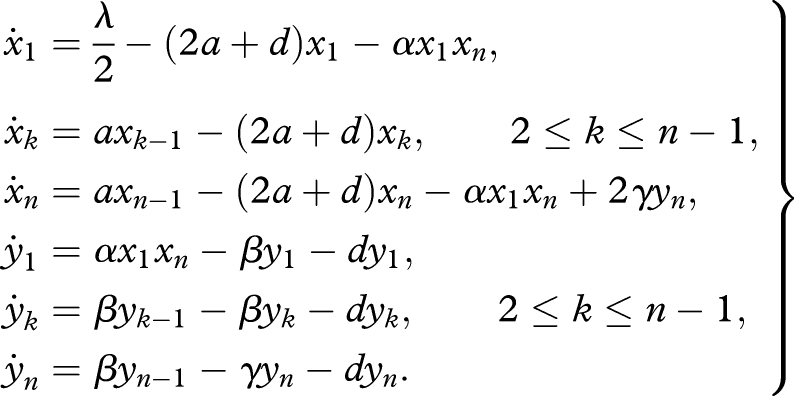 |
C1 |
Remember that the stationary density of activated monomers is subsumed in the rate constants, λ, a and β. We assume that the rate of template-based elongation, β, is much faster than other rate constants such as a, d and γ. For the quasi-equilibrium abundance of full double strands, we obtain
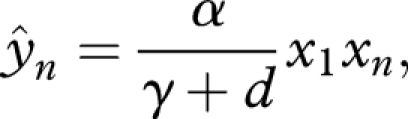 |
C2 |
and therefore
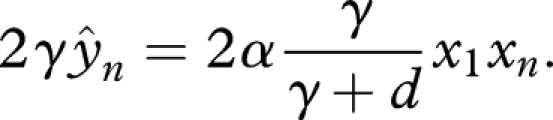 |
C3 |
Substituting equation (C 3) into the first three lines of equation (C 1) yields
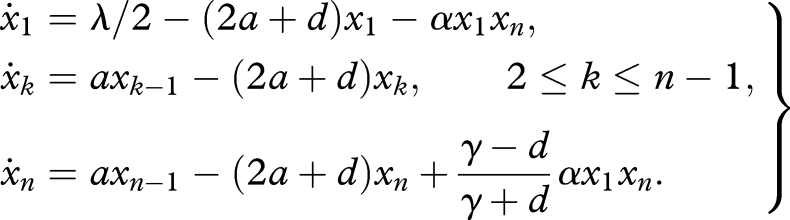 |
C4 |
Rewriting parameters as r = α and δ = (γ − d)/(γ + d) reproduces equation (4.1) in the main text.
We note that the assumption of fast elongation (large β) is entirely consistent with our model for prelife catalysis, which also contains an implicit assumption of a fast ‘elongation’ step. The prelife catalyst, 0n, binds its target sequence, 0k, to form a complex [0n 0k]. This complex reacts very fast with an activated monomer, 0*, to give rise to [0n 0k+1]. Subsequently, the complex dissociates into 0n and 0k+1. Equation (3.1) assumes that the elongation reaction is not rate limiting. Therefore, a replicator with a fast elongation reaction is the proper comparison for the prelife catalyst described by equation (3.1). The difference between the replicator and the prelife catalyst is that the catalytic activity of the replicator ‘walks along’ the sequence, whereas the prelife catalyst can accelerate only a single elongation step and dissociates subsequently.
References
- Allen W. V., Ponnamperuma C.1967A possible prebiotic synthesis of monocarboxylic acids. Curr. Mod. Biol. 1, 24–28 [DOI] [PubMed] [Google Scholar]
- Bartel D. P., Szostak J. W.1993Isolation of new ribozymes from a large pool of random sequences. Science 261, 1411–1418 (doi:10.1126/science.7690155) [DOI] [PubMed] [Google Scholar]
- Benner S. A., Ricardo A.2005Planetary systems biology. Mol. Cell 17, 471–472 (doi:10.1016/j.molcel.2005.02.005) [DOI] [PubMed] [Google Scholar]
- Benner S. A., Caraco M. D., Thomson J. M., Gaucher E. A.2002Planetary biology-paleontological, geological, and molecular histories of life. Science 296, 864–868 (doi:10.1126/science.1069863) [DOI] [PubMed] [Google Scholar]
- Cech T. R.1993The efficiency and versatility of catalytic RNA: implications for an RNA world. Gene 135, 33–36 (doi:10.1016/0378-1119(93)90046-6) [DOI] [PubMed] [Google Scholar]
- Chen I. A.2006The emergence of cells during the origin of life. Science 314, 1558–1559 (doi:10.1126/science.1137541) [DOI] [PubMed] [Google Scholar]
- Chen I. A., Szostak J. W.2004aA kinetic study of the growth of fatty acid vesicles. Biophys. J. 87, 988–998 (doi:10.1529/biophysj.104.039875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A., Szostak J. W.2004bMembrane growth can generate a transmembrane pH gradient in fatty acid vesicles. Proc. Natl Acad. Sci. USA 1017965–7970 (doi:10.1073/pnas.0308045101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A., Roberts R. W., Szostak J. W.2004The emergence of competition between model protocells. Science 305, 1474–1476 (doi:10.1126/science.1100757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. A., Salehi-Ashtiani K., Szostak J. W.2005RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 127, 13 213–13 219 (doi:10.1021/ja051784p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H.1968The origin of the genetic code. J. Mol. Biol. 38, 367–379 (doi:10.1016/0022-2836(68)90392-6) [DOI] [PubMed] [Google Scholar]
- de Duve C.2005The onset of selection. Nature 433, 581–582 (doi:10.1038/433581a) [DOI] [PubMed] [Google Scholar]
- de Duve C.2007Chemistry and selection. Chem. Biodivers. 4, 574–583 (doi:10.1002/cbdv.200790051) [DOI] [PubMed] [Google Scholar]
- Dyson F. J.1982A model for the origin of life. J. Mol. Evol. 18, 344–350 (doi:10.1007/BF01733901) [DOI] [PubMed] [Google Scholar]
- Dyson F.1999Origins of life. Cambridge, UK: Cambridge University Press [Google Scholar]
- Eigen M.1971Molecular self-organization and the early stages of evolution. Q. Rev. Biophys. 4, 149–212 (doi:10.1017/S0033583500000627) [PubMed] [Google Scholar]
- Eigen M., Schuster P.1977The hyper cycle. A principle of natural self-organization. Part A: emergence of the hyper cycle. Naturwissenschaften 64, 541–565 (doi:10.1007/BF00450633) [DOI] [PubMed] [Google Scholar]
- Eigen M., Schuster P.1979The hypercycle, a principle of natural self-organization. Berlin, Germany: Springer-Verlag; [DOI] [PubMed] [Google Scholar]
- Eigen M., Schuster P.1982Stages of emerging life: five principles of early organization. J. Mol. Evol. 19, 47–61 (doi:10.1007/BF02100223) [DOI] [PubMed] [Google Scholar]
- Eigen M., McCaskill J., Schuster P.1989The molecular quasi-species. Adv. Chem. Phys. 75, 149–263 (doi:10.1002/9780470141243.ch4) [Google Scholar]
- Ellington A. D., Szostak J. W.1990In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (doi:10.1038/346818a0) [DOI] [PubMed] [Google Scholar]
- Farmer J. D., Kauffman S. A., Packard N. H.1986Autocatalytic replication of polymers. Phys. D 22, 50–67 (doi:10.1016/0167-2789(86)90233-2) [Google Scholar]
- Fontana W., Buss L. W.1994aWhat would be conserved if ‘the tape were played twice’? Proc. Natl Acad. Sci. USA 91, 757–761 (doi:10.1073/pnas.91.2.757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana W., Buss L. W.1994b‘The arrival of the fittest’: toward a theory of biological organization. Bull. Math. Biol. 56, 1–64 [Google Scholar]
- Fontana W., Schuster P.1998Continuity in evolution: on the nature of transitions. Science 280, 1451–1455 (doi:10.1126/science.280.5368.1451) [DOI] [PubMed] [Google Scholar]
- Hanczyc M. N., Fujikawa S. M., Szostak J. W.2003Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science 302, 618–622 (doi:10.1126/science.1089904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves W. R., Mulvihill S., Deamer D. W.1977Synthesis of phospholipids and membranes in prebiotic conditions. Nature 266, 78–80 (doi:10.1038/266078a0) [DOI] [PubMed] [Google Scholar]
- Hughes R. A., Robertson M. P., Ellington A. D., Levy M.2004The importance of prebiotic chemistry in the RNA world. Curr. Opin. Chem. Biol. 8, 629–633 (doi:10.1016/j.cbpa.2004.09.007) [DOI] [PubMed] [Google Scholar]
- Johnston W. K., Unrau P. J., Lawrence M. S., Glasner M. E., Bartel D. P.2001RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 (doi:10.1126/science.1060786) [DOI] [PubMed] [Google Scholar]
- Joyce G. F.1989RNA evolution and the origins of life. Nature 338, 217–224 (doi:10.1038/338217a0) [DOI] [PubMed] [Google Scholar]
- Joyce G. F.2002The antiquity of RNA-based evolution. Nature 418, 214–221 (doi:10.1038/418214a) [DOI] [PubMed] [Google Scholar]
- Kauffman S. A.1986Autocatalytic sets of proteins. J. Theor. Biol. 119, 1–24 (doi:10.1016/S0022-5193(86)80047-9) [DOI] [PubMed] [Google Scholar]
- Kuppers B.1983Molecular theory of evolution: outline of a physico-chemical theory of the origin of life Berlin, Germany: Springer-Verlag [Google Scholar]
- Lifson S., Lifson H.1999A model of prebiotic replication: survival of the fittest versus extinction of the unfittest. J. Theor. Biol. 199, 425–433 (doi:10.1006/jtbi.1999.0969) [DOI] [PubMed] [Google Scholar]
- Luther A., Brandsch R., von Kiedrowski G.1998Surface-promoted replication and exponential amplification of DNA analogues. Nature 396, 245–248 (doi:10.1038/24343) [DOI] [PubMed] [Google Scholar]
- Manapat M., Ohtsuki H., Bürger R., Nowak M. A.2009Originator dynamics. J. Theor. Biol. 256, 586–595 (doi:10.1016/j.jtbi.2008.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J.1984A stochastic theory of macromolecular evolution. Biol. Cybern. 50, 63–73 (doi:10.1007/BF00317940) [Google Scholar]
- Miller S. L.1953A production of amino acids under possible primitive earth conditions. Science 117, 528–529 (doi:10.1126/science.117.3046.528) [DOI] [PubMed] [Google Scholar]
- Miller S. L., Orgel L. E.1974The origins of life on the Earth Englewood Cliffs, NJ: Prentice-Hall [Google Scholar]
- Morowitz H. J., Heinz B., Deamer D. W.1988The chemical logic of a minimum protocell. Orig. Life Evol. Biosph. 18, 281–287 (doi:10.1007/BF01804674) [DOI] [PubMed] [Google Scholar]
- Nowak M. A.1992What is a quasispecies? Trends Ecol. Evol. 7, 118–121 (doi:10.1016/0169-5347(92)90145-2) [DOI] [PubMed] [Google Scholar]
- Nowak M. A., May R. M.1992Evolutionary games and spatial chaos. Nature 359, 826–829 (doi:10.1038/359826a0) [Google Scholar]
- Nowak M. A., Ohtsuki H.2008Prevolutionary dynamics and the origin of evolution. Proc. Natl Acad. Sci. USA 105, 14 924–14 927 (doi:10.1073/pnas.0806714105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. A., Schuster P.1989Error thresholds of replication in finite populations. Mutation frequencies and the onset of Muller's ratchet. J. Theor. Biol. 137, 375–395 (doi:10.1016/S0022-5193(89)80036-0) [DOI] [PubMed] [Google Scholar]
- Ohtsuki H., Hauert C., Lieberman E., Nowak M. A.2006A simple rule for the evolution of cooperation on graphs and social networks. Nature 441, 502–505 (doi:10.1038/nature04605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparin A. I.1953The origin of life. New York, NY: Dover [Google Scholar]
- Orgel L. E.1968Evolution of the genetic apparatus. J. Mol. Biol. 38, 381–393 (doi:10.1016/0022-2836(68)90393-8) [DOI] [PubMed] [Google Scholar]
- Orgel L. E.1986RNA catalysis and the origins of life. J. Theor. Biol. 123, 127–149 (doi:10.1016/S0022-5193(86)80149-7) [DOI] [PubMed] [Google Scholar]
- Orgel L. E.1992Molecular replication. Nature 358, 203–209 (doi:10.1038/358203a0) [DOI] [PubMed] [Google Scholar]
- Rao M., Eichberg J., Oro J.1982Synthesis of phosphatidylcholine under possible primitive Earth conditions. J. Mol. Evol. 18, 196–202 (doi:10.1007/BF01733046) [DOI] [PubMed] [Google Scholar]
- Ricardo A., Carrigan M. A., Olcott A. N., Benner S. A.2004Borate minerals stabilize ribose. Science 303, 196 (doi:10.1126/science.1092464) [DOI] [PubMed] [Google Scholar]
- Rousset F.2004Genetic structure and selection in subdivided populations Princeton, NJ: Princeton University Press [Google Scholar]
- Rushdi A. I., Simoneit B. R.2001Lipid formation by aqueous Fischer–Tropsch-type synthesis over a temperature range of 100 to 400°C. Orig. Life Evol. Biosph. 31, 103–118 (doi:10.1023/A:1006702503954) [DOI] [PubMed] [Google Scholar]
- Segre D., Lancet D., Kedem O., Pilpel Y.1998Graded autocatalysis replication domain (GARD): kinetic analysis of self-replication in mutually catalytic sets. Orig. Life Evol. Biosph. 28, 501–514 (doi:10.1023/A:1006583712886) [PubMed] [Google Scholar]
- Segre D., Ben-Eli D., Lancet D.2000Compositional genomes: prebiotic information transfer in mutually catalytic noncovalent assemblies. Proc. Natl Acad. Sci. USA 97, 4112–4117 (doi:10.1073/pnas.97.8.4112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R.1984The improbability of prebiotic nucleic acid synthesis. Orig. Life Evol. Biosph. 14, 565–570 (doi:10.1007/BF00933705) [DOI] [PubMed] [Google Scholar]
- Shapiro R.2006Small molecule interactions were central to the origin of life. Q. Rev. Biol. 81, 105–125 (doi:10.1086/506024) [DOI] [PubMed] [Google Scholar]
- Shapiro R.2007A simpler origin for life. Sci. Am. 296, 46–53 [DOI] [PubMed] [Google Scholar]
- Sievers D., von Kiedrowski G.1994Self-replication of complementary nucleotide-based oligomers. Nature 369, 221–224 (doi:10.1038/369221a0) [DOI] [PubMed] [Google Scholar]
- Stein D. L., Anderson P. W.1984A model for the origin of biological catalysis. Proc. Natl Acad. Sci. USA 81, 1751–1753 (doi:10.1073/pnas.81.6.1751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Moore P. B.2003RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 28, 411–418 (doi:10.1016/S0968-0004(03)00169-5) [DOI] [PubMed] [Google Scholar]
- Szathmary E., Demeter L.1987Group selection of early replicators and the origin of life. J. Theor. Biol. 128, 463–486 (doi:10.1016/S0022-5193(87)80191-1) [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Bartel D. P., Luisi P. L.2001Synthesizing life. Nature 209, 387–390 (doi:10.1038/35053176) [DOI] [PubMed] [Google Scholar]
- Tarnita C. E., Antal T., Ohtsuki H., Nowak M. A.2009Evolutionary dynamics in set structured populations. Proc. Natl Acad. Sci. USA 106, 8601–8604 (doi:10.1073/pnas.0903019106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. D., Day T., Wild G.2007Evolution of cooperation in a finite homogeneous graph. Nature 447, 469–472 (doi:10.1038/nature05784) [DOI] [PubMed] [Google Scholar]
- Traulsen A., Nowak M. A.2006Evolution of cooperation by multilevel selection. Proc. Natl Acad. Sci. USA 103, 10 952–10 955 (doi:10.1073/pnas.0602530103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G.2007On the chemistry and evolution of the pioneer organism. Chem. Biodivers. 4, 584–602 (doi:10.1002/cbdv.200790052) [DOI] [PubMed] [Google Scholar]