Abstract
Seed germination is the first adaptive decision in the development of many land plants. Advances in genetics and molecular physiology have taught us much about the control of germination using the model plant Arabidopsis thaliana. Here we review the current state of the art with an emphasis on mechanistic considerations and explore the potential impact of a systems biology approach to the problem.
Keywords: seed germination, systems biology, Arabidopsis
1. Systems biology of seed germination control
Seed dormancy is an important developmental checkpoint, allowing plants to regulate when and where they grow. The germination of dormant seeds is promoted by environmental signals that allow plants to restrict the timing of their establishment to certain seasons, rainfall or to the removal of vegetation cover. Dormancy is also important for the spatial control of seedling establishment, as germination generally terminates dispersal: one example is that a seed that requires light for germination has a mechanism to restrict germination if too far below the soil surface. Seed dormancy is therefore one of the most important adaptive traits in plants.
Dormancy is usually defined as failure of a viable seed to germination in conditions favourable for germination to proceed (Bewley 1997), which under experimental conditions usually means that ambient temperature is suitable for germination and enough water is available. Dormancy is also separated from germination sensu stricto by seed biologists, because dormancy states can change in the absence of germination. This is known as dormancy cycling, and is well characterized in agricultural systems where the seed dormancy of weeds makes them hard to eradicate. In this case dormancy states other than that present at maturity are usually defined collectively by the term ‘secondary dormancy’, and these can be further sub-divided by the environmental conditions that are successful in forcing germination of those seeds. The factors controlling cycling between different dormant states in buried seeds are still poorly understood.
While seeds of some species require very specific treatments to break dormancy, the dormancy of most seeds can be broken in a number of different ways, using different combinations of signals. For this reason the control of dormancy and germination is a key model for understanding the integration of environmental signalling in plants. The most important environmental signals are light quality (sensed primarily by the phytochromes), temperature, nutrient availability and dry after-ripening. After-ripening is the process through which dormancy is lost simply through increasing duration of storage of seeds in a low hydrated state, through a mechanism that remains largely unknown. This is essentially a response to prolonged drought that promotes germination as soon as water is available.
Another interesting feature of seed dormancy is that plants have evolved different mechanisms for inducing dormancy. The most important of these are seeds whose dormancy requires the presence of the seed coat (coat-imposed dormancy), and those whose dormancy persists even if the embryo is excised from the seed (embryo dormancy). Those interested in more detail are referred to the detailed classification of Baskin & Baskin (2004), and the neat recent summary by Finch-Savage & Leubner-Metzger (2006).
2. Seed dormancy in arabidopsis
The model plant Arabidopsis is suitable for understanding the control of seed dormancy, and various ecotypes exhibit differing levels of primary dormancy (Alonso-Blanco et al. 2003). Cape Verde Islands is the most useful more dormant model ecotype, but even genes discovered in CVI, such as DELAY OF GERMINATION1 (DOG1), have obvious phenotypes when knocked out in the more commonly used rapid cycling ecotypes (Bentsink et al. 2006), suggesting that any Arabidopsis ecotype can be used for dormancy research. Depending on the background, Arabidopsis seeds will germinate in the presence of light coupled with a dormancy-breaking signal (DBS): these include cold stratification, dry after-ripening or exogenous nitrate. Under field conditions dormancy is expected to cycle in the absence of germination in buried seeds, and in this situation dormancy loss is seasonal and uncoupled from the switch to germination. In order to understand how we might apply the principles of systems biology to the problem of seed germination control, key known players in germination potential in the imbibed seed will first be considered, and how these are known to link with environmental signalling pathways. This knowledge, coupled with the necessary features of a biological switch, will be used to arrive at a static model encompassing various known features of the process, which can be tested and refined by experiment.
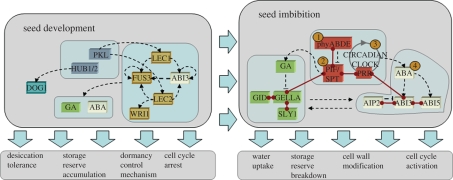
The regulatory network controlling germination is established during seed development (figure 1): readers interested in this establishment phase are directed to a recent review (Holdsworth et al. 2008). In the following sections the key players in the dormancy/germination-controlling network will be outlined, along with their key properties. Then the process of how we can assemble the players into a switching mechanism that conforms to the key constraints known from experiments will be explored.
Figure 1.
A gene network implicated in the seed dormancy in Arabidopsis. Dormancy is established during seed development when a transcription factor network (orange) and chromatin modifications (blue) programme pathways associated with seed development. In the imbibed seed, the interaction of the gibberellin signalling pathway (green) with the abscisic acid signalling pathway (yellow) and an environmental signalling network (red) control germination potential. The collective action of the regulatory genes controls events before or after germination. Connections in red indicate known direct protein–protein interactions in the network, black-dashed lines indicate regulation. Numbers in orange represent the first points at which environmental information modulating dormancy enters the network: 1, light quality and fluence; 2, temperature; 3, dry after-ripening; 4, nitrate.
3. Abscisic acid and seed dormancy and germination
The phytohormone abscisic acid (ABA) is essential for dormancy, as it is severely reduced in Arabidopsis mutants that lack ABA, and in some that lack the correct response to ABA (Koornneef et al. 1982, 1984, 1989). Much of the ABA present in mature dry seeds is eliminated in a similar manner in both dormant and non-dormant seeds (Ali-Rachedi et al. 2004). However, non-dormant seeds are characterized by the transient expression shortly after imbibition of an enzyme important for ABA catabolism, CYP707A2, whereas this expression is not observed in primary dormant seeds (Kushiro et al. 2004; Millar et al. 2006). The expression of CYP707A2 is promoted by the dormancy breaking treatments including dry after-ripening and cold, and requires GIGANTEA (GI) expression, a circadian clock-associated protein (Millar et al. 2006; Penfield & Hall in press), and is one of the first obvious differences between dormant and germinating seeds upon imbibition. These observations suggest that there is a role for active ABA breakdown in dormancy loss.
The effectiveness of inhibitors of the carotenoid pathway (from which ABA is derived) in breaking dormancy in imbibed seeds demonstrates that ABA must be synthesized de novo in imbibed seeds to maintain dormancy. This new ABA synthesis is accompanied by the increased expression of NCED genes and ABA1, both of which are controlled by the circadian clock (Michael et al. 2008; Penfield & Hall 2009; see later sections). Although the expression of these genes is apparent in dormant seeds 24 h after imbibition (Penfield & Hall 2009), detectable levels of ABA do not increase until as late as the third day after imbibition (Ali-Rachedi et al. 2004). This, together with the observation that most ABA present in dry seeds disappears even in dormant seeds upon imbibition, suggests that only a very low level of ABA is essential for dormancy maintenance, or that this ABA is highly localized. Light quality has an important role in the control of ABA synthesis. Red light inhibits ABA1 and NCED expression, while far-red light has the opposite effect (Seo et al. 2006).
Much is known of the ABA signal transduction pathway in seeds, but it is clear that ABA INSENSITIVE3 (ABI3) is a central player in dormant seeds (Giraudat et al. 1992). Abi3 mutants are non-dormant, and ABI3 protein levels are promoted by exogenous ABA (Lopez-Molina et al. 2001). ABI5 and related proteins may also have a role in dormancy control that is not obvious in the single mutants owing to overlapping functions with other ABI5-related genes (Finkelstein et al. 2005). The ABI5 protein interacts in vivo with ABI3 and is important for ABI3 action (Lopez-Molina et al. 2001).
4. Gibberellin signalling and seed dormancy and germination
The plant hormone gibberellin (GA) was well known as a germination promoter before it was demonstrated genetically that GAs are absolutely required for Arabidopsis seed germination (Koornneef et al. 1982). GAs act by destabilizing the growth-repressing DELLA proteins (Peng et al. 1997; McGinnis et al. 2003), and several of these are required for seed dormancy (Penfield et al. 2006a,b). Most authors have in the past associated GA action with germination sensu stricto rather than dormancy loss, but the low primary dormancy of DELLA mutants strongly indicates that GAs have a role in dormancy control as well as germination (Penfield et al. 2006a,b). DELLA protein turnover is required for normal germination as sleepy1 (sly1) mutants, deficient in the F-Box protein required for ubiquitination of DELLAs, exhibit a poor germinating phenotype reminiscent of strong GA-deficient mutants (Steber et al. 1998). Although SLY1 and the GA receptors (GA-INDEPENDENT DWARF1, 2 and 3; GIDs) are important for germination control by GAs, there is as yet no evidence that DBSs modify the levels of these proteins, so for now we will ignore them for the purposes of constructing a preliminary model. Environmental signals promoting germination, most notably light quality and temperature, affect the GA content of seeds by promoting GA synthesis and inhibiting GA degradation (Yamaguchi et al. 1998; Ogawa et al. 2003; Yamauchi et al. 2004). Germination in Arabidopsis is accompanied by a strong increase in GIBBERELLIC ACID 3-OXIDASE (GA3OX) expression, the last step in active GA biosynthesis, which may have a role in regulating cell elongation during radicle emergence, and in endosperm loosening. Environmental signals that prevent germination, such as darkness and high temperature, generally result in the promotion of GA-catabolizing enzymes, the GA 2-oxidases (GA2OX). Dormant seeds also express GA2OXs, suggesting that the switch from dormancy to germination is accompanied by a switch in the expression of genes encoding enzymes that metabolize GAs.
Another important facet of GA signalling is the presence of a mechanism for feedback regulation. This was first noticed in Arabidopsis when it was discovered that the dominant gain-of function DELLA mutant ga insensitive (gai) accumulates higher levels of active GAs than wild-type. So high levels of DELLA signalling induce GA biosynthesis, and this, in turn, in a wild-type plant, would be expected to cause DELLA protein abundance to tend towards equilibrium with GA levels. The same phenomenon can also be observed in GA-deficient mutants (Olszewski et al. 2002), and in PAC-treated seeds (Penfield et al. 2006a,b). It is interesting to consider seed dormancy control in the context of feedback regulation. Because GA deficiency and PAC are potent inducers of GA biosynthetic gene expression in seeds, there must be sufficient GA present to prevent this from happening. Therefore, wild-type seeds contain plenty of GA shortly after imbibition that does not require the subsequent transcription of GA3OXs for its formation. This, in part, explains the need for the expression of GA-catabolizing genes in non-germinating seeds.
Interestingly, after-ripened sly1 mutant seeds germinate at a low frequency, even in the presence of high RGL2 and RGA levels (Ariizumi & Steber 2007). These authors hypothesize that the binding of the GA receptor (GID) to DELLAs is sufficient to inactivate them in the absence of turnover by the 26S proteasome. It would be interesting to see whether these seeds contained high levels of GA owing to feedback up-regulation by high DELLA levels or whether this too was abolished by GID-binding.
5. The integration of gibberellin and abscisic acid signalling
A key question in dormancy control is how the GA and ABA pathways interact: how is the information from both hormones integrated to control germination? One theory is that the role of ABA is to act directly and negatively on GA synthesis (e.g. Seo et al. 2006). Key evidence for this is that ABA-deficient mutants show elevated expression of the GA3OXs (Seo et al. 2006). This theory suggests that once ABA levels fall below a certain threshold, GA synthesis can begin and germination follows. However, this can only be part of the story because abi3 mutants can germinate in the absence of GA (Nambara et al. 1991), placing ABI3 action downstream, or equivalent to GA action. In addition, della mutants show ABA-resistant germination in the absence of GA synthesis, suggesting that DELLAs have a role in ABA responses that is distinct from the regulation of GA metabolism (Penfield et al. 2006a,b). ABA can also affect DELLA protein levels in a GA-independent manner (Achard et al. 2006). Modifying the levels of ABI5 also affects the PAC-sensitivity of germination (Piskurewicz et al. 2008), suggesting that the roles of ABI5, ABI3 and DELLAs act in parallel at the same point to control dormancy and germination. Taken together, these data suggest that simultaneous action of ABI3/ABI5 and DELLAs are necessary for dormancy, and that these act on a common set of targets to prevent germination, as well as influencing each other's protein levels. Also important is the observation that if DELLA proteins bind the GA receptors they can be prevented from inhibiting growth, without being turned over by the SLY1 complex (Ariizumi et al. 2008). This shows that DELLA inhibition of germination requires a protein partner that competes with the GA receptor for DELLA binding.
Other evidence suggests that DELLAs can promote ABA levels in seeds. The XERICO gene has been shown to be induced in seeds by a DELLA protein-containing complex, and in turn XERICO can promote ABA synthesis in leaves, leading to drought-tolerant phenotypes (Zentella et al. 2007). Together, all this evidence shows that ABA and GA levels in seeds are subject to a complex regulatory relationship with each hormone subject to regulation of its levels by the other.
6. Phytochrome-interacting factors
Phytochrome-interacting factors are a subfamily of bHLH transcription factors, first known from the characterization of PIF3 (Ni et al. 1998). Mutants lacking one of these genes, pif3-like 5 (pil5, otherwise known as pif1), germinate after a normally inhibitory far-red treatment (Oh et al. 2004), and in the absence of light (Penfield et al. 2005). PIL5 binds phytochrome A and B proteins directly and is stabilized in the absence of red light (Shen et al. 2005). Another PIF-family transcription factor, SPATULA (SPT), is involved in establishing the sensitivity of dormancy loss to cold. In the Ler ecotype STP functions as a germination promoter (S. Penfield, unpublished data), the absence of which attenuates the response to cold stratification. SPT is unusual in the family in that it lacks the motifs believed to be important in phytochrome binding, and does not bind phyA or phyB in vitro (Khanna et al. 2004). Further, PIF transcription factors are also expressed in seeds, either during dormancy induction or during and after germination.
Important in the consideration of PIFs is that they are known to compete with the GA receptors for DELLA binding: hence PIFs can stabilize DELLAs in the presence of GA (de Lucas et al. 2008; Feng et al. 2008). This activity may account for the described role of PIL5 in modifying GA sensitivity in seeds (Oh et al. 2007), rather than the control of RGA and GAI transcription. PIFs also act upstream of GA and ABA metabolism, and are required for the normal induction of hormone-related gene expression associated with the germinating state by environmental cues (Penfield et al. 2005; Oh et al. 2006), and also affect GAI and RGA transcript levels to a small degree (Oh et al. 2007).
7. The circadian clock and seed germination control
This is a relative newcomer to the germination scene but central to the regulation of both dormancy and germination. Current models of the circadian clock invoke a series of interlocking transcriptional loops involving at least five pseudoresponse regulator (PRR) proteins (Locke et al. 2006; Fujiwara et al. 2008), timing of CAB expression 1 (TOC1, otherwise known as PRR1), PRR7, PRR9, PRR5 and PRR3. The PRR proteins are expressed sequentially during the afternoon and in the evening, and their protein abundance is regulated post-translationally: in the case of TOC1 it has been shown that GI stabilizes TOC1 indirectly by competing for F-box protein ZEITLUPE (ZTL; Kim et al. 2007) in the presence of blue light. Loss of both GI and ZTL has a similar effect on primary seed dormancy, reducing the effectiveness of dormancy-breaking treatments such as cold and after-ripening (Penfield & Hall 2009). This suggests that these genes either establish the dormancy level during seed development, or are involved in dormancy loss in the imbibed seed. In contrast, loss of TOC1 has little effect on seed germination, suggesting that the gi and ztl phenotypes are explained in part by the interaction with the functions of the other PRR proteins, or that the PRR proteins can act redundantly in dormancy control.
Oliverio et al. (2007) showed that if you promote germination by a far-red pulse, the extent of the promotion varies over a 24 h cycle, suggesting gating by the circadian clock and a role for the clock in imbibed seeds. Recently, we have shown that mutations in multiple clock-associated genes modify the seed's response to after-ripening and cold stratification (Penfield & Hall 2009). Dormancy loss is associated with increased amplitude of CCA1 expression on the first day and decreased amplitude of PRR and GI gene expression on the second day. Interestingly, this induction of CCA1 expression by after-ripening is not dependent on light signalling, showing that after-ripening or dormancy state directly influences circadian transcription. Another recent publication links a third Arabidopsis DBS, nitrate, to the circadian clock (Gutiérrez et al. 2008). These authors report that organic and inorganic nitrogen levels affect clock activity, and that CCA1 is important in the regulation of nitrate metabolism. Therefore, it seems possible that circadian clock genes respond to, and may have a role in the transduction of DBSs.
The TOC1 protein has been shown to interact with several PIFs, providing a potential mechanism through which light signalling and dormancy-breaking treatments can be integrated (Yamashino et al. 2003). What is clear is that two mutants we have shown to affect cold responses in seeds strongly, the PIF transcription factor mutant spt-2 (Penfield et al. 2005) and the ‘evening’ clock gene mutant gi-11 (Penfield & Hall 2009), are linked by their potential role in regulating PRR gene function, although it remains to be demonstrated that these genes are important in imbibed seeds. TOC1 has also been shown to interact with the ABI3 protein in vivo (Kurup et al. 2000), suggesting that PRR genes could have a central role in hormone balance control.
Interestingly, it is now known that the transcription of a large subsection of hormone metabolism is also controlled by the circadian clock, including many genes important in germination control, suggesting that this is the mechanism through which the clock controls germination (Michael et al. 2008; Penfield & Hall 2009; fig. S1 in the electronic supplementary material). It has also recently been shown that there is a strong circadian influence on ABA and GA signalling, with GA-repressed and ABA-induced gene expression biased towards dawn, and ABA-repressed and GA-induced gene expression biased towards dusk (Covington et al. 2008). Genes important to germination such as GA3OX1 and CYP707A2 belong to the GA-repressed and ABA-induced morning set, respectively, while genes associated with dormancy, including GA2OX2 and ABA1, belong to the GA-induced and ABA-repressed morning set. Hence, a shift from strong promotion of dawn transcription to strong promotion of dusk transcription alone could have a strong effect on germination potential, and after-ripening appears to have precisely this effect (Penfield & Hall 2009).
8. How do the components of the germination-controlling network constitute a switch?
At this point it is instructive to consider not the biology of seed germination control, but the necessary architecture of a switch and the key features. The switch to germination represents a transition to or from one stable non-germinating state to another germinating state. As such, germination control can be viewed as a classical bifurcating system with two stable attracting states: non-germination and germination (Tyson et al. 2003). In-between lies a critical unstable transition that is passed as the system flips from the unstable state that provides the borderline (and thus quantifies the critical point for transition) between the two stable ones. The role of dormancy is to modify the sensitivity of seeds to signals that flip the switch. A central point is that simple linear causative systems do not provide a mechanism for such a bifurcation, the attracting states being stabilized by feedback mechanisms that drive the system away from the unstable state. Central questions for a systems biologist are then what species constitute the components of the switch, and how they are arranged to generate the nonlinear positive or negative feedback necessary to drive a switch between two stable states. At this point we have to accept that some assumptions are going to be necessary that are not yet justified by experiment. This does not in the long term deplete the value of the model, because the model itself can then be used to design experiments specifically to test the justification of these assumptions, and can then be improved accordingly. This iteration itself forms an intrinsic part of systems biology (Kitano 2004). Modelling biological switches has led to improved understanding of similar phenomena, such as the cell cycle, transcription and apoptosis (Novick & Weiner 1957; Novak & Tyson 1993; Albeck et al. 2008). So here we will make a first attempt to define a static model describing the switch between dormancy and germination, and try to encapsulate as many of the known constraints as possible.
9. Hormone balance from protein–protein interactions
A key message from the literature on seed germination control is that it is the balance between ABA and GA signalling that underpins germination potential, rather than one or the other alone. Furthermore, if we perturb genetically either ABA or GA signalling, there would be a collateral effect on the other (Nambara et al. 1991; Achard et al. 2006; Penfield et al. 2006a,b; Seo et al. 2006; Zentella et al. 2007; Piskurewicz et al. 2008). Any model must account for this as a central feature. To start with, let us assume that the stabilization of ABI3 by ABA and the destabilization (or at least stimulation of GID-binding and deactivation) of DELLAs by GA are central processes in hormone balance. One possibility is as follows: germination is promoted by an unknown heterodimeric complex containing two proteins, X and Y. The important features of X and Y are that they can interact with each other, and each with one of either ABI3 or DELLA, but that heterotrimeric complexes are contraindicated (e.g. by the stereochemistry of overlapping binding sites). Let us assume that X interacts with DELLA and Y with ABI3 (figure 2). In a dormant seed, the concentration of the germination-promoting X–Y complex is low because X remains bound to DELLA and Y to ABI3 (figure 2b). In a germinating seed, free DELLA levels are low (because DELLAs are either turned over or are prevented from binding X by GIDs), as are those of ABI3. This allows the formation of X–Y and subsequent germination (figure 2c). This logic also applies in reverse. In a germinating seed X and Y are prised from DELLA and ABI3, respectively, allowing both to be degraded by the GA or lack of ABA. In this simple model, manipulating GA or ABA levels changes the amount of the X–Y complex a small amount, but simultaneous manipulation of ABA and GA produces massive swings and the formation of a large amount of X–Y. Thus, this simple scheme accounts for the key feature of hormone balance, that the balance of GA and ABA signalling is important for the dormancy state. Note that it can be postulated that X and Y promote germination by an unidentified GA- and ABA-independent pathway. However, the same scheme works equally well if we assume that downstream targets inhibited by ABI3 and DELLA promote germination. In this situation the role of X–Y is to achieve hormone balance, to link the responses of the DELLA- and ABI3-dependent pathways.
Figure 2.
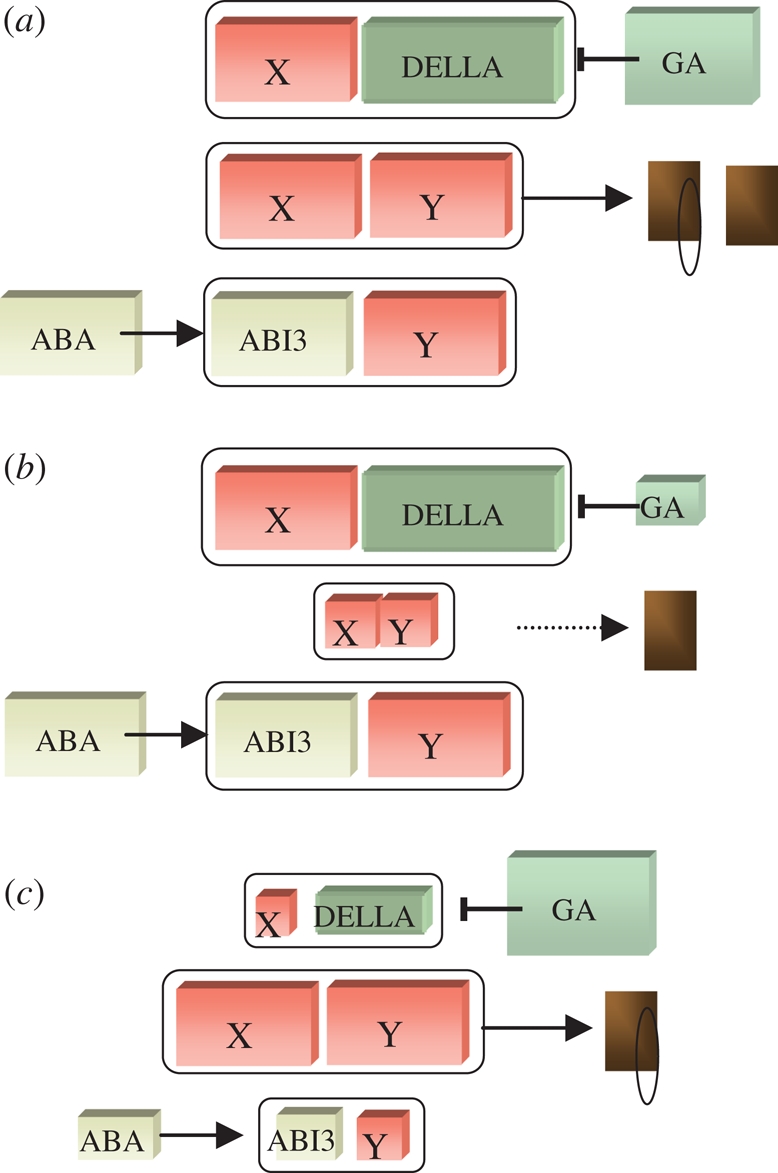
A theoretical scheme for the control of hormone balance. Coloured boxes represent variables (proteins or hormones) with outlines in black denoting protein complexes. ABI3 levels are promoted by abscisic acid (ABA), DELLA levels decreased by gibberellin (GA). In (b) and (c) the size of the boxes provides a measure of the amount of the variable present. (a) Depiction of the general scheme whereby DELLA and ABI3 compete with X and Y, respectively, and prevent formation of the X–Y complex, which promotes germination. (b) Representation of a seed with high ABA and low GA levels. The combined action of ABA plus low GA action collectively results in low levels of the X–Y complex. (c) A representation of a seed with high GA and low ABA levels. This situation results in high concentrations of the germination-promoting X–Y complex.
10. Phytochrome-interacting factors and pseudoresponse regulators are candidates for X and Y
Known protein–protein interactions suggest that PIFs and PRRs are good candidates for X and Y. Two recent papers show that PIFs bind DELLAs using the basic helix-loop-helix DNA-binding domain (de Lucas et al. 2008; Feng et al. 2008). ABI3 is known to bind at least one PRR protein, TOC1, in yeast two-hybrid assays (Kurup et al. 2000). The final piece of the jigsaw is that PIFs can also bind PRR proteins (Yamashino et al. 2003). Hence, a model that includes PIFs and PRRs instead of X and Y is supported by our current understanding of protein–protein interactions in seeds and seedlings.
The observation that GI, ZTL, LHY and CCA1 all influence dormancy and are all regulators of PRR genes is good circumstantial evidence for the role for PRRs themselves in dormancy control and possibly also in germination. At this point, it is interesting to consider the phenotype of the TOC1 minigene line (TMG; Más et al. 2003), which carries an extra copy of the TOC1 genomic fragment fused to YFP under its own promoter. TMG seeds show a previously unreported class of germination phenotype in which their PAC-sensitivity is decreased and their ABA-sensitivity increased (Penfield & Hall 2009). Mutants in GI have opposite effects, in which ABA signalling is inhibited and GA signalling is slightly increased. Gi mutants show low-amplitude TOC1 protein oscillations (Kim et al. 2007). Together, these results show that manipulating PRR protein levels can have equal and opposite effects on ABA and GA signalling, suggesting that they may have a role in hormone balance.
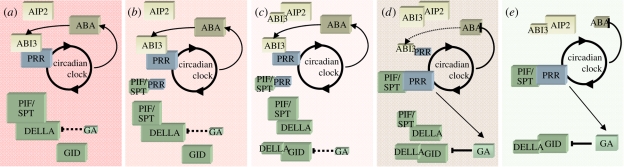
For now we will go ahead and replace X and Y by PIF and PRR in our scheme (figure 3). Note, though, that this scheme predicts that individual PIFs and PRRs can be either inhibitors or promoters of germination, depending on their relative affinities for each other, DELLAs and ABI3. PIL5 would appear to be unsuited for this ‘PIF’ role because it is a germination repressor, and because pil5 mutants show normal dormancy in the presence of light (Penfield et al. 2005). So if this model is to reflect reality then PIFs other than PIL5 must perform the crucial pivotal role in the seed. One possibility is that PIL5 inputs light into the system by preventing the PRR-binding of other PIFs. However, there may be other proteins that can also perform the functions of X and Y. DBSs are also sensed very close to the proposed PIF–PRR complex in the network (figure 1), suggesting that a mechanism in which DBSs affect this complex is plausible.
Figure 3.
The transition of the network from a dormant to a germinating state. In (a) the seed is dormant with high abscisic acid (ABA) and low gibberellin (GA) levels. In (b) an unspecified environmental perturbation permits the formation of a small amount of phytochrome-interacting factor–pseudoresponse regulator (PIF–PRR) complexes. This releases some ABI3 and DELLA to bind with their complexes, which control degradation (c). Decreasing free ABI3 and DELLA allow the formation of more PIF–PRR complexes (d). These switch on GA production and ABA degradation. The increased GA levels, coupled with the release of DELLA from PIF, result in states with very low DELLA-PIF levels (e). Low ABA coupled with the release of ABI3 from PRR results in low ABI3 levels. From genetic experiments we know that loss of either ABI3 or DELLA leads to low dormancy.
11. Adding hormones creates positive feedback in the system
One puzzling aspect of the signalling regulating seed germination is the need for hormones at all. To explain why this should be a puzzle, consider the following. It seems that light signalling in seeds requires normal PIF action (Oh et al. 2004). We know that PIFs interact with DELLAs (de Lucas et al. 2008), so if we require a mechanism through which PIFs transduced an environmental signal to the growth-regulatory DELLA proteins then we already have it, without invoking the much-studied environmental regulation of GA metabolism. In the presence of a constant level of GA, the lack of PIF-binding could free DELLAs to bind GID and be degraded (or be prevented from inhibiting growth). What then is the real role of changing hormone levels? One possibility is that GA enables coordination of responses within or between tissues, such as the embryo and endosperm. A second possibility is that the hormones have a necessary role in the stable-switching mechanism, namely to add the necessary feedback that can stabilize both the dormant and germinating states. The simple signal transduction from PIFs to DELLAs creates no feedback so the new low-DELLA state could be inherently unstable, or sensitive to noise in the system (Kitano 2002). However, if at the same time PIFs or a protein complex in our system that is also responsive to DBSs can also regulate GA levels, we can start to see how feedback might be incorporated into the system (figure 3). In this scenario, DBSs trigger the formation of a small amount of X and Y or PIF–PRR. This frees some DELLA to bind GID and be degraded, but this degradation is further potentiated by GA promoted by the action of PIF that is not bound to DELLAs. This frees more PIF, leading to more GA and more DELLA turnover, through a positive feedback loop. Evidence that PIFs have a role in the regulation of GA metabolism is well known, but there is no evidence that PIFs directly bind these promoters (Seo et al. 2009). Instead, there are many known ways in which PIFs can indirectly control transcription, either via phytochrome or through clock genes (Michael et al. 2008; Penfield & Hall 2009). A third alternative clear in our model is that PIFs may sequester PRRs from ABI3, and that the ABI3 or the ABI3–PRR complex regulates GA metabolism. This may also free ABI3 to interact with a partner that controls its degradation, such as ABI3-INTERACTING PROTEIN2 (AIP2; Kurup et al. 2000; Zhang et al. 2005; figure 3), and thus relieve a repression of GA metabolism. In terms of the logic of the system, all these alternatives are broadly equivalent, in that GA synthesis becomes dependent, directly or indirectly, on the PIF–PRR complex. In fact, the wiring can be completed in a number of ways that still lead to a similarly functioning germination control network.
An interesting side-effect of the scheme now becomes apparent. It has been shown that exogenous ABA represses the expression of GA3OX1 in seeds and that the levels of bioactive GAs are elevated in aba2 mutants (Seo et al. 2006). It has also been shown that ABA can affect DELLA protein abundance directly (Achard et al. 2006; Penfield et al. 2006a,b). Our scheme apparently has the potential to capture both of these effects. ABA inhibits GA synthesis indirectly in our model by inhibiting the formation of the PIF–PRR complex. Similarly, ABA can influence DELLA protein levels independently of GA by affecting the abundance of a free DELLA-binding protein that competes with GID. As we have seen, PIF is a candidate for this protein and ABA can modulate free PIF levels according to our scheme through an indirect effect of competing with PIF for PRR binding (figure 3). All these possibilities are experimentally testable. An interesting feature of this model is that certain complexes are likely to be abundant at different time-points, because of circadian regulation of the PRR protein levels.
12. Conclusions
A simple model has been described that accounts for many, but not all, of the well-known features of hormone balance. The central feature of the model is that a heterodimeric complex exists that promotes germination, and that the abundance of one monomer is influenced by ABA and the other by GA. This complex also regulates GA and ABA metabolism in seeds, and this creates feedback necessary to stabilize the dormant and germinating states. Evidence suggests that PIF and PRR proteins are candidates for the components of the germination-promoting complex, but further characterization of the role of PIF and PRR proteins in seeds is required to support or refute this hypothesis. In this scheme, the plant hormones GA and ABA, long considered to be key outputs of dormancy-modifying treatments, take on a new role as amplifying signals that reinforce either the dormant or germinating states. Further work is now required to analyse the properties of the model and to elucidate the functions of the relevant genes not yet characterized with respect to their role in germination control. It will also be important to integrate other genes into the scheme which are known or suspected to have a role in dormancy control in the imbibed seed, such as DELAY OF GERMINATION1 (Bentsink et al. 2006).
Acknowledgements
The second author gratefully acknowledges the funding of the BBSRC/EPSRC and of the Royal Society and Wolfson Foundation. We are grateful for stimulating discussions with the participants at the Inaugural Mathematics in the Plant Sciences Study Group at the University of Nottingham, 17–20 December 2007, some preliminary modelling work undertaken there being recorded in Penfield et al. (2008).
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N. P.2006Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (doi:10.1126/science.1118642) [DOI] [PubMed] [Google Scholar]
- Albeck J. G., Burke J. M., Spencer S. L., Lauffenburger D. A., Sorger P. K.2008Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 6, 2831–2852 (doi:10.1371/journal.pbio.0060299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Rachedi S., Bouinot D., Wagner M. H., Bonnet M., Sotta B., Grappin P., Jullien M.2004Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219, 479–488 (doi:10.1007/s00425-004-1251-4) [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Bentsink L., Hanhart C. J., Blankestijn-de Vries H., Koornneef M.2003Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164, 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Steber C. M.2007The germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19, 791–804 (doi:10.1105/tpc.106.048009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Murase K., Sun T. P., Steber C. M.2008Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20, 2447–2459 (doi:10.1105/tpc.108.058487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M., Baskin C. C.2004A classification system for seed dormancy. Seed Sci. Res. 14, 1–16 (doi:10.1079/SSR2003150) [Google Scholar]
- Bentsink L., Jowett J., Hanhart C. J., Koornneef M.2006Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 17 042–17 047 (doi:10.1073/pnas.0607877103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J. D.1997Seed germination and dormancy. Plant Cell 9, 1055–1066 (doi:10.1105/tpc.9.7.1055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M. F., Maloof J. N., Straume M., Kay S. A., Harmer S. L.2008Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9, R130 (doi:10.1186/gb-2008-9-8-r130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J. M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., Fankhauser C., Blázquez M. A., Titarenko E., Prat S.2008A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 (doi:10.1038/nature06520) [DOI] [PubMed] [Google Scholar]
- Feng S., et al. 2008Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 (doi:10.1038/nature06448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W. E., Leubner-Metzger G.2006Seed dormancy and the control of germination. New Phytol. 171, 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S. S., Lynch T. J., Thomas T. L., Rock C. D.2005Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59, 253–267 (doi:10.1007/s11103-005-8767-2) [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S. S., Salomé P. A., McClung C. R., Somers D. E.2008Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23 073–23 083 (doi:10.1074/jbc.M803471200) [DOI] [PubMed] [Google Scholar]
- Giraudat J., Hauge B. M., Valon C., Smalle J., Parcy F., Goodman H. M.1992Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261 (doi:10.1105/tpc.4.10.1251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R. A., et al. 2008Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl Acad. Sci. USA 105, 4939–4944 (doi:10.1073/pnas.0800211105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M. J., Finch-Savage W. E., Grappin P., Job D.2008Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 13, 7–13 (doi:10.1016/j.tplants.2007.11.002) [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E. A., Al-Sady B., Lanzatella C., Quail P. H.2004A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16, 3033–3044 (doi:10.1105/tpc.104.025643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Fujiwara S., Suh S. S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H. G., Somers D. E.2007ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360 (doi:10.1038/nature06132) [DOI] [PubMed] [Google Scholar]
- Kitano H.2002Computational systems biology. Nature 420, 206–210 (doi:10.1038/nature01254) [DOI] [PubMed] [Google Scholar]
- Kitano H.2004Biological robustness. Nat. Rev. Genet. 5, 826–837 (doi:10.1038/nrg1471) [DOI] [PubMed] [Google Scholar]
- Koornneef M., Jorna M. L., Brinkhorst-van der Swan D. L. C., Karssen C. M.1982The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana L. Heynh. Theor. Appl. Genet. 61, 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Reuling G., Karssen C. M.1984The isolation and characterisation of abscisic acid insensitive mutants of Arabidopsis thaliana. Physiol. Plant 61, 377–383 (doi:10.1111/j.1399-3054.1984.tb06343.x) [Google Scholar]
- Koornneef M., Hanhart C. J., Hilhorst H. W., Karssen C. M.1989In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 90, 463–469 (doi:10.1104/pp.90.2.463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S., Jones H. D., Holdsworth M. J.2000Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21, 143–155 (doi:10.1046/j.1365-313x.2000.00663.x) [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E.2004The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656 (doi:10.1038/sj.emboj.7600121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J. C., Kozma-Bognár L., Gould P. D., Fehér B., Kevei E., Nagy F., Turner M. S., Hall A., Millar A. J.2006Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2, 59 (doi:10.1038/msb4100102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N. H.2001A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl Acad. Sci. USA 98, 4782–4787 (doi:10.1073/pnas.081594298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Alabadí D., Yanovsky M. J., Oyama T., Kay S. A.2003Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236 (doi:10.1105/tpc.006734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K. M., Thomas S. G., Soule J. D., Strader L. C., Zale J. M., Sun T. P., Steber C. M.2003The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130 (doi:10.1105/tpc.010827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T. P., Breton G., Hazen S. P., Priest H., Mockler T. C., Kay S. A., Chory J.2008A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6, e225 (doi:10.1371/journal.pbio.0060225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. A., Jacobsen J. V., Ross J. J., Helliwell C. A., Poole A. T., Scofield G., Reid J. B., Gubler F.2006Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8'-hydroxylase. Plant J. 45, 942–954 (doi:10.1111/j.1365-313X.2006.02659.x) [DOI] [PubMed] [Google Scholar]
- Nambara E., Akazawa T., McCourt P.1991Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol. 97, 736–738 (doi:10.1104/pp.97.2.736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Tepperman J. M., Quail P. H.1998PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667 (doi:10.1016/S0092-8674(00)81636-0) [DOI] [PubMed] [Google Scholar]
- Novak B., Tyson J. J.1993Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J. Cell Sci. 106, 1153–1168 [DOI] [PubMed] [Google Scholar]
- Novick A., Weiner M.1957Enzyme induction as an all-or-none phenomenon. Proc. Natl Acad. Sci. USA 43, 553–566 (doi:10.1073/pnas.43.7.553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi Y.2003Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604 (doi:10.1105/tpc.011650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J. I., Kang C., Choi G.2004PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16, 3045–3058 (doi:10.1105/tpc.104.025163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W. I., Choi G.2006Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139 (doi:10.1111/j.1365-313X.2006.02773.x) [DOI] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H. S., Sun T. P., Kamiya Y., Choi G.2007PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208 (doi:10.1105/tpc.107.050153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio K. A., Crepy M., Martin-Tryon E. L., Milich R., Harmer S. L., Putterill J., Yanovsky M. J., Casal J. J.2007GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 144, 495–502 (doi:10.1104/pp.107.097048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Sun T. P., Gubler F.2002Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl.), S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Hall A. J.In press A role for multiple circadian clock genes in the response to seed dormancy-breaking signals. Plant Cell (doi:10.1105/tpc.108.064022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E. M., Kannangara R., Gilday A. D., Halliday K. J., Graham I. A.2005Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006 (doi:10.1016/j.cub.2005.11.010) [DOI] [PubMed] [Google Scholar]
- Penfield S., Gilday A. D., Halliday K. J., Graham I. A.2006aDELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16, 2366–2370 (doi:10.1016/j.cub.2006.10.057) [DOI] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A. D., Graham S., Graham I. A.2006bArabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18, 1887–1899 (doi:10.1105/tpc.106.041277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Bastow R., Fozard J., King J., Savage N.2008Network properties underlying seed germination control. MPSSG 2007 Report, CPIB [Google Scholar]
- Peng J., Carol P., Richards D. E., King K. E., Cowling R. J., Murphy G. P., Harberd N. P.1997The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205 (doi:10.1101/gad.11.23.3194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L.2008The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745 (doi:10.1105/tpc.108.061515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., et al. 2006Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366 (doi:10.1111/j.1365-313X.2006.02881.x) [DOI] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S.2009Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69, 463–472 (doi:10.1007/s11103-008-9429-y) [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E.2005PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44, 1023–1035 (doi:10.1111/j.1365-313X.2005.02606.x) [DOI] [PubMed] [Google Scholar]
- Steber C. M., Cooney S. E., McCourt P.1998Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. J., Chen K. C., Novak B.2003Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell. Biol. 15, 221–231 (doi:10.1016/S0955-0674(03)00017-6) [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Smith M. W., Brown R. B., Kamiya Y., Sun T. P.1998Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126 (doi:10.1105/tpc.10.12.2115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T.2003A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44, 619–629 (doi:10.1093/pcp/pcg078) [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S.2004Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16, 367–378 (doi:10.1105/tpc.018143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., et al. 2007Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 19, 3037–3057 (doi:10.1105/tpc.107.054999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N. H.2005The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543 (Mol. Cell. 2008;30:11–25). (doi:10.1101/gad.1318705) [DOI] [PMC free article] [PubMed] [Google Scholar]