Abstract
There are many well-studied examples of human phenotypes resulting from nonsense or frameshift mutations that are modulated by Nonsense-Mediated mRNA Decay (NMD), a process that typically degrades transcripts containing premature termination codons (PTCs) in order to prevent translation of unnecessary or aberrant transcripts. Different types of germline mutations in the VHL gene cause the von Hippel-Lindau disease, a dominantly inherited familial cancer syndrome with a marked phenotypic variability and age-dependent penetrance. By generating the Drosophila UAS:Upf1D45B line we showed the possible involvement of NMD mechanism in the modulation of the c.172delG frameshift mutation located in the exon 1 of Vhl gene. Further, by Quantitative Real-time PCR (QPCR) we demonstrated that the corresponding c.163delG human mutation is targeted by NMD in human HEK 293 cells. The UAS:Upf1D45B line represents a useful system to identify novel substrates of NMD pathway in Drosophila melanogaster. Finally, we suggest the possible role of NMD on the regulation of VHL mutations.
1. Background
Germline mutations in the VHL gene cause the von Hippel-Lindau disease (VHL; MIM# 193300), a dominantly inherited familial cancer syndrome with retinal and central nervous system hemangioblastomas, renal cell carcinoma, pheochromocytoma, pancreatic endocrine tumors, and endolymphatic sac tumors [1–4]. The VHL mutation pattern includes missense, nonsense, frameshift, and splice site mutations. Genotype-phenotype correlation studies showed that the incidence of renal involvement in VHL disease was increased in families with nonsense or frameshift mutations that disrupted the structural integrity of VHL protein, whereas missense mutations associated with a higher risk of pheochromocytoma [5–7].
We hypothesize that the genic localization of VHL variations and nonsense mutations of VHL activating NMD pathway may play an important role in the determination of a specific phenotype.
NMD is an evolutionarily conserved mRNA surveillance pathway that protects cells from potentially harmful effects of truncated proteins that would otherwise be translated from mRNAs bearing PTC. The process serves as a general surveillance mechanism to abolish aberrant transcripts resulting not only from rare mutations but also from mistakes in RNA processing [8], regulating the expression of about 3%–10% of the transcriptome in S. cerevisiae, D. melenogaster, and human cells. These natural NMD targets play a role in different biological processes such as transcription, cell proliferation, cell cycle, telomere maintenance, cellular transport and organization, and metabolism [9]. The high evolutionary conserved Upf proteins, Upf1, Upf2, and Upf3, constitute the core of NMD machinery [10–14]. The key molecular component is Upf1, an RNA helicase that recognizes aberrant translation termination events [15].
Although conserved in all eukaryotes that have been analysed so far, NMD employs different molecular mechanisms, depending on the species, to discriminate between natural and premature stop codons and to degrade the targeted mRNAs. In mammalian cells, termination codons that lie upstream of an exon-exon boundary are generally recognized as premature and target the mRNA for degradation by NMD.
The Drosophila melanogaster intron-less Vhl gene maps at polytene chromosomal position 47E5-6 (http://flybase.org/). The human and fly proteins show a high degree of amino acid similarity spread throughout the entire length of VHL with 67% and 76% of similarity in the functional domains of PKCl and elongin C binding domains, respectively.
Here, by establishing a Drosophila NMD mutant, we showed the involvement of NMD mechanism in the modulation of a novel human VHL frameshift mutation and we confirmed this data in the HEK 293 human cell line using a molecular strategy based on the minigene constructs.
2. Methods
2.1. Fly Strains and Culture Conditions
Flies were cultured at 25°C on standard cornmeal-sucrose-yeast-agar medium containing propionic acid as mold inhibitors. Detailed description of mutations and chromosome rearrangements used in the present study could be found at FlyBase: http://flybase.bio.indiana.edu. The stocks used in the present work were supplied by Bloomington Stock Center.
2.2. Northern Blotting Assays
Total RNAs from testes and ovaries of adult flies were isolated using RNeasy Kit (Qiagen) and poly(A)+ RNAs were prepared with oligo(dT)-coupled beads (Oligotex, Qiagen). RNAs were separated in denaturing formaldehyde agarose gel (5–20 μg/lane) and blotted onto positively charged nylon membranes (Amersham). Upf1 and Rp49 32P-labeled probes were generated by random priming using standard methods. Hybridization was carried out overnight at 65°C in hybridization solution (formamide 50%, SSC 5x, Denhardt's 5x, SDS 0.5x, EDTA pH 8.0 10 mM, Salmon Sperm DNA 100 g/mL). After hybridization, the membranes were washed four times at 65°C. Autoradiography was carried out for both 16 and 48 hours at −80°C using intensifying screens. Filters were stripped and hybridization was repeated with a Rp49 specifc probe.
2.3. Genetics Mutants
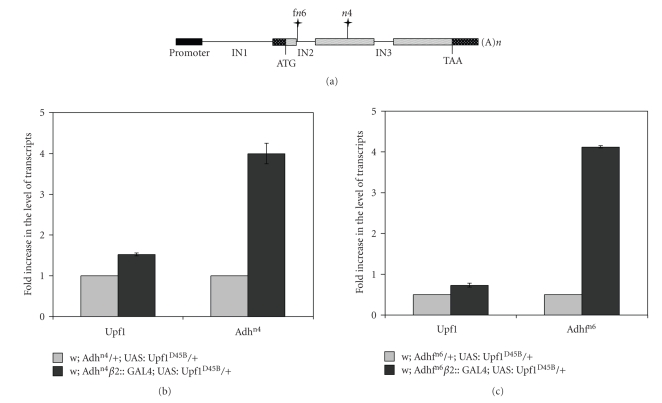
The Adh alleles were previously characterized by Brogna [16]. The Adhn4 allele contains a premature stop codon located 258 bp upstream from the boundary between exon 3 and exon 4 while Adhfn6 is a small deletion that eliminates the 5′ splicing signal of intron 2 leading to an in-frame premature stop codon (Figure 1(a)).
Figure 1.
Schematic map of the Adh gene. (a) Rectangles represent exons; black box represents promoter region; hatched boxes indicate coding regions; punctated boxes represent untranslated regions. The horizontal lines represent introns, IN1-IN3. The position of the mutations are indicated with vertical lines marked with a star; Adhfn6 is the mutation affecting splicing while Adhn4 is a nonsense mutation. (b) and (c) Compared levels of Adh and Upf1 transcripts in flies that were heterozygous for Adh alleles and for Upf1D45B. A 3.8- and 8.8-fold accumulation of nonsense-containing Adh mRNA was observed in w1118; Adhn4/β2::GAL4; UAS:Upf1D45B/+ and w1118;Adhfn6/β2::GAL4; UAS:Upf1D45B/+ mutant flies with respect to the endogenous levels of Adh and Upf1 control mRNAs. The experiment was repeated three times.
2.4. Isolation of Full Length Upf1 cDNA
Drosophila genomic DNA was isolated from adults using Genomic DNA Extraction Kit (Qiagen). As probe for subsequent screens, a fragment of 700 bp was PCR amplified from genomic DNA as template (Upf1F and Upf1R primers in Table 1). The identity of cloned PCR product was confirmed by sequencing. For the isolation of the Upf1 cDNA full-length, a Drosophila melanogaster ovaric cDNA library was screened using that Upf1-probe. Hybridization of the Hybond-N+ filters (Amersham) was carried out at 65°C according to the manufacturer's instructions. The positive Upf1 cDNA clone was cloned into the pGEM-T-Easy vector (Promega) and verified by DNA sequencing.
Table 1.
Primer's list used in this study.
| Name | Sequence 5′-3′ | Utilization |
|---|---|---|
| Upf1F | TTGGAATCATCACGCCTTACGA | Upf1 probe |
| Upf1R | CATGCCAACCGGAACTGGCATG | Upf1 probe |
| Upf1Fmut | ATGTCTTGCGTGTGTTCTAACGAACGT | Mutagenesis |
| Upf1Rmut | ACGTTCGTTAGAACACACGCAAGACAT | Mutagenesis |
| Vhln1F | CCCTCAAGCCCTTCAGGAGGTGCGGGTGAAC | Vhln1 mutagenesis |
| Vhln1R | GGGAGTTCGGGAAGTCCTCCACGCCCACTTG | Vhln1 mutagenesis |
| Vhln2F | GGATGCACGTACGCTGCAGAGGATCTTTCA | Vhln2 mutagenesis |
| Vhln2R | TGAAAGATCCTCTGCAGCGTACGTGCATCC | Vhln2 mutagenesis |
| Vhln1F construct1(EcoRI) | CGCGCGGAATTCATGCCCCGGAGGGCGGAGAACT | Construct1 |
| Vhln1R construct1(XhoI) | CGCGCGCTCGAGATGGTGAAACCCCGTCTCTACT | Construct1 |
| Vhln1F1 construct1(XhoI) | CGCGCGCTCGAGTCAGGGGAAATGGAGAAAATAG | Construct1 |
| Vhln1R1 construct1(XbaI) | CGCGCGTCTAGAGAGAATAGGATACAAAAAGATTGGA | Construct1 |
| c.163delGF | ACTGGGCGCCGAGGAGGAGATGAGGCCGGGCGGCCGC | Construct1 mutagenesis |
| c.163delGR | GCGGCCGCCCGGCCTCATCTCCTCCTCGGCGCCCAGT | Construct1 mutagenesis |
| Vhln2F construct2(EcoRI) | CGCGCGGAATTCATGCCCCGGAGGGCGGAGAACT | Construct2 |
| Vhln2R construct2(XhoI) | CGCGCGCCTCGAGTCAATCTCCCATCCGTTGATGTGC | Construct2 |
| c.172delGF | CAGGTCATCTTCTGCAATCGCATCCGCGCGTCGTGCT | Construct2 mutagenesis |
| c.172delGR | AGCACGACGCGCGGATGCGATTGCAGAAGATGACCTG | Construct2 mutagenesis |
| pVHLF | GGCCGCCGCATCCA | VHL QPCR |
| pVHL R | CATCGTGTGTCCCTGCATCTC | VHL QPCR |
| pGFP F | GCAACTACAAGACCCGC | GFP QPCR |
| pGFP R | GTCGGCCATGATATAGACG | GFP QPCR |
| rp49_F | CACACCGGAAACTCAATGGAT | rp49 QPCR |
| rp49_R | GGTCATCTTGAAGCTGGAAGG | rp49 QPCR |
| Act4A_F | GCTTCGCTGTCTACTTTCCA | Act4A QPCR |
| Act4A_R | CAGCCCGACTACTGCTTAGA | Act4A QPCR |
| Adh_RT_F | GGCGGTCCCGGTGGTA | RT-PCR, Adh QPCR |
| Adh_RT_R | CTGGTAGATGGCATTGAATCC | RT-PCR, Adh QPCR |
2.5. UAS:Upf1D45B Negative Dominant and UAS:Vhln1 and UAS:Vhln2 Nonsense Mutant Lines
The entire coding sequence of the Vhl gene was amplified by PCR with Pfu Polymerase (Promega) using cDNA obtained by reverse transcription of total RNA extracted from adult flies. The PCR fragment was inserted into the pcDNA3 vector and verified by direct sequencing. The Upf1 and Vhl mutations were introduced in the cloned cDNAs by site-directed mutagenesis with the QuickChange II kit (Stratagene) using the following oligonucleotides: Upf1Fmut and Upf1Rmut for Upf1, Vhln1F and Vhln1R for Vhln1, and Vhln2F and Vhln2R for Vhln2 mutations (Table 1), respectively. Subsequently, mutated Upf1 and Vhl cDNAs were sequenced and inserted into pUAST vector by site directed cloning.
P element transformation was performed by microinjection of pUAST:Upf1 or pUAST:Vhl together with a Δ2-3 transposase containing plasmid into a w1118 Drosophila melanogaster strain [17]. Multiple lines were obtained for each injected construct. The expression of different upstream activating sequences (UASs) constructs was tested using a pGAL4 line that drives ubiquitous expression.
The Upf1 dominant negative activity of the transgenic lines selected was tested by crossing virgin females w1118;β2::GAL4 with w1118; Adhn4/Cy; UAS:Upf1D45B/TM3 and w1118; Adhfn6/Cy; UAS:Upf1D45B/TM3, respectively. The β2::GAL4 line drives the transgene expression in the testis. The levels of Adh and Upf1 mRNA from testes of w1118; Adhn4/β 2::GAL4; UAS:Upf1D45B/+ and w1118; Adhfn6/β2::GAL4; UAS:Upf1D45B/+ flies and the levels of Vhl from testes of w1118; UAS:Vhln1/β2::GAL4; UAS:Upf1D45B/+ and w1118; UAS:Vhln2/β2::GAL4; UAS:Upf1D45B/+ flies were analysed by QPCR.
2.6. Reverse Transcription PCR (RT-PCR) and Quantitative Real-Time PCR (QPCR)
Total RNA from testis of 50 adult flies was obtained using RNeasy Mini Kit and reverse transcripted using QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. RT-PCR was performed using pVHLF and pVHLR and Adh_RT_F and Adh_RT_R primers for Vhl and Adh, respectively (Table 1). QPCR was carried out in triplicates using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and run on ABI 7900HT Sequence Detection System with defaults parameters. The geometric mean of two reference genes (Rp49 and Act4A) was used to normalize the relative quantities. The calculations were made using the Comparative CT method as reported (User Bulletin #2, Applied Biosystems).
2.7. Minigenes Construct
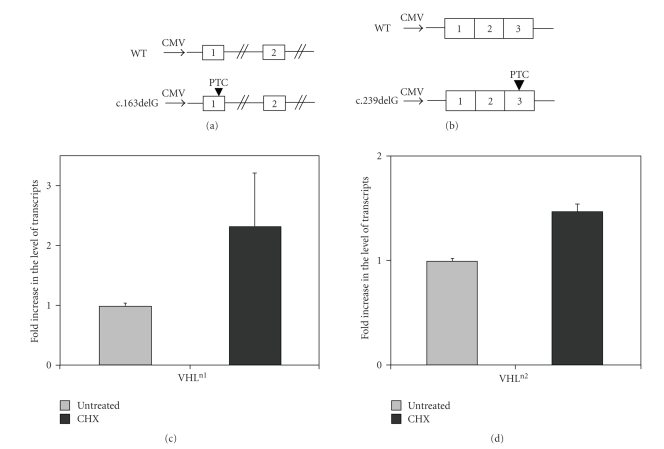
Construct 1 (Figure 3(a)). We ligated two PCR fragments from DNA of healthy individual carrying wild type VHL alleles. The PCR fragments contained exon 1 and part of its downstream intronic sequence (700 bp) and exon 2 and part of its flanking introns (390 bp at intron 1 and 134 bp at intron 2). The PCR fragments were amplified using the primers listed in Table 1. The different constructs were cloned into the pcDNA3.1 vector (Invitrogen). The c.163delG mutation was introduced by site-directed mutagenesis using the QuickChange II kit (Stratagene) with the generation of a stop codon in exon1 (V66X).
Figure 3.
Effect of CHX treatment on the level of the c.163delG and c.239delG VHL mRNA. (a) Scheme of the WT (upper panel) and c.163delG constructs, which contained the exons 1-2 (marked in the boxes by numbers), a part of the intronic sequence between exons 1-2 and a piece of intron downstream the exon 2. The CMV promoter is marked by a thick horizontal arrow. (b) Scheme of the WT and c.239delG construct containing the entire coding sequence of the VHL gene. (c) and (d) QPCR analysis of VHL transcripts before and following CHX treatment. The level of mRNA transcribed from VHL construct carrying either the wild type sequence or the c.163delG and c.239delG mutations was normalized to the mRNA level of GFP. The ratio between these normalized levels following CHX treatment was calculated and compared with the ratio in untreated cells. The fold increase in the level of VHL c.163delG and c.239delG transcripts is shown as mean ± SEM.
Construct 2 (Figure 3(b)). The entire coding sequence of the VHL gene was amplified by PCR using as template the cDNA obtained by reverse transcription of RNA extracted from HEK293 cells. The c.239delG stop codon mutation was introduced by site-directed mutagenesis.
2.8. Transfection, RNA Extraction, and QPCR on HEK 293 Cell Lines
HEK 293 cells were grown at 37°C in DMEM supplemented with 10% fetal calf serum and 1% penicillin-streptomycin (Invitrogen). They were seeded at 1 × 106 cells per 100 mm diameter petri dish 24 hours before transfection, performed by Fugene HD (Roche) with either 1 μg of VHL wt or mutant constructs. A GFP plasmid was used as a reference for transfection efficiency in each cell line. The mRNA levels of the different constructs were normalized to the mRNA level of GFP. Then, the ratio between the normalized mRNA levels transcribed from the mutant and the wt constructs following CHX treatment was calculated and compared with this ratio in untreated cells. The experiments were repeated at least three times.
3. Results
3.1. RT-PCR Analysis Reveals that Upf1, Adh, and Vhl Are Expressed in Ovaries and Testes of Adult Flies
To test whether Drosophila testis expresses detectable levels of the endogenous Upf1, Adh, and Vhl genes, we performed Northern blot and RT-PCR analysis on poly(A)+ and total RNA extract from ovaries and testes of adult flies. As shown in Figure 4 these analyses revealed that Upf1, Adh, and Vhl are normally expressed in ovaries and testes of adult flies.
Figure 4.
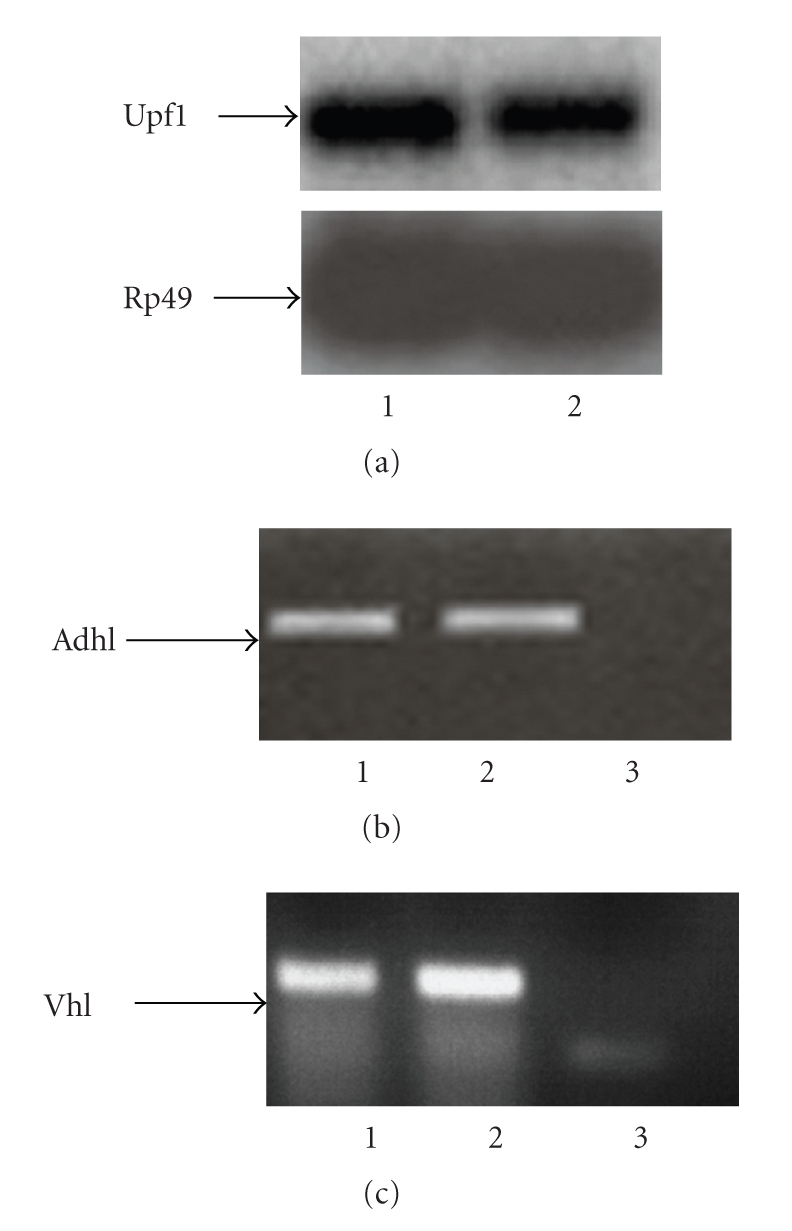
Upf1, Adh, and Vhl expression in Drosophila testes and ovaries. (a) Northern blot analysis of poly (A+) RNAs isolated from wild type adult flies. Lane 1: poly(A+) RNA from ovaries of adult females; lane 2: poly(A+) RNA from testes of adult males. Arrowheads on the left mark the position of the detected mRNAs. (b) RT-PCR analysis of Adh transcripts in wild type adult flies. Lane 1: total RNA from ovaries of adult females; lane 2: total RNA from testes of adult males; lane 3: negative control. (c) RT-PCR analysis of Vhl transcripts in wild type adult flies. Lane 1: total RNA from testes of adult males; lane 2: total RNA from ovaries of adult females; lane 3: negative control.
3.2. Upf1 Dominant-Negative Mutant Abolish Degradation of Nonsense Transcripts in Drosophila melanogaster
The yeast R779C and mammals R844C Upf1 mutations convert the conserved arginine to cysteine at residue 799 and 844 respectively, within the RNA helicase domain, conferring a dominant-negative effect on yeast and human Upf1p activity in nonsense-mediated mRNA decay pathway [18]. Because of the high identity of Drosophila UPF1 protein with the human and yeast Upf1 protein (67% and 53%, resp.) we introduced the same mutation into the fly Upf1 cDNA and we tested whether this mutation in vivo exerts a dominant negative effect on the regulation of alcohol dehydrogenase gene, Adh, a specific substrate of NMD, by utilizing the heterologous GAL4-UAS binary expression system [19].
By screening of a Drosophila melanogaster adult ovaric cDNA library with a Upf1 probe we isolated one positive clone that consisted of an Upf1 full-length containing an open reading frame of 3530 bp with a 5′-untranslated region of 372 bp and a 3′-untranslated region of 802 bp, respectively.
Using directed mutagenesis we generated Upf1 cDNA that carries the R822C substitution that mimics the yeast R779C and mammals R844C Upf1 mutations. This mutated Upf1 cDNA was cloned into P element expression vectors under the control of yeast GAL4-UAS. P element-mediated germ line transformation was used to generate an UAS:Upf1 transgenic fly line that we called UAS:Upf1D45B. We observed that the ubiquitous expression of the dominant-negative UPF1D45B protein, driven by actin-GAL4 driver line (P{Act5C − GAL4}17bFO1), caused 100% larval lethality (0/1445). Consistently, the observed phenotype and the efficacy of UAS:Upf1D45B transgene were confirmed by isolation and characterization of a loss-of-function mutation in the Drosophila Upf1 gene that causes lethality during larval development [14].
To verify whether Upf1D45B is able to modulate NMD pathway, we used QPCR to test its effect on mRNA levels of Adhn4 and Adhfn6, two nonsense mutations of the alcohol dehydrogenase gene (Adh) (Figure 1(a)), known to be targeted by NMD in S2 cells and in vivo [14, 20]. First, we measured the levels of Adh mRNA in both Adh mutant strains and we detected, in agreement with previous data [20], that they were 25% and 10% lower compared to the wild type strain (data not shown), because of increased turnover of the mutant transcript by the NMD pathway.
Since the ubiquitous expression of UPF1D45B protein caused larval lethality, we reasoned that the choice of a tissue in which the absence of the NMD pathway is not essential for Drosophila viability or development is fundamental to carry out functional analysis on NMD targets.
Interestingly, we observed that the expression of UPF1D45B protein in Drosophila testis, driven by a testis-specific GAL4 driver line (β2::GAL4), had no effect on testis development and/or larval lethality. Thus, we retained that the use of β2::GAL4 driver line could allow a detailed analysis of adult-onset phenotypes induced by loss of UPF1 activity.
Further, we tested whether the Upf1 mutation has a dominant-negative effect on the regulation of Adhn4 and Adhfn6transcripts by inducing the expression of UAS:Upf1D45B transgene in Drososphila testis. w1118; Adhn4/Cy; UAS:Upf1D45B/TM3 and w1118; Adhfn6/Cy; UAS:Upf1D45B/TM3 males were crossed to β2::GAL4 females and the levels of Adh and Upf1 mRNAs from testes of w1118; Adhn4/β2::GAL4; UAS:Upf1D45B/+ and w1118; Adhfn6/β2::GAL4; UAS:Upf1D45B/+ flies were measured by using QPCR. Consistently with [11] we observed a 3.8- and 8.8-fold accumulation of Adh mRNA when compared to the control (Figures 1(b) and 1(c)), implying that the expression of UPF1D45B mutation could abolish NMD pathway function.
Together these results strong point out that Upf1D45Bline represents a useful system to conduct a functional study to identify possible substrates of NMD pathway in Drosophila melanogaster.
3.3. Vhl Gene Is a Target of NMD in Drosophila melanogaster
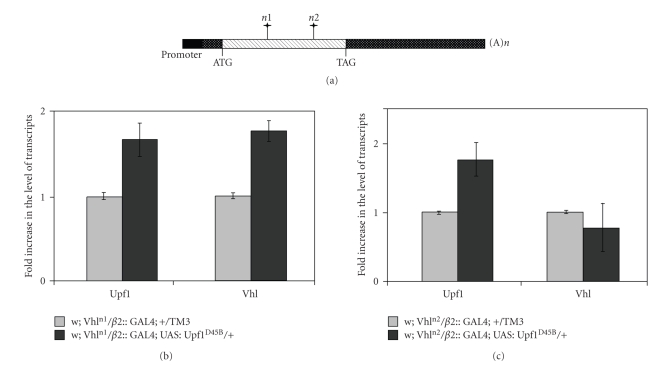
The UAS:Upf1D45Bline was used to investigate whether Vhl alleles carrying PTCs are degraded by NMD pathway. We constructed two different Vhl nonsense mutants by using site-directed mutagenesis strategy. The Vhln1allele contains the c.172delG frameshift mutation located in the exon 1 of Vhl gene. This mutation corresponds to the human c.163delG pathogenic mutation that we recently identified in a sporadic case of human cerebellar hemangioblastomas (Muscarella, submitted). The Vhln2 allele contains the c.254delC mutation corresponding to the c.239delG in human VHL gene (Figure 2(a)) [21].
Figure 2.
QPCR to measure the levels of transgenic Vhl lines. (a) Schematic map of the Vhl gene. Black rectangles represent the promoter region; hatched box represents the coding region; punctated boxes represent untranslated regions. The position of the point mutations n1 and n2 are indicated with vertical lines marked with a star. (b) and (c) We compared the levels of Vhl and Upf1 transcripts in flies that were heterozygous for Vhl nonsense alleles and for Upf1D45B. No increase of Vhl transcript in transgenic UAS:Vhln2 line was observed under the inhibition of NMD pathway whereas the UAS:Vhln1 transgenic line showed a 1.8 accumulation of nonsense-containing Vhl mRNA. The experiment was repeated three times.
P element-mediated germ line transformation was used to generate two independent transgenic fly lines carrying Vhl mutations, UAS:Vhln1 and UAS:Vhln2.
To verify whether the two different Vhl mutant transcripts were targeted by NMD pathway, w1118; UAS:Vhln1/Cy; UAS:Upf1D45B/TM3 and w1118; UAS:Vhln2/Cy; UAS:Upf1D45B/TM3 males were crossed to w1118;β2::GAL4 females. The mRNA levels of both wild type and mutated forms of Vhl and Upf1 from testis of w1118; UAS:Vhln1/β2::GAL4; UAS:Upf1D45B/+ and w1118; UAS:Vhln2/β2::GAL4; UAS:Upf1D45B/+ flies were measured by QPCR. We observed an increased of Vhl transcript in UAS:Vhln1/β2::GAL4; UAS:Upf1D45B/+ transgenic line. Conversely, the UAS:Vhln2/β2::GAL4;UAS:Upf1D45B/+ flies did not show any difference in Vhl expression compared to the Upf1 wild type line (Figures 2(b) and 2(c)).
3.4. VHL Mutations Are Targeted by NMD in Human Cell Lines
To investigate the stability of VHL wild type and certain human nonsense transcripts, we measured by QPCR the mRNA levels of constructs carrying wild type, c.163delG and c.239delG (Figures 3(a) and 3(b)) VHL mutations transfected into HEK 293 cells in absence and presence of cycloheximide (CHX), a widely used inhibitor of NMD. The analysis showed that the fold increase differs between the two mutants, with a modest increase in the level observed (1.4 ± 0.10) for c.239delG and a higher increase of 2.0 (2.4 ± 0.8) for c.163delG (Figures 3(c) and 3(d)).
4. Discussion
In humans, the role of NMD as a modifier of the phenotypic consequences of PTC is becoming more apparent. There are a consistent number of genetic diseases in which NMD partially mitigates the consequences of mutation owing to phenotypic variability.
The VHL is a well-known tumor suppressor gene, involved in cell cycle, regulation of hypoxia inducible genes and proper fibronectin assembly in extracellular matrix [22]. Germline mutations of the VHL gene lead to the development of the von Hippel-Lindau disease, a rare dominantly inherited familial cancer syndrome with a marked phenotypic variability and age-dependent penetrance.
The number of mutations in VHL gene is enormous and includes missense, nonsense, frameshift, and splice site mutations. In the past few years many different approaches using several molecular gene parameters have been used to make a possible correlation between VHL gene mutation and tumor phenotype [5–7, 23].
In the present study we generated Upf1 mutant fly line, UAS:Upf1D45B, to investigate whether two nonsense alleles of the Vhl gene are NMD targets. In agreement with others we observed that the ubiquitous expression of UPF1D45B protein, induced by a specific actin-GAL4 driver line, causes 100% larval lethality [14].
NMD pathway modulates the activity of specific native transcripts, whose misregulation would perturb the development or function of select cells or tissues and leading to lethality. Since Upf1 gene is broadly expressed and active throughout development, identification of the tissue target of Upf1 lethality will be an important first step to select the cellular substrates of NMD gene regulation.
Since the expression of UAS-transgene can be targeted to a specific tissue using the GAL4/UAS binary expression system, for our experiments we have chosen the β2::GAL4 line that drives the transgene expression in a region of testis in which the perturbation of NMD pathway is not essential for Drosophila viability and development.
The observed abolishing effect of degradation of Adh nonsense transcript, an NMD substrate, by Upf1 dominant-negative mutant suggested that the UAS:Upf1D45B line could be used as genetic system to screen nonsense mutations substrate of NMD pathway.
We investigated if the expression of two specific Vhl nonsense transcripts, Vhln1 and Vhln2, is modulated by NMD mechanism. The observed effect of NMD on the degradation of these two Vhl nonsense transcripts was different. In particular we showed the involvement of NMD in the degradation of Vhln1 transcript, whereas Vhln2 transgenic line showed no difference in Vhl expression compared to the wild type. These results suggest that the NMD of Vhl transcript carrying the c.172delG mutation is effective, whereas the c.254delC is not.
Similar results were observed in HEK293 cell line transfected with VHL constructs harboring the corresponding human VHL mutations after CHX treatment. Indeed while the c.239delG mutation transcript level showed only a marginal increase following the NMD inhibition, a fairly good increase of mRNA expression of c.163delG mutant transcript was observed.
Our data confirm that some frameshift mutations located in the last portion of VHL gene could escape NMD. In fact in both mammalian cells and Drosophila the c.239delG and the c.254delC transcripts, respectively, are immune to NMD.
Not all nonsense codons trigger NMD. In mammalian cells a splicing-dependent signal seems to be involved in PTC definition. Remarkably, stop codons located at least 50–55 nt upstream of an exon-exon boundary are generally defined as premature, whereas most PTCs downstream of this point do not elicit decay [24]. Notably, although the splicing and NMD machineries are conserved in D. melanogaster, the NMD in this organism was shown to occur independently of EJC components and PTC-containing transcripts that derive from intronless genes were found to be NMD-competent [11, 25].
Recent data demonstrated that the position of nonsense codons relative to the cytoplasmic poly(A)-binding protein 1 (PABPC1) is also a critical determinant for PTC definition both in Drosophila and in human [26]. Interestingly, if PABPC1 is in close proximity to the PTC, it seems to function as an NMD repressor. The observation that the Vhln1 but not the Vhln2 mRNA abundance is subject to NMD regulation might be attributable to the spatial rearrangements of the 3′UTR that close PABPC1 to the PTC. Probably, proximity of the PABPC1 protein to the PTC generated by c.172delG mutation inhibits NMD.
Another possible explanation for the difference observed between the two mutants should be that whereas the c.163delG mutation is located sufficiently upstream of an exon-exon junction and therefore is able to trigger NMD, the c.239delG mutation is located in the 3′-most exon. Therefore it is not surprising that this mutation is not able to trigger NMD in mammalian cells.
The possibility that VHL truncating mutations may recognised by NMD was recently verified [6]. In that study, inhibition of NMD in two sporadic RCC cell lines (786O and KTCL26) with endogenous frameshift VHL mutations that generated stop codons at residues 104 and 147 amino acidic composition residues, respectively, did not produce major changes in VHL mRNA expression (1.6- and 0.4-fold increase following emetine treatment, unpublished observations). The authors concluded that the NMD is not effective on the modulation of VHL because of the small length of VHL gene that consists of only three exons [6].
5. Conclusions
Our experiments confirm the utility of Drosophila melanogaster as an easy experimental system for understanding the NMD mechanism with a relevant potential applicability. Further molecular investigations on a greater number of Drosophila transgenic lines harbouring mutations that result in truncated proteins in different regions of Vhl gene or whatever also gene will be need to get more indications on the correlations between mutation position, activation of NMD in Drosophila, and specification of a definite phenotype.
Finally, a larger number of mutations need to be tested to definitely establish whether the NMD is involved in the pathogenesis of von Hippel Lindau disease.
Acknowledgments
The authors thank T. Kaufman for providing the strain β2::GAL4 which was originated in his laboratory. Any information about this strain should be directed to Professor. T. Kaufman (kaufman@indiana.edu). This work was supported by Grants from the Italian Ministry of Health (Ricerca Corrente 2006-07), the Fondazione Banca del Monte di Foggia “Domenico Siniscalco Ceci” to G. Merla. This work contributed equally to L. Micale and L. A. Muscarella.
References
- 1.Choyke PL, Glenn GM, Wagner JP, et al. Epididymal cystadenomas in von Hippel-Lindau disease. Urology. 1997;49(6):926–931. doi: 10.1016/s0090-4295(97)00074-5. [DOI] [PubMed] [Google Scholar]
- 2.Maher ER, Yates JRW, Harries R, et al. Clinical features and natural history of von Hippel-Lindau disease. Quarterly Journal of Medicine. 1990;77(283):1151–1163. doi: 10.1093/qjmed/77.2.1151. [DOI] [PubMed] [Google Scholar]
- 3.Manski TJ, Heffner DK, Glenn GM, et al. Endolymphatic sac tumors: a source of morbid hearing loss in von Hippel-Lindau disease. Journal of the American Medical Association. 1997;277(18):1461–1466. doi: 10.1001/jama.277.18.1461. [DOI] [PubMed] [Google Scholar]
- 4.Shehata BM, Stockwell CA, Castellano-Sanchez AA, Setzer S, Schmotzer CL, Robinson H. Von Hippel-Lindau (VHL) disease: an update on the clinico-pathologic and genetic aspects. Advances in Anatomic Pathology. 2008;15(3):165–171. doi: 10.1097/PAP.0b013e31816f852e. [DOI] [PubMed] [Google Scholar]
- 5.Gallou C, Chauveau D, Richard S, et al. Genotype-phenotype correlation in von Hippel-Lindau families with renal lesions. Human Mutation. 2004;24(3):215–224. doi: 10.1002/humu.20082. [DOI] [PubMed] [Google Scholar]
- 6.Kai RO, Woodward ER, Killick P, Lim C, Macdonald F, Maher ER. Genotype-phenotype correlations in von Hippel-Lindau disease. Human Mutation. 2007;28(2):143–149. doi: 10.1002/humu.20385. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Llorente S, Bravo J, Cebrian A, et al. Genetic characterization and structural analysis of VHL Spanish families to define genotype-phenotype correlations. Human Mutation. 2004;23(2):160–169. doi: 10.1002/humu.10309. [DOI] [PubMed] [Google Scholar]
- 8.Pulak R, Anderson P. mRNA Surveillance by the Caenorhabditis elegans smg genes. Genes and Development. 1993;7(10):1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends in Biochemical Sciences. 2006;31(11):639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes and Development. 1995;9(4):423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 11.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. The EMBO Journal. 2003;22(15):3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Molecular and Cellular Biology. 1997;17(3):1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J, Shu M-D, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when downstream of a termination codon. Cell. 2000;103(7):1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 14.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genetics. 2006;2(12, article e180) doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432(7013):112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 16.Brogna S. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA. 1999;5(4):562–573. doi: 10.1017/s1355838299981359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(17):10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 20.Brogna S, Bourtzis K, Gomulski LM, et al. Genomic organization and functional characterization of the alcohol dehydrogenase locus of Ceratitis capitata (Medfly) Insect Molecular Biology. 2006;15(3):259–268. doi: 10.1111/j.1365-2583.2006.00642.x. [DOI] [PubMed] [Google Scholar]
- 21.Stebbins CE, Kaelin WG, Jr., Pavletich NP. Structure of the VHL-elonginC-elonginB complex: implications for VHL tumor suppressor function. Science. 1999;284(5413):455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 22.Nyhan MJ, O’Sullivan GC, McKenna SL. Role of the VHL (von Hippel-Lindau) gene in renal cancer: a multifunctional tumour suppressor. Biochemical Society Transactions. 2008;36(3):472–478. doi: 10.1042/BST0360472. [DOI] [PubMed] [Google Scholar]
- 23.Maranchie JK, Afonso A, Albert PS, et al. Solid renal tumor severity in von Hippel Lindau disease is related to germline deletion length and location. Human Mutation. 2004;23(1):40–46. doi: 10.1002/humu.10302. [DOI] [PubMed] [Google Scholar]
- 24.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends in Biochemical Sciences. 1998;23(6):198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 25.Silva AL, Romão L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? The FEBS Letters. 2009;583(3):499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 26.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. The EMBO Journal. 2007;26(6):1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]