Abstract
Trypanosomes encode a family of proteins known as Major Surface Metalloproteases (MSPs). We have identified six putative MSPs encoded within the partially sequenced T. congolense genome. Phylogenic analysis indicates that T. congolense MSPs belong to five subfamilies that are conserved among African trypanosome species. Molecular modeling, based on the known structure of Leishmania Major GP63, reveals subfamily-specific structural variations around the putative active site despite conservation of overall structure, suggesting that each MSP subfamily has evolved to recognize distinct substrates. We have cloned and purified a protein encoding the amino-terminal domain of the T. congolense homologue TcoMSP-D (most closely related to Leishmania GP63). We detect TcoMSP-D in the serum of T. congolense-infected mice. Mice immunized with the amino-terminal domain of TcoMSP-D generate a persisting IgG1 antibody response. Surprisingly, a low-dose challenge of immunized mice with T. congolense significantly increases susceptibility to infection, indicating that immunity to TcoMSP-D is a factor affecting virulence.
1. Introduction
African trypanosomes cause devastating disease within humans and livestock. Trypanosoma brucei gambiense and T. b. rhodesiense cause sleeping sickness in humans. African trypanosomiasis in domestic animals is caused by infections with T. congolense, T. vivax, or T. b. brucei, of which T. congolense is the most important pathogen for livestock [1]. Antiparasite drugs are available but far from ideal [2]. Vaccination has not been effective as the periodic switching among genes encoding for the immunodominant variable surface glycoprotein (VSG) coat of the parasite precludes use of this surface molecule as a candidate [3–6].
Given the variability in the VSG coat we are examining the feasibility of generating pre-existing immunity to invariant surface proteins or secreted virulence factors as a means of controlling trypanosome infection. Our model organism is T. congolense, a causative agent of the chronic wasting disease of livestock, Nagana. T. congolense remains in the blood stream throughout the infection. Thus generation of a protective immune response to surface-exposed or secreted virulence factors of the blood stream form should have the potential to prevent or limit disease.
Among the African trypanosomes, T. brucei has previously been demonstrated to encode Major Surface Proteases (MSPs) with homology to MSPs of Leishmania [7]. In addition, the blood stream form T. brucei has been shown to be killed in vitro by a peptidomimetic inhibitor that specifically blocks L. major MSP activity [8]. Together, these observations suggest that at least one MSP homologue is expressed and essential for survival in the blood stream form of T. brucei. Based on this, and the findings that in Leishmania this family of proteins is important for virulence and is surface-exposed [9], we were encouraged to pursue whether T. congolense also encoded MSP homologues, and whether immunity to a member(s) of this family could attenuate T. congolense infection.
To date, there have been no reports of MSP homologues found in T. congolense. Fortunately, the African trypanosome sequencing project undertaken by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/) has now generated a significant amount of sequence information for T. congolense, though it has yet to be annotated. We performed searches of the available T. congolense sequence for predicted homologues to known MSPs of T. brucei and Leishmania sp. We found six potential MSP encoding ORFs. Two of the potential MSPs are >95% identical whereas the remaining four are <40% identical to each other, suggesting that we had tentatively identified five subfamilies of MSPs within T. congolense. A phylogenic analysis between the T. congolense MSPs and homologues found in other African and new world trypanosomes revealed that they indeed are representatives of five subfamilies that are shared among African trypanosomes. We subsequently cloned and expressed a region encoding the amino-terminal domain of the putative MSP ORF from T. congolense whose subfamily was found to group the closest with MSPs from Leishmania sp. Antisera against this domain revealed one major band in a Western blot of lysates of blood stream forms of T. congolense, strongly suggesting that this subfamily is expressed during infection. We describe the immunization with this purified partial protein and its effects on a subcutaneous infection of mice with a low dose of T. congolense.
2. Materials and Methods
2.1. Parasites
T. congolense, Trans Mara strain, variant antigenic types TC13 and TC14 were used in this study. The origin of the parasite strain [10] and clone TC13 [11] has been described previously. T. congolense clone TC14 was obtained from a C57BL/6 mouse infected with 103 T. congolense clone TC13. It was cloned from a blood sample collected 14 days after the infection. The variant surface glycoprotein of clone TC14 is different from that of clone TC13.
2.2. Sequence Analysis and Molecular Modeling
T. congolense DNA sequences encoding potential MSP homologues were identified using TBLASTN via the Trypanosoma Blast Server web page (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/t_brucei/), using the published amino acid sequences for T. brucei MSP-A, MSP-B, and MSP-C [7]. Potential open reading frames were identified using ORF Finder on the NCBI server (http://www.ncbi.nlm.nih.gov/projects/gorf/) and MSP family homology confirmed by independent BLAST analysis of each identified ORF against the NCBI database. ORFs showing homology to annotated MSPs from the NCBI database were collected. The closest homologues to these T. congolense putative MSPs in other individual Trypanosome species were identified using TBLASTN via the Trypanosoma Blast server for T. b. gambiense and T. vivax and via the NCBI Blast Server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for T. brucei TREU927, T.b. rhodesiense, and T. cruzi, as were homologues in Leishmania (taxid:5658). Full amino acid sequences for each protein analyzed are provided in the Supplementary Material available online at doi: 10.1155/2010/418157. Protein sequence alignments were generated and prepared for presentation using ClustalX ver 2.0.7 [12]. Phylogenic trees were generated by ClustalX using the bootstrapped N-J method with 1000 iterations. The unrelated E. coli protease HflB was included as an outgroup for alignment. Trees were prepared for presentation using PhyloDraw ver 0.8 as radial trees with terminal leaf extension for clarity of labeling, and Njplot ver 2.3 for display of bootstrap values (Supplementary Figure 1S). Three-dimensional structures for the predicted amino acid sequences of the putative MSPs were generated via the web-based server 3D-JIGSAW (http://bmm.cancerresearchuk.org/~3djigsaw/) [13], using the provided default settings. Figures were generated from the resulting structural files using PyMol1.1 [14]. Predicted amino acid sequences for the MSPs were submitted to PredGPI (http://gpcr2.biocomp.unibo.it/predgpi/pred.htm) [15] to predict the presence of a potential GPI anchor site and to Signal 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) [16] to predict the presence of a signal peptide indicative of secretion.
2.3. Cloning, Expression, and Purification of the Putative TcoMSP-D
The DNA sequence encoding the amino-terminal domain of the MSP homologue from clone TC13 (predicted residues 14–248), which we named TcoMSP-D (see below), was amplified via PCR from T. congolense chromosomal DNA. The primers used for PCR were as follows: amino end, 5′ CTGGGTACCGCAACAGTTTTGCTGCTGAC; carboxyl end, 5′ GTGCTCGAGTTAGGCCACATGTTCACTAAGG. Restriction enzyme recognition sites for KpnI (amino end primer) and XhoI (carboxyl end primer) used for directional cloning are underlined. The resulting PCR product was digested with KpnI and XhoI and ligated into similarly digested pET30a expression vector (Novagen). The protein construct is expressed under the control of the T7 promoter and contains 39 additional residues (MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTA) including a six residue histidine tag at the N-terminal. The plasmid was transformed into competent cells of the E. coli BL21(DE3) strain for protein expression. The amino-terminal domain of TcoMSP-C was similarly prepared but will be described in full elsewhere (H. Bull, G. Wei, V. Marcoux, H. Tabel unpublished).
Protein expression was induced by adding 0.4 mM isopropyl 1-thio-β-D-galactopyranoside when the cells reached an OD600 of 0.5–0.7 absorbance units, followed by a 2-3-hour incubation period at 37°C. The cells were then harvested by centrifugation, and the resulting pellet suspended in buffer A prior to lyses by French press. The resulting lysate was centrifuged at 15,000 g for 20 minutes and supernatant and pellet assayed by SDS PAGE for the presence of the expressed protein. The protein was found to remain with the cell debris pellet, likely as an inclusion body. The pellet was dissolved in 6M guanidine hydrochloride and the his-tagged protein was subsequently purified using nickel affinity chromatography in the presence of 6M guanidine hydrochloride throughout. The elution buffer was exchanged through extensive dialysis with PBS at pH 7.4. The protein yield was 2 mg mL−1 with an estimated 95% purity as determined by SDS-polyacrylamide gel electrophoresis and staining with coomassie brilliant blue.
2.4. Immunization and Antibody Production
Each of two female eight-week-old New Zealand White rabbits was injected subcutaneously at multiple sites with 1 mg purified amino-terminal domain of TcoMSP-D emulsified in TiterMax Gold adjuvant (Sigma chemicals). Rabbits were boosted at four weeks with identically prepared inoculums. Blood was collected at eight weeks, and separated serum was stored at –20°C without further purification. Groups of 5 six- to eight-week-old female BALB/c AnNCrlBR (BALB/c) mice were injected subcutaneously (s.c.) with TiterMax alone (Control), 5 μg, or 50 μg of N-terminal fragment of TcoMSP-D in TiterMax at 4 different sites (25 μL/site). Blood was collected at two, four, and six weeks and checked for serum antibody titers to purified amino-terminal domain of TcoMSP-D by ELISA. Rabbits and mice were kept in polycarbonate cages on sawdust and allowed free access to food and water throughout the experiments, according to the recommendation of the Canadian Council of Animal Care. All animal experiments were conducted in accordance with the standards of the University Committee on Animal Care and Supply of the University of Saskatchewan (Protocol numbers 20070043 and 19920139).
2.5. Western Analysis
Purified blood-stream T. congolense TC13 [17] was solubilized in 2X-SDS loading buffer (95°C for two minutes) and the equivalent of 5 million parasites per lane were separated by SDS PAGE in a 12% slab gel. Separated proteins were transferred to a Hybond-P (GE Healthcare) nylon membrane using a Minitrans-Blot cell (Bio-Rad) at 100 V for 1 hour. After transfer, membranes were blocked for one hour with TBS/0.2% Tween-20/2.5% skim milk, followed by one-hour incubation in the presence of immune sera (1/500 dilution for rabbit anti-amino-terminal domain of TcoMSP-D). Bound antibody was detected by incubation with HRP-conjugated secondary goat antirabbit antibody at 1/500 in TBS/0.2%Tween-20/2.5% skim milk. For detection of TcoMSP-D in serum of infected BALB/c mice the equivalent of 0.5 μL of cell-free serum was loaded per lane of a 12% SDS PAGE gel and detected as above.
2.6. ELISA for TcoMSP-D Amino-Terminal Domain-Specific Antibodies
Immulon 4HBX (Thermo) ELISA plates were coated overnight at 4°C with purified TcoMSP-D amino-terminal domain (1 μg/well in 100 μL of coating buffer). The plates were washed twice with PBS/Twen-20 (PBST) and blocked with 5% skim milk in PBST, 200 μL/well, for 2 hours at 37°C. After 4 washes, twofold serial diluted normal or test mouse sera were carried out in antibody diluents (PBST containing 2.5% skim milk), 100 μL/well. The plates were incubated for 2 hours at 37°C and then washed four times. HRP-conjugated goat antimouse IgG1 or IgG2a (Southern Biotechnology Associates) in antibody diluents was added and incubated for one hour at 37°C. After six washes with PBST, color development was achieved by adding 100 μL of TMB peroxidase substrate (KPL) and incubating the plates at room temperature in the dark for 10 to 20 minutes. The reaction was stopped by adding 50 μL of 1 M H2SO4 to each well. OD450 was read in a microtiter plate ELISA reader.
2.7. Mouse Infection Challenge
Mice immunized as described above were infected with 2 × 103 T. congolense clone TC14 s.c. in a hind footpad (i.f.p.). A drop of blood was taken from the tail vein of each infected mouse. The parasitemia was estimated by counting the number of parasites present in at least 10 fields at X400 magnification by phase-contrast microscopy. A count of 256 parasites per field is equivalent to ~109 parasites/mL [18]. The survival time was defined as the number of days post infection during which the infected mice remained alive. Moribund mice were euthanized.
2.8. Statistical Analysis
Data are represented as means ± SE. Significance of differences was determined by ANOVA using StatView SE1988 Software (Abacus Concepts) or a log-rank test for curve comparison using a GraphPad Prism computer program (BD Biosciences).
3. Results
3.1. Identifying MSP Homologues in T. congolense
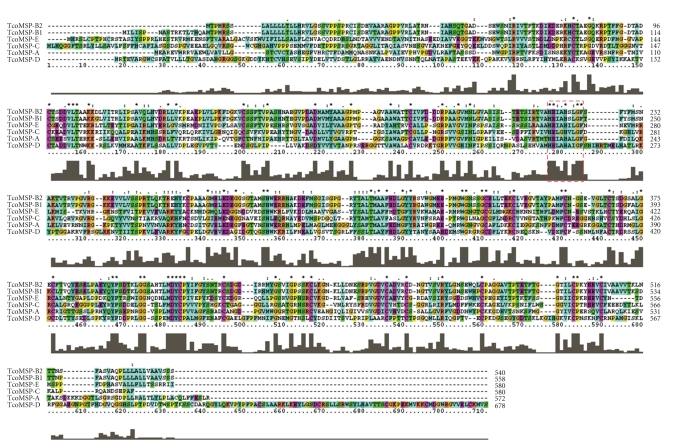
To identify potential MSP homologues encoded by T. congolense, we performed a TBLASTN analysis against the partially completed and nonannotated T. congolense genome sequencing database currently being compiled by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/). We searched for proteins with significant homology to previously characterize MSP-A, MSP-B, and MSP-C from T. brucei [7] as well as to the highly conserved nine amino acid motif thought to be important in Zn2+ binding at the catalytic active site. Both approaches yielded similar results. Our searches revealed a number of DNA sequence contigs that potentially encode MSP homologues. Each contig was examined for potential open reading frames (ORFs). The predicted protein sequence from each detected ORF was independently subjected to a BLAST homology search of the NCBI public databases to reveal those with high homology to previously identified members of the MSP family in related species. Using this approach we have identified six full length ORFs encoding putative MSP homologues in T. congolense. Four of the six full-length homologues are notably diverse in their predicted amino acid sequences (Figure 1). On the basis of phylogenic analysis described in detail below we have named the putative MSPs TcoMSP-A, TcoMSP-B1, TcoMSP-B2, TcoMSP-C, TcoMSP-D, and TcoMSP-E. Interestingly, the two members showing high homology to each other are located adjacent to each other on the same DNA contig, perhaps suggesting a tandem array with multiple members of this particular subfamily. Notably, others have found a family of MSP-B homologues to be arranged as a tandem array in T. brucei [19]. The remaining four genes identified are not flanked by MSP homologues within their respective assembled contigs.
Figure 1.
Multiple sequence alignment of T. congolense MSP homologues. The predicted amino acid sequences for each T. congolense MSP homologue were aligned to each other using the ClustalW alignment tool. Output was directly generated using ClustalX for Windows. The dashed red box indicates a conserved motif shown to be essential for Zn2+ binding in related proteins.
The alignment shown in Figure 1 reveals that there is considerable variation among the proteins, with extensive variation at both the amino and carboxyl ends. Remarkably however, there are 18 conserved cysteine residues interspersed throughout the proteins as well as a lesser number of conserved glycine and proline residues. This finding, in conjunction with the conservation of the putative active site, strongly suggests that this family of diverse proteins has retained a common set of folds, and thus an overall conservation of tertiary structure.
Given the potential retention of tertiary structure, we attempted molecular modeling of the putative MSPs using the 3DjigSaw protein modeling server (http://bmm.cancerresearchuk.org/~3djigsaw/). The program was able to generate predicted structure files for each MSP. The first column of Figure 2 presents a cartoon model for each predicted structure as well as the solved structure for L. major MSP GP63 (1LML.pdb) [20]. Despite some individual loop deviations the overall three domain core structure is remarkably well retained through all six structures. The overlays between each T. congolense homologue and 1LML structure in the center column reveal that the key folds around the putative active site (centered around the zinc ion shown in grey) are very highly conserved. This supports the interpretation that these proteins are homologues of L. major MSP GP63, which is a virulence factor [9]. However, the third column reveals key topological and surface charge differences between GP63 and between each of the predicted T. congolense MSPs around the putative active site. If these proteins retain protease activity, it is evident that they are very likely to have distinctly different substrate specificities.
Figure 2.
Predicted structures of T. congolense MSP homologues. Predicted amino acid sequences for each homologue were submitted for molecular modeling to the 3D-JigSaw comparative modeling web server (http://bmm.cancerresearchuk.org/~3djigsaw/). The solved Leishmania GP63 crystal structure 1LML.pdb was chosen by the program (independently of the user) as the best fit as a modeling template for each protein. The resulting generated pbd structure files were visualized using PyMOL Molecular Graphics System Version 1.1r1. Individual structures on the left side of the figure are as follows: (a) Leishmania GP63 (1LML.pdb), (b) TcoMSP-D, (c) TcoMSP-B1, (d) TcoMSP-A, (e) TcoMSP-C, and (f) TcoMSP-E. The center column depicts an overlay of that structure with the structure of Leishmania GP63. The third column depicts a surface representation of each predicted structure and Leishmania GP63 on the face containing the putative active site. Red-orange regions denote oxygen atoms, blue regions denote nitrogen atoms, green denotes carbon and hydrogen atoms, and yellow denotes sulfur atoms. The light grey sphere indicates the position of the Zn2+ ion in the solved Leishmania GP63 structure. It has been modeled into the predicted structures as an aid to orientation.
3.2. Identifying Subfamilies among MSP Homologues
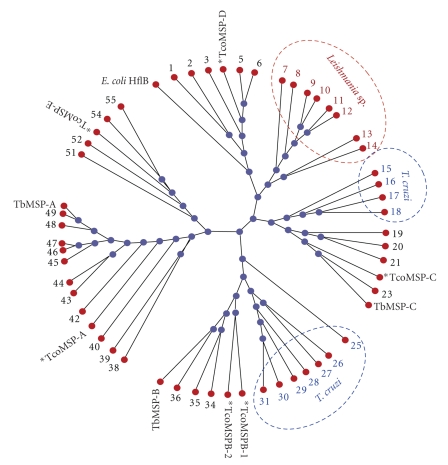
Given the observed diversity between the putative MSPs, we were interested to determine if they were representative members of conserved subfamilies in African trypanosomes, or if they were specific to T. congolense. To address this question, we queried the databases for predicted proteins showing the highest homology to each T. congolense MSP independently. To ensure sufficient depth to our analysis, we retained the predicted amino acid sequences for the three most homologous to each T. congolense MSP from each of T. brucei TREU 927, T. brucei rhodesience, T. brucei gambiense, and T. vivax as well as from the Central and South American trypanosome species T. cruzi. We also performed queries against the related Leishmania taxid. In cases where individual T. congolense MSPs retrieved the identical protein from a queried species, duplicates were removed. The amino acid sequences described for T. brucei MSP-A, MSP-B, and MSP-C were also included for comparison [7]. Figure 3 shows the radial phylogenic tree generated from comparison of this resulting group of 55 predicted protein sequences.
Figure 3.
Clustal-W generated dendogram comparing T. congolense putative MSPs with predicted homologues from T. brucei TREU927, T. brucei gambiense, T. vivax, T. brucei rhodesiense, T. cruzi, and Leishmania sp. (taxid 5658). Predicted protein sequences were obtained in a TBLASTN search against the translated genome database for each species indicated. The top three homologues were obtained for each T. congolense putative MSP independently for each species indicated. Redundant retrievals were eliminated. Protein sequence alignments were generated and prepared for presentation using ClustalX ver 2.0.7 [12]. Phylogenic trees were generated by ClustalX using the bootstrapped N-J method with 1000 iterations. The unrelated E. coli protease HflB was included as an outgroup for alignment. Trees were prepared for presentation using PhyloDraw ver 0.8 as radial trees. Note that the terminal arms have been extended for clarity of presentation and labeling and therefore should not be considered in interpreting phylogenetic distance. T. congolense putative MSP proteins are indicated in bold lettering. The T. brucei homologues previously characterized by LaCount et al. [7] are indicated for comparison. Nodes populated solely from individual proteins from T. cruzi and Leishmania sp. are indicated by blue and red dashed circles, respectively. The individual putative proteins represented by numbers in the tree are listed by number in Table 1. Full amino acid sequences for the proteins represented here are available in the supplementary information as is a phylogenetic tree with bootstrap values and genetic distance indicated.
A number of interesting features are revealed by the analysis. It is evident that indeed the six T. congolense MSPs we have identified do fall into five discrete subfamilies. The previously described T. brucei MSP-A, -B, and -C fall into discrete subfamilies and colocalize with TcoMSP-A, TcoMSP-B1 and 2, and TcoMSP-C, respectively. Notably, TcoMSP-D and TcoMSP-E each map to individual subfamilies that are clearly distinct from those containing T. brucei MSP-A, -B, and -C. This finding indicates that the African trypanosomes share, at a minimum, five distinct subfamilies of MSPs, each with closer homologues in related species than within their own species. This is expected for proteins that evolved and were selected in a common ancestor prior to speciation. This also very likely implies subfamily specific functions/targets.
In an attempt to further classify the MSPs examined in Figure 3, we submitted each of the proteins to predictions of secretion signals (signal peptides) and GPI anchors. Table 1 lists each of the MSP proteins analyzed and indicates the presence or absence of predicted signal peptides and GPI anchor signals. The homologues examined fall into three categories. Most (32 of the 55) are predicted to have a signal peptide and a moderate or high probability of having a GPI anchor. 19 are predicted to have a signal peptide and not have a GPI anchor, suggesting that they are either actively secreted, or anchored by another mechanism. Interestingly, the remaining four homologues are not predicted to encode either a signal peptide or a GPI anchor. If the motif predictions reflect biological function, this indicates that this subset of “Major Surface Proteases” may actually function within the cytoplasm.
Table 1.
Major surface protease homologues.
| Number | Species | Identifier* | Probability of a signal peptide | Probability of a GPI anchor |
|---|---|---|---|---|
| 1 | T. cruzi | XM_808234 | High (29-30) | moderate |
| 2 | T. cruzi | XM_806897 | Not probable | not probable |
| 3 | T. vivax | 1037d01.p1k | High (36-37) | not probable |
| 4 | T. congolense | TcoMSP-D | High (23-24) | not probable |
| 5 | T.b. gambiense | 11_v2.orf-3 | High (23-24 | not probable |
| 6 | T. brucei TREU927 | XM_823726 | High (23-24) | not probable |
| 7 | L. brazilliensis | XM_001562766 | High (30-31) | not probable |
| 8 | L. guyanensis | LEIGP63X | High (41-42) | High |
| 9 | L. mexicana | X64394 | High (41-42) | not probable |
| 10 | L. donovani | AJ495007 | High (41-42) | not probable |
| 11 | L. major | XM_001681325 | High (41-42) | High |
| 12 | L. major | XM_001681324 | High (41-42) | High |
| 13 | L. major | XM_001684283 | High (24-25) | High |
| 14 | L. infantum | XM_001470056 | High (24-25) | High |
| 15 | T. cruzi | XM_802134 | High (28-29) | not probable |
| 16 | T. cruzi | XM_807916 | High (30-31) | not probable |
| 17 | T. cruzi | XM_801120 | High (29-30) | not probable |
| 18 | T. cruzi | XM_799438 | High (29-30) | not probable |
| 19 | T. vivax | 942f04.q1k | High (30-31) | low |
| 20 | T. vivax | 1924b07.q1k | High (36-37) | High |
| 21 | T. vivax | 1240d01.p1k | High (36-37) | High |
| 22 | T. congolense | TcoMSP-C | High (30-31) | not probable |
| 23 | T.b. gambiense | 10_v2 | High (24-25) | not probable |
| 24 | T.b TREU927 | TbMSP-C=XM_817402 | High (24-25) | not probable |
| 25 | T. cruzi | XM_799266 | High (27-28) | High |
| 26 | T. cruzi | XM_802023 | High (59-60) | moderate |
| 27 | T. cruzi | XM_798784 | High (59-60) | moderate |
| 28 | T. cruzi | XM_815532 | High (22-23) | High |
| 29 | T. cruzi | XM_815930 | Poor (24-25) | moderate |
| 30 | T. cruzi | XM_812094 | High (20-21) | High |
| 31 | T. cruzi | XM_798806 | High (22-23) | High |
| 32 | T. congolense | TcoMSP-B1 | High (21-22) | High |
| 33 | T. congolense | TcoMSP-B2 | High (21-22) | High |
| 34 | T. brucei TREU927 | XM_841902 | High (58-59) | High |
| 35 | T.b. gambiense | 08_v2 | High (58-59) | High |
| 36 | T. brucei TREU927 | XM_841905 | High (64-65) | moderate |
| 37 | T.brucei TREU927 | TbMSP-B=XM_841904 | High (64-65) | moderate |
| 38 | T. vivax | 1393e12.p1k | High (20-21) | not probable |
| 39 | T. vivax | 797h07.q1k | High (22-23) | not probable |
| 40 | T. vivax | 899g05.p1k | High (19-20) | not probable |
| 41 | T. congolense | TcoMSP-A | High (27-28) | moderate |
| 42 | T. brucei TREU927 | XM_823758 | High (26-27) | High |
| 43 | T.b. gambiense | 1167d01.p1k | High (26-27) | High |
| 44 | T.b. rhodesiense | MSP-A2 | High (26-27) | High |
| 45 | T. brucei TREU927 | XM_823757 | High (26-27) | High |
| 46 | T. b. gambiense | 11_v2-orf2 | High (26-27) | High |
| 47 | T.b. gambiense | 2537b02.p1k | High (26-27) | High |
| 48 | T. brucei TREU927 | XM_823755 | High (26-27) | High |
| 49 | T.b. rhodesiense | MSP-A1 | High (26-27) | High |
| 50 | T.b. rhodesiense | TbMSP-A=U86345 | High (26-27) | High |
| 51 | T. vivax | 1124g05.p1k | High (28-29) | low |
| 52 | T. vivax | 1764g03.p1k | High (23-24) | moderate |
| 53 | T. congolense | TcoMSP-E | not probable | not probable |
| 54 | T. brucei TREU927 | XM_823843 | not probable | not probable |
| 55 | T.b. gambiense | 11_v2-orf1 | not probable | low |
*See supplementary data for full protein sequences.
Predictions of localization for each of the T. congolense MSPs can be made based on these motif patterns. TcoMSP-D and neighboring proteins 3–7 have strong signal peptide motifs and are not predicted to be GPI anchored. This suggests that these proteins are actively secreted. Interestingly, TcoMSP-C and its two closest predicted neighbors, including the previously described TbMSP-C protein, are also predicted to be actively secreted. Conversely, TcoMSP-B-1 and TcoMSP-B-2 and their MSP-B grouping homologues are predicted to be exported and surface linked via a GPI anchor. This prediction is in agreement with the known function of TbMSP-B in life-cycle specific cleavage of VSG from procyclic T. brucei [19]. It is very probable that each member of this subfamily provides the same function in each of the species represented. Of note, the T. cruzi specific subgrouping of MSPs (proteins 25–31) also universally encodes both signal peptides and GPI anchors. This commonality with the TbMSP-B subfamily is surprising given that T. cruzi does not express VSG but rather produces a variable mucin coat [21]. It is possible that this subgroup of T. cruzi MSPs plays an analogous role to TbMSP-B in the T. cruzi life cycle, perhaps in shedding bloodstream-expressed mucins. Similarly to the MSP-B subfamily, TcoMSP-A and the grouping homologues from 42–50 also are predicted to be exported and surface linked via a GPI anchor. The role and targets of the MSP-A homologues have yet to be elucidated. Unexpectedly, TcoMSP-E, and three out of four of its closest homologues appear to lack both a leading transport signal sequence and a GPI anchor motif indicating that they are retained within the cytoplasm.
The Leishmania MSPs are all predicted to encode a signal sequence but are mixed with regard to encoding predicted GPI anchors. It is possible that some MSP members failed to be recognized as having GPI anchors when in fact they do, as others have found that some variants of Leishmania MSP contain a variant GPI anchor signal [22]. Alternatively, it is likely that some subsets of Leishmania MSPs are actively secreted [23].
3.3. Cloning, Expression, and Purification of Amino-Terminal Domain of TcoMSP-D and Generation of Specific Antisera
Given the conservation of the MSP-D subfamily, even among the new world trypanosome T. cruzi, the known role of Leishmania MSPs as virulence factors, and the possibility that TcoMSP-D may be secreted, we elected to further examine TcoMSP-D as a potential target for attempted attenuation of T. congolense infection. Indeed, the three MSP-D homologues encoded by T. congolense, T. brucei TREU97, and T. gambiense display a remarkably high level of identity (Figure 4). It is very probable that these homologues target the identical substrate in vivo. It is therefore likely that conditions or treatments that are found to disrupt TcoMSP-D function will also prove effective against homologues in these related species.
Figure 4.
Multiple sequence alignment of MSP putative functional homologues: and T. congolense TcoMSP-D, T. brucei TREU927 XM_823726, T. gambiense chromosome 11 MSP ORF3. The predicted amino acid sequences for each homologue were aligned using the ClustalW alignment tool. Output was directly generated using ClustalX for Windows. The dashed red box indicates a conserved motif shown to be essential for Zn2+ binding in related proteins.
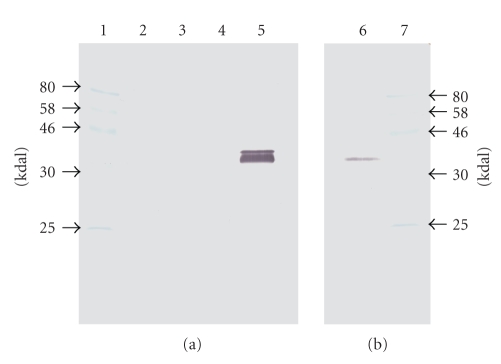
The amino-terminal domain from the ORF predicted to encode TcoMSP-D was amplified by PCR and cloned into the pET30a expression vector such that the expressed protein would encode a poly-histidine (6-his) tag on the amino end. The recombinant protein was purified following induction from the expression vector via nickel affinity chromatography. Purified protein was used to inoculate rabbits for polyclonal antibody production using a standard immunization protocol. Rabbit antiserum was used as the primary antibody for Western analysis of whole cell lysates of blood stream forms of T. congolense separated by SDS PAGE. Figure 5(b) reveals that the rabbit antiserum raised against the purified TcoMSP-D amino-terminal domain reacts with a protein with an apparent molecular weight of about 40,000 daltons, which is considerably less than the predicted molecular weight of 74,538 daltons for the full length nascent form. Not visible on the scanned image but visible to the naked eye are two faint larger molecular weight bands of about 50,000 and 70,000 daltons. The relative intensities of the observed bands are consistent both with rapid and significant posttranslational processing and with degradation of the full length protein, neither of which can be conclusively proven from these data. We have independently raised antiserum against the amino-terminal domain of TcoMSP-C and find that this serum detects a single protein of approximately 120,000 daltons in lysates of blood-form T. congolense (Bull, Wei, and Tabel, unpublished) suggesting that there is little cross-reaction between expressed MSPs in T. congolense (consistent with the low level of identity among the putative MSP members (Figure 1)), and that more than one subfamily is expressed in the blood stream form.
Figure 5.
Demonstration of TcoMSP-D in whole cell lysates of T. congolense and in cell-free serum of infected mice. Cell-free serum (equivalent to 0.2 μL of undiluted serum) from control and infected mice and T. congolense whole cell lysates were separated by SDS PAGE and transferred to nylon membrane for Western analysis. The membrane was probed with rabbit α-TcoMSP-D (NH3 domain) at 1/500 dilution and visualized with goat α-rabbit HRP conjugated secondary antibody at 1/500 dilution. Lane 1: molecular weight standards; lane 2: normal mouse serum; lane 3: serum from T. congolense-infected mouse at 3 days post infection; lane 4: empty lane; lane 5: serum from T. congolense-infected mouse at 6 days post infection; lane 6: lysate from equivalent of 5 million blood-form T. congolense parasites; lane 7: molecular weight standards.
In an attempt to resolve the localization of TcoMSP-D we tried to visualize TcoMSP-D on the surface of fixed blood stream forms of the parasite with the use of fluorescent antibodies. The attempts by both direct microscopic examination and by FACS were unsuccessful. However, we did detect TcoMSP-D in the serum of BALB/c mice following intraperitoneal infection with 103 T. congolense, clone TC13. No antigen was found in normal serum or serum collected at three days postinfection (DPI) when the parasitemia was less than 105 parasites per mL of blood. TcoMSP-D was detected as a strong double band in serum at six DPI (Figure 5(a)) when the parasitemia was 5 × 108 parasites per mL. The TcoMSP-D detected in the serum of infected mice had apparent molecular weights of about 40,000 and 43,000 daltons. While this result is in agreement with the prediction that TcoMSP-D is secreted and further processed and not surface-bound, we cannot rule out the possibility that the observed protein is simply a degradation product released from in vivo disrupted trypanosomes, or during serum preparation. The relative abundance of the detected protein band(s) in the 0.2 μL serum sample (equivalent to 1 × 105 parasites) versus the obviously reduced amount seen in 5 × 106 lysed purified blood-stream parasites suggests that during the course of infection TcoMSP-D accumulates in the blood to a level in excess of 50-fold greater than can be accounted for by intact parasites. This indicates either that TcoMSP-D is being actively secreted and is stable or that in vivo destruction of the parasites by day six is very prominent and released TcoMSP-D accumulates. Future studies will be required to determine whether the detected form of the protein has any function.
3.4. Effect of TcoMSP-D Immunization on Infection by T. congolense
We wished to address whether prior immunization of mice with recombinant TcoMSP-D amino-terminal domain provides protection against a T. congolense infection. BALB/c mice were immunized with purified protein in adjuvant and sera were collected at various times post immunization. Figure 6(a) shows that immunization generated a strong IgG1 antibody response to TcoMSP-D amino-terminal domain as measured by ELISA. There was also a detectable but modest IgG2a antibody response (Figure 6(b)). When we challenged the immunized mice with a dose of 104 T. congolense TC14 or higher, all control mice as well as all mice immunized with the amino-terminal domain of TcoMSP-D developed parasitemia (data not shown). Thus, at high-dose challenge the immunization had no observable effect.
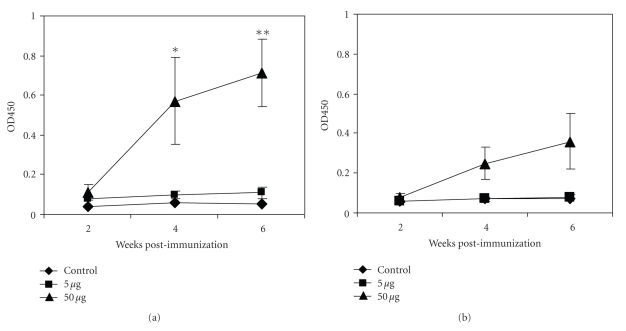
Figure 6.
Immunization with a high dose (50 μg/mouse) of TcoMSP-D resulting in an increased antigen specific IgG1 antibody response. Groups of 5 BALB/c mice were immunized subcutaneously with TiterMax alone (Control), 5 μg, or 50 μg of antigen TcoMSP-D (amino-terminal domain) in TiterMax at 4 different sites (25 μL/site). Serum was collected at weeks 2, 4, and 6 postimmunization. Sera IgG1 (a) and IgG2a (b) specific for TcoMSP-D were determined by ELISA. The 50 μg group showed a significantly higher IgG1 antibody titer than the control group at week 4 and week 6 postimmunization. *P ≤ .05; **P ≤ .005.
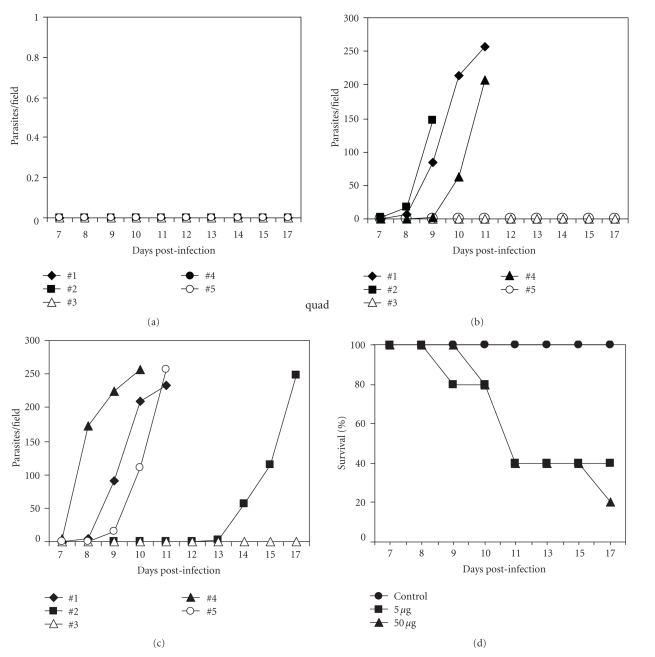
To determine if immunized mice had gained increased protection to a lower dose challenge, we challenged immunized and control (adjuvant only) BALB/c mice with a subcutaneous low-dose infection in the hind footpad. This route and challenge size was chosen as it provides a border line infection probability. The results of a typical challenge assay are shown in Figure 7. BALB/c mice that had received adjuvant only and were infected with 2 × 103 T. congolense clone TC14 did not develop parasitemia (Figure 7(a)) and remained entirely healthy until the end of the observation period of 30 days (Figure 7(d)). More than 50% of the mice immunized with either 5 μg or 50 μg of the amino-terminal domain of TcoMSP-D, however, developed parasitemia (Figures 7(b) and 7(c)) and had a mean 50% survival time of only 11 days (Figure 7(d)). Thus unexpectedly, immunization with TcoMSP-D amino-terminal domain results in a significant increase in susceptibility to infection with T. congolense.
Figure 7.
Immunization with TcoMSP-D amino-terminal domain resulted in increased susceptibility to infection: enhanced parasitemia and shorter survival time. Groups of 5 BALB/c mice were injected subcutaneously (s.c.) with TiterMax alone (Control), 5 μg, or 50 μg of the amino-terminal domain of TcoMSP-D in TiterMax at 4 different sites (25 μL/site). Six weeks postimmunization, all mice were infected with 2 × 103 T. congolense clone TC14 s.c. in a hind footpad. Parasitemia of the individual mice in each group [(a), control; (b), 5 μg; (c), 50 μg] and survival (d) were monitored after the infection. Mice which did not show any detectable parasitemia lived normally without any sign of disease until the termination of the experiment on day 30 postinfection. The results are representative of two different experiments.
4. Discussion
Kinetoplastid protozoa have been known to encode a conserved family of zinc metalloproteases on their surface, collectively called Major Surface Proteases (MSPs) [7, 9]. In vitro treatment of T. brucei with a peptidomimetic inhibitor, which has been demonstrated to inhibit a purified predominant L. major MSP in vitro, also results in active killing of T. brucei [8]. This finding strongly suggests that at least one MSP protein in T. brucei plays a vital role in cell survival. To date, there has not been any report of a member of this family of proteins in T. congolense. Given the expected conservation of this family of potentially surface-exposed proteins in T. congolense, and the potential for targeting this group of proteins as invariant surface antigens and key virulence factors, we have searched for potential homologues predicted to be encoded within the partially completed T. congolense genomic DNA sequence. We were successful in identifying six potential homologues. The large degree of variation among all but two of the predicted proteins suggested to us that these homologues were likely to have differing roles in the T. congolense life cycle. To date, three subtypes of MSPs have been described in the related T. brucei. These have been named TbMSP-A, TbMSP-B, and TbMSP-C [7]. Of these only TbMSP-B has been characterized to a level whereby its expression pattern and target substrate have been demonstrated, indicating that there is still very much to learn about the roles these proteins play in trypanosome biology [7, 19]. As a starting point to understanding the observed divergence among the T. congolense MSPs we performed a phylogenic analysis including the closest homologues from related trypanosomes and Leishmania sp. Our finding of five conserved subfamilies among the African trypanosomes clearly indicates that we have identified diverse, conserved functional subfamilies within the MSP family of proteins. Not surprisingly, the TbMSP-A, -B, -C proteins each fall in a separate subfamily. In addition, we have identified two novel subfamilies which we have named MSP-D and MSP-E. We believe this to be a major finding. However it is important to stress that, as was the case for the initial report of T. brucei MSP subfamilies [7], additional subfamilies may become apparent as additional complete genomic sequences become available. Indeed, T. congolense may likely be shown to harbor additional MSPs that have yet to be sequenced.
Notably, the less related New World trypanosome T. cruzi has members in the MSP-D (proteins 1 and 2 in Figure 3 and Table 1) and MSP-C (proteins 15–18 in Figure 3 and Table 1) subfamilies as well as a grouping of six related proteins in what appears to be their own subfamily that maps near the MSP-B subfamily (Figure 3). This may suggest that the MSP-D and MSP-C subfamilies are more ancient than, for example, the MSP-A subfamily, or alternatively, that this subfamily was lost in the T. cruzi species after species separation. As a consequence, the MSP-A subfamily may be uniquely required for an aspect of the African trypanosome life cycle that is not shared with T. cruzi. Similarly, even though the protein homologues most similar to each individual T. congolense MSP were independently obtained from the database, the entirety of unique Leishmania homologues groups together in the phylogenic tree. This indicates that each is more similar to other Leishmania proteins than to any Trypanosome protein included in the analysis. This may indicate separation of Leishmania from trypanosomes prior to the generation of the subfamilies or species-specific loss of all but one of the subfamilies in Leishmania species. Notably, Leishmania MSPs have recently been shown to fall into three distinct clades as well [24]. It may be of interest to determine if the homologues retrieved here are restricted to one clade or represent multiple clades. Of particular interest is the finding that the Leishmania MSP subfamily branches out closest to the MSP-D subfamily, which, as argued above, may be one of the more ancient subfamilies.
To test the potential usefulness of immunization against members of a given MSP subfamily, we chose to concentrate on TcoMSP-D. This member had a number of perceived characteristics that attracted our attention. The MSP-D subfamily falls in a node that is closest to the MSPs of Leishmania. A Leishmania MSP, called GP63 or leishmanolysin, is an important virulence factor that has been demonstrated, among other functions, to protect L. major against complement-mediated lysis [25]. GP63 can cleave complement component C3 and in particular cleave C3b into iC3b [25], which, in turn, prevents complement-mediated lysis of L. major but favors its CR3-mediated phagocytosis by macrophages [25].
T. congolense infections lead to profound and persistent hypocomplementia in cattle [26], sheep [27], and mice [28]. Homogenates of T. congolense, and particulate as well as supernatant fractions thereof, activate bovine complement via the alternative pathway [10]. Binding of IgM antibodies to the variant surface glycoprotein of T. congolense activates complement, mediates binding of C3b to the trypanosomes, and leads to phagocytosis of T. congolense by macrophages via complement receptor 3 (CR3) [29]. Lysis of T. congolense by antibody and complement is inefficient [30, 31]. Since we found that TcoMSP-D appears to be most closely related to Leishmania GP63 (Figure 3), we are considering that TcoMSP-D may have enzyme activities similar to those of Leishmania GP63. Our demonstration that TcoMSP-D accumulates in mouse serum to greater than 50-fold higher levels than can be accounted for by intact live trypanosomes by day 6 post infection (Figure 5) is consistent with an extracellular role. It is intriguing to speculate that the inefficiency of complement-mediated lysis of T. congolense might be mediated by the effects of TcoMSP-D. Definitive testing of this hypothesis will require purification of the active form of the protein.
Our finding that immunization against TcoMSP-D amino-terminal domain significantly enhanced the infectivity of a low dose of T. congolense was unanticipated. We expected that the binding of the antibody to the MSP would inhibit the function of this MSP and either would lead to an attenuated infection, or would have no detectable effect. The demonstration of an effect of enhanced infectivity (Figure 7) indicates that immune responses against this MSP protein can somehow downregulate the resistance to infection. CD4+ CD25+ Foxp3+ T regulatory cells are involved in immune suppression in T. congolense infections [32]. When mice were infected subcutaneously with a low dose of T. congolense clone TC13 (102), they controlled the infection but showed enhanced susceptibility upon subcutaneous challenge with a different clone of T. congolense (Wei and Tabel, unpublished). Though this finding very closely mirrors the increased susceptibility seen in TcoMSP-D immunized mice, we presently have no information on the potential mechanism(s) responsible. Perhaps significantly, the amino-terminal domain of TcoMSP-D, used for immunization, did not include the potential catalytic site (Figure 1). How could one envision an increase of infectivity of T. congolense by a specific antibody response to the N-terminal fragment of the putative enzyme? It is conceivable that, rather than neutralizing enzyme activity, the binding of antibodies to the N-terminal portion of the putative enzyme might actually stimulate its activity, as has been demonstrated for antibody binding to horse radish preoxidase [33]. The propeptide of Leishmania GP63 inhibits the activation of the proenzyme [34]. It is conceivable that the antibody, when binding to the N-terminal domain of the putative proenzyme form of TcoMSP-D, might alter the structure of the protein in such a way that it is able to exert enzyme activity.
In summary, we have cloned, expressed, and purified a partial protein encoding the amino-terminal domain of TcoMSP-D, the T. congolense MSP homologue found to be most closely related to Leishmania GP63. Whatever the mechanism might be, our observations have led us to conclude that TcoMSP-D is a virulence factor of T. congolense.
Supplementary Material
Supplementary figure contains Clustal-W generated bootstrapped phylogenetic tree comparing T. congolense putative MSPs with predicted homologues from T. brucei TREU927, T. brucei gambiense, T. brucei vivax, T. brucei rhodesiense, T. cruzi, and Leishmania sp. (taxid 5658).
Acknowledgments
V. Marcoux was supported by an undergraduate research fellowship from the Immunology and Infectious Disease Research Group, University of Saskatchewan, and a Dean's Project fellowship, College of Medicine, University of Saskatchewan. H. J. Bull received start-up funding from the College of Medicine, University of Saskatchewan. Contributions by G. Wei and H. Tabel were supported by CIHR/RPP Grant no. 85167. The authors thank P. Bretscher and J. Hill for helpful discussion.
References
- 1.Mulligan HW, Potts WH, editors. The African Trypanosomiases. New York, NY, USA: Wiley-InterScience; 1970. [Google Scholar]
- 2.Barrett MP, Burchmore RJ, Stich A, et al. The trypanosomiases. The Lancet. 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 3.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Advances in Parasitology. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 4.Cross GAM. Cellular and genetic aspects of antigenic variation in trypanosomes. Annual Review of Immunology. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 5.McCulloch R. Antigenic variation in African trypanosomes: monitoring progress. Trends in Parasitology. 2004;20(3):117–121. doi: 10.1016/j.pt.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends in Parasitology. 2005;21(11):517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 7.LaCount DJ, Gruszynski AE, Grandgenett PM, Bangs JD, Donelson JE. Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. Journal of Biological Chemistry. 2003;278(27):24658–24664. doi: 10.1074/jbc.M301451200. [DOI] [PubMed] [Google Scholar]
- 8.Bangs JD, Ransom DA, Nimick M, Christie G, Hooper NM. In vitro cytocidal effects on Trypanosoma brucei and inhibition of Leishmania major GP63 by peptidomimetic metalloprotease inhibitors. Molecular and Biochemical Parasitology. 2001;114(1):111–117. doi: 10.1016/s0166-6851(01)00244-4. [DOI] [PubMed] [Google Scholar]
- 9.Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Molecular and Biochemical Parasitology. 2003;132(1):1–16. doi: 10.1016/s0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- 10.Tabel H. Activation of the alternative pathway of bovine complement by Trypanosoma congolense. Parasite Immunology. 1982;4(5):329–335. doi: 10.1111/j.1365-3024.1982.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 11.Wei G, Qualtiere L, Tabel H. Trypanosoma congolense: complement independent immobilization by a monoclonal antibody. Experimental Parasitology. 1990;70(4):483–485. doi: 10.1016/0014-4894(90)90133-w. [DOI] [PubMed] [Google Scholar]
- 12.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 13.Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;45(supplement 5s):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 14.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, Calif, USA: DeLano Scientific; 2008. [Google Scholar]
- 15.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9, article 392 doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 17.Lanham SM, Godfrey DG. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Experimental Parasitology. 1970;28(3):521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- 18.Herbert WJ, Lumsden WHR. Trypanosoma brucei: a rapid “matching” method for estimating the host’s parasitemia. Experimental Parasitology. 1976;40(3):427–431. doi: 10.1016/0014-4894(76)90110-7. [DOI] [PubMed] [Google Scholar]
- 19.Grandgenett PM, Otsu K, Wilson HR, Wilson ME, Donelson JE. A function for a specific zinc metalloprotease of African trypanosomes. PLoS Pathogens. 2007;3(10):1432–1445. doi: 10.1371/journal.ppat.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlagenhauf E, Etges R, Metcalf P. The crystal structure of the Leishmania major surface proteinase leishmanolysin (GP63) Structure. 1998;6(8):1035–1046. doi: 10.1016/s0969-2126(98)00104-x. [DOI] [PubMed] [Google Scholar]
- 21.Buscaglia CA, Campo VA, Frasch AC, Di Noia JM. Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nature Reviews Microbiology. 2006;4(3):229–236. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 22.Voth BR, Kelly BL, Joshi PB, Ivens AC, McMaster WR. Differentially expressed Leishmania major GP63 genes encode cell surface leishmanolysin with distinct signals for glycosylphosphatidylinositol attachment. Molecular and Biochemical Parasitology. 1998;93(1):31–41. doi: 10.1016/s0166-6851(98)00013-9. [DOI] [PubMed] [Google Scholar]
- 23.Halle M, Gomez MA, Stuible M, et al. The Leishmania surface protease GP63 cleaves multiple intracellular proteins and actively participates in p38mitogen-activated protein kinase inactivation. Journal of Biological Chemistry. 2009;284(11):6893–6908. doi: 10.1074/jbc.M805861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauricio IL, Gaunt MW, Stothard JR, Miles MA. Glycoprotein 63 (GP63) genes show gene conversion and reveal the evolution of Old World Leishmania. International Journal for Parasitology. 2007;37(5):565–576. doi: 10.1016/j.ijpara.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Brittingham A, Morrison CJ, McMaster WR, McGwire BS, Chang K-P, Mosser DM. Role of the Leishmania surface protease GP63 in complement fixation, cell adhesion, and resistance to complement-mediated lysis. The Journal of Immunology. 1995;155(6):3102–3111. [PubMed] [Google Scholar]
- 26.Rurangirwa FR, Tabel H, Losos G, Tizard IR. Hemolytic complement and serum C3 levels in zebu cattle infected with Trypanosoma congolense and Trypanosoma vivax and the effect of trypanocidal treatment. Infection and Immunity. 1980;27(3):832–836. doi: 10.1128/iai.27.3.832-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malu MN, Tabel H. The alternative pathway of complement in sheep during the course of infection with Trypanosoma congolense and after Berenil treatment. Parasite Immunology. 1986;8(3):217–229. doi: 10.1111/j.1365-3024.1986.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 28.Otesile EB, Lee M, Tabel H. Plasma levels of proteins of the alternative complement pathway in inbred mice that differ in resistance to Trypanosoma congolense infections. Journal of Parasitology. 1991;77(6):958–964. [PubMed] [Google Scholar]
- 29.Pan W, Ogunremi O, Wei G, Shi M, Tabel H. CR3 (CD11b/CD18) is the major macrophage receptor for IgM antibody-mediated phagocytosis of African trypanosomes: diverse effect on subsequent synthesis of tumor necrosis factor α and nitric oxide. Microbes and Infection. 2006;8(5):1209–1218. doi: 10.1016/j.micinf.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Tabel H, Kaushik RS, Uzonna JE. Susceptibility and resistance to Trypanosoma congolense infections. Microbes and Infection. 2000;2(13):1619–1629. doi: 10.1016/s1286-4579(00)01318-6. [DOI] [PubMed] [Google Scholar]
- 31.Tabel H, Wei G, Shi M. T cells and immunopathogenesis of experimental African trypanosomiasis. Immunological Reviews. 2008;225(1):128–139. doi: 10.1111/j.1600-065X.2008.00675.x. [DOI] [PubMed] [Google Scholar]
- 32.Wei G, Tabel H. Regulatory T cells prevent control of experimental African trypanosomiasis. The Journal of Immunology. 2008;180(4):2514–2521. doi: 10.4049/jimmunol.180.4.2514. [DOI] [PubMed] [Google Scholar]
- 33.Ermolenko DN, Zherdev AV, Dzantiev BB, Popov VO. Antiperoxidase antibodies enhance refolding of horseradish peroxidase. Biochemical and Biophysical Research Communications. 2002;291(4):959–965. doi: 10.1006/bbrc.2002.6544. [DOI] [PubMed] [Google Scholar]
- 34.Macdonald MH, Morrison CJ, McMaster WR. Analysis of the active site and activation mechanism of the Leishmania surface metalloproteinase GP63. Biochimica et Biophysica Acta. 1995;1253(2):199–207. doi: 10.1016/0167-4838(95)00155-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure contains Clustal-W generated bootstrapped phylogenetic tree comparing T. congolense putative MSPs with predicted homologues from T. brucei TREU927, T. brucei gambiense, T. brucei vivax, T. brucei rhodesiense, T. cruzi, and Leishmania sp. (taxid 5658).