Abstract
Olfaction is a primitive sense in organisms. Both vertebrates and insects have receptors for detecting odor molecules in the environment, but the evolutionary origins of these genes are different. Among studied vertebrates, mammals have ∼1,000 olfactory receptor (OR) genes, whereas teleost fishes have much smaller (∼100) numbers of OR genes. To investigate the origin and evolution of vertebrate OR genes, I attempted to determine near-complete OR gene repertoires by searching whole-genome sequences of 14 nonmammalian chordates, including cephalochordates (amphioxus), urochordates (ascidian and larvacean), and vertebrates (sea lamprey, elephant shark, five teleost fishes, frog, lizard, and chicken), followed by a large-scale phylogenetic analysis in conjunction with mammalian OR genes identified from nine species. This analysis showed that the amphioxus has >30 vertebrate-type OR genes though it lacks distinctive olfactory organs, whereas all OR genes appear to have been lost in the urochordate lineage. Some groups of genes (θ, κ, and λ) that are phylogenetically nested within vertebrate OR genes showed few gene gains and losses, which is in sharp contrast to the evolutionary pattern of OR genes, suggesting that they are actually non-OR genes. Moreover, the analysis demonstrated a great difference in OR gene repertoires between aquatic and terrestrial vertebrates, reflecting the necessity for the detection of water-soluble and airborne odorants, respectively. However, a minor group (β) of genes that are atypically present in both aquatic and terrestrial vertebrates was also found. These findings should provide a critical foundation for further physiological, behavioral, and evolutionary studies of olfaction in various organisms.
Keywords: olfactory receptor, multigene family, molecular evolution, chordate, vertebrate, amphioxus
Introduction
In vertebrates, odor molecules in the environment are detected by olfactory receptors (ORs) that are predominantly expressed in the main olfactory epithelium in the nasal cavity (Buck and Axel 1991; for review, see Niimura and Nei 2006; Nei et al. 2008). To distinguish among tens of thousands of different odorants, the vertebrate genome contains numerous OR genes, which form the largest multigene family in vertebrates. Vertebrate ORs are G protein–coupled receptors (GPCRs) having seven transmembrane α-helical regions. GPCRs can be classified into five groups by sequence similarities (Fredriksson et al. 2003), and OR genes belong to the largest group of them, the rhodopsin-like GPCR superfamily. Ligands for the rhodopsin-like GPCRs are highly diverse and include photons (for opsin genes), neurotransmitters, peptide hormones, chemokines, lipids, and nucleotides, in addition to odorants. Insects also have OR genes in their genomes, but insect and vertebrate OR genes share no sequence similarity (see Nei et al. 2008 and the references therein). Also, although insect ORs contain seven transmembrane α-helical regions, their membrane topology is inverted compared with that of rhodopsin-like GPCRs. Therefore, vertebrate and insect OR genes are thought to have different evolutionary origins.
Entire repertoires of OR genes have been studied in humans (Glusman et al. 2001; Zozulya et al. 2001; Niimura and Nei 2003), mice (Young et al. 2002; Zhang and Firestein 2002; Niimura and Nei 2005a), dogs (Quignon et al. 2003; Olender et al. 2004), and other mammals (Niimura and Nei 2007). These studies have revealed that the numbers of OR genes in mammals vary extensively, ranging from <400 in higher primates or platypuses to ∼1,200 in rats or opossums (Niimura and Nei 2007; Go and Niimura 2008). On the other hand, whole-genome analyses of OR gene families in nonmammalian vertebrates are relatively limited (Alioto and Ngai 2005; Niimura and Nei 2005b). It is generally thought that teleost fishes have much smaller numbers of OR genes than mammals (∼100, Ngai et al. 1993). Fish detect mainly four groups of water-soluble molecules as odorants: amino acids, gonadal steroids, bile acids, and prostaglandins. These odorants are nonvolatile, so humans cannot smell them (Laberge and Hara 2001).
Previously, we identified the entire sets of OR genes from draft genome sequences of the zebra fish, fugu, western clawed frog, and chicken (Niimura and Nei 2005b). Phylogenetic analyses showed that fish OR genes are more diverse than mammalian OR genes despite the smaller repertoires in fish compared with mammals. The analyses also indicated that the entire set of vertebrate OR genes can be classified into two groups of genes, named Type 1 and Type 2 genes. Mammalian OR genes are known to be clearly classified into class I and class II (Glusman et al. 2000), and both classes of genes belong to Type 1 (Niimura and Nei 2005b, 2006). Moreover, it was suggested that the most recent common ancestor (MRCA) between teleost fishes and tetrapods had at least nine ancestral OR genes, but only two of them (named groups α and γ) were dramatically expanded in the tetrapod lineage (Niimura and Nei 2005b).
Prior to the advent of whole-genome sequences, several OR genes were identified from a jawless vertebrate, the lamprey. Berghard and Dryer (1998) and Freitag et al. (1999) reported putative OR genes that are expressed in the olfactory epithelia of river lampreys. However, only the genes identified in Freitag et al. (1999) showed significant sequence similarities to known vertebrate OR genes. Later, Liberles and Buck (2006) reported that trace amine–associated receptors (TAARs), which were initially identified as receptors to a specific group of biogenic amines in the brain, are actually expressed in the olfactory epithelia in mice and are able to be regarded as a second class of ORs. Phylogenetic studies indicated that the genes reported by Berghard and Dryer (1998) belong to a TAAR gene family rather than an OR gene family (Hashiguchi and Nishida 2007). Satoh (2005) reported one OR-like gene from amphioxus, the most basal chordate species and showed it to be expressed in the rostral epithelia of the adult amphioxus.
Recently, whole-genome sequences of several key organisms in chordate evolution have become available. Chordates include cephalochordates, urochordates, and vertebrates. Now the genome sequences of the amphioxus Branchiostoma floridae (Putnam et al. 2008), two species of ascidians, Ciona intestinalis (Dehal et al. 2002) and Ciona savignyi (Vinson et al. 2005), and the larvacean Oikopleura dioica (Seo et al. 2001) are available: Amphioxus is a cephalochordate, and ascidians and larvaceans are urochordates. As representatives of jawless vertebrates and cartilaginous fishes, two early-diverging lineages in vertebrates, the genome sequences of the sea lamprey Petromyzon marinus and the elephant shark Callorhinchus milii (Venkatesh et al. 2007), respectively, have been determined. Moreover, the draft genome sequences of five teleost fishes are also available: zebra fish, medaka (Kasahara et al. 2007), stickleback, fugu (Aparicio et al. 2002), and spotted green puffer fish (Jaillon et al. 2004). To investigate the early evolution of vertebrate OR gene families and to obtain insights into the origin of this tremendous gene family, in the present study, I identified the OR gene repertoires using the genome sequences of 14 nonmammalian chordate species and conducted a large-scale phylogenetic analysis together with mammalian OR genes.
Materials and Methods
Data
In this study, I analyzed the whole-genome sequences of 14 nonmammalian chordate species and the sea urchin. In addition, nine mammalian genome sequences were used for the search of Type 2 genes (see below). The draft genome sequence of amphioxus (B. floridae, Assembly v2.0; Putnam et al. 2008) was obtained from the Joint Genome Institute Web site (http://genome.jgi-psf.org/euk_home.html). The genome sequences of sea squirts (C. intestinalis, version 2.0, released in March 2005; Dehal et al. 2002, and C. savignyi, CSAV 2.0, released in October 2005; Vinson et al. 2005), zebra fish (Danio rerio, Zv7, released in April 2007), and opossum (Monodelphis domestica, monDom4, released in January 2006; Mikkelsen et al. 2007) were retrieved from the Ensembl Genome Browser (http://www.ensembl.org). The larvacean genome (O. dioica, Assembly v3, released in February 2007; Seo et al. 2001) was obtained from the Genoscope Web site (http://www.genoscope.cns.fr/spip/Projects.html). The sea lamprey (P. marinus, Petromyzon_marinus-3.0, released in February 2007) and platypus (Ornithorhynchus anatinus, Ornithorhynchus_anatinus-5.0, released in December 2005; Warren et al. 2008) genome sequences were downloaded from the Genome Sequencing Center at Washington University School of Medicine (http://genome.wustl.edu). The elephant shark (C. milii) genome sequence was obtained from the Elephant Shark Genome Project Web site (http://esharkgenome.imcb.a-star.edu.sg/, 1.4× assembly; Venkatesh et al. 2007). The genome sequences of medaka (Oryzias latipes, oryLat1, released in April 2006; Kasahara et al. 2007), stickleback (Gasterosteus aculeatus, gasAcu1, released in February 2006), fugu (Takifugu rubripes, fr2, released in October 2004; Aparicio et al. 2002), tetraodon (Tetraodon nigroviridis, tetNig1, released in Feb. 2004; Jaillon et al. 2004), western clawed frogs (Xenopus tropicalis, xenTro2, released in August 2005), lizard (Anolis carolinensis, anoCar1, released in January 2007), chicken (Gallus gallus, galGal3, released in May 2006; International Chicken Genome Sequencing Consortium 2004), dog (Canis familiaris, canFam2, released in May 2005; Lindblad-Toh et al. 2005), cow (Bos taurus, bosTau3, released in August 2006), mouse (Mus musculus, mm9, released in July 2007; Mouse Genome Sequencing Consortium 2002), rat (Rattus norvegicus, rn4, released in November 2004; Rat Genome Sequencing Project Consortium 2004), rhesus macaque (Macaca mulatta, rheMac2, released in January 2006; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), chimpanzee (Pan troglodytes, panTro2, released in March 2006; Chimpanzee Sequencing and Analysis Consortium 2005), and human (Homo sapiens, hg18, released in Mach 2006; International Human Genome Sequencing Consortium 2001) were downloaded from the University of California Santa Cruz Genome Bioinformatics Site (http://genome.ucsc.edu). The sea urchin genome (Strongylocentrotus purpuratus, Spur_2.1, released in September 2006; Sea Urchin Genome Sequencing Consortium 2006) was obtained from the Web site of the Human Genome Sequencing Center at Baylor College of Medicine (http://www.hgsc.bcm.tmc.edu/projects/). The C. intestinalis gene Ci0100130320 was obtained from the database ANISEED (Ascidian Network for InSitu Expression and Embryological Data, http://crfb.univ-mrs.fr/aniseed/).
Identification of OR-Like genes
Here, “OR-like genes” include amphioxus OR genes and Type 1 and Type 2 genes in vertebrates, though some Type 2 genes are suggested to be non-OR genes (see Results). The method for identifying OR-like genes is essentially the same as that described in a previous paper (Niimura and Nei 2007) but was slightly modified. TBlastN (Altschul et al. 1997) searches were conducted against genome sequences of 14 nonmammalian chordate species using known OR genes as queries. The query genes included an OR-like gene from amphioxus (GenBank accession number, AB182635; Satoh 2005) and two OR genes from river lampreys (AJ012708 and AJ012709; Freitag et al. 1999) as well as zebra fish, fugu, western clawed frog, chicken, mouse, and human OR genes that had been previously identified (Niimura and Nei 2003, 2005a, 2005b). From the sequences detected by the TBlastN searches, functional OR genes were identified by the method in Niimura and Nei (2007). To identify Type 2 genes from mammalian genomes, TBlastN searches were conducted against the platypus, opossum, cow, dog, mouse, rat, macaque, chimpanzee, and human genome sequences using nonmammalian Type 2 genes identified in this study as queries. Because Type 2 genes and amphioxus OR genes are more diverse than mammalian OR genes, I conducted TBlastN searches iteratively using functional Type 2 genes and amphioxus OR genes identified above as queries and confirmed that no new genes were detected. The functional genes identified were classified into groups α–λ on the basis of phylogenetic trees (see Results).
Truncated genes and pseudogenes were detected by conducting TBlastN searches against the genome sequences with the cutoff E value of 1 × 10−20 using the functional OR-like genes identified above as queries (for details, see Niimura and Nei 2007). The truncated genes and pseudogenes were classified into groups α–λ in the following way. Suppose that, for a given sequence A (a truncated gene or a pseudogene), a query (functional) gene B showed the lowest E value among all queries. In this case, the sequence A was assigned to the group to which the gene B belongs. Amino acid sequences of all OR-like genes identified in this study are available in supplementary data sets 1 and 2 (Supplementary Material online). The names of genes that belong to each group are provided in supplementary data set 3 (Supplementary Material online).
Phylogenetic Tree Construction
Translated amino acid sequences of OR genes were aligned by the program E-INS-i in MAFFT version 5.8 (Katoh et al. 2005). Poisson correction distances were calculated after all alignment gaps were eliminated. A phylogenetic tree was constructed from these distances using the Neighbor-Joining method (Saitou and Nei 1987) by the program LINTREE (Takezaki et al. 1995) available at http://www.bio.psu.edu/People/Faculty/Nei/Lab.
Results
OR Genes in 14 Nonmammalian Chordate Species
Table 1 shows the numbers of OR genes in 14 nonmammalian chordate species for which the draft genome sequences are available. The numbers of functional (intact) genes, truncated genes, and pseudogenes are shown separately. Truncated genes are sequences that are located at contig ends and do not contain any disruptive (nonsense or frameshift) mutations or long deletions (Niimura and Nei 2007). All genes identified in this study form a monophyletic clade and are clearly distinguishable from other non-OR rhodopsin-like GPCR genes (supplementary fig. 1, Supplementary Material online).
Table 1.
Numbers of OR Genes in 14 Nonmammalian Chordate Species
| Common Name | Species Name | Fa | Ta | Pa | Total |
| Amphioxus | Branchiostoma floridae | 31 | 3 | 9 | 43 |
| Ascidian | Ciona intestinalis, Ciona savignyi | 0 | 0 | 0 | 0 |
| Larvacean | Oikopleura dioica | 0 | 0 | 0 | 0 |
| Sea lamprey | Petromyzon marinus | 32 | 8 | 27 | 67 |
| Elephant shark | Callorhinchus milii | 1 | 1 | 0 | 2 |
| Zebra fish | Danio rerio | 154 | 1 | 21 | 176 |
| Medaka | Oryzias latipes | 68 | 6 | 24 | 98 |
| Stickleback | Gasterosteus aculeatus | 102 | 5 | 52 | 159 |
| Fugu | Takifugu rubripes | 47 | 39 | 39 | 125 |
| Spotted green puffer fish | Tetraodon nigroviridis | 11 | 4 | 19 | 34 |
| Western clawed frog | Xenopus tropicalis | 824 | 200 | 614 | 1638 |
| Lizard | Anolis carolinensis | 112 | 4 | 30 | 146 |
| Chicken | Gallus gallus | 211 | 89 | 133 | 433 |
F, T, and P indicate the numbers of functional genes, truncated genes, and pseudogenes, respectively. A functional gene is a sequence that does not contain any nonsense or frameshift mutations or long deletions and has initiation and stop codons at proper positions. A truncated gene is a part of an intact sequence that is located at a contig end. These numbers do not include group θ1, θ2, κ, and λ genes (see Results).
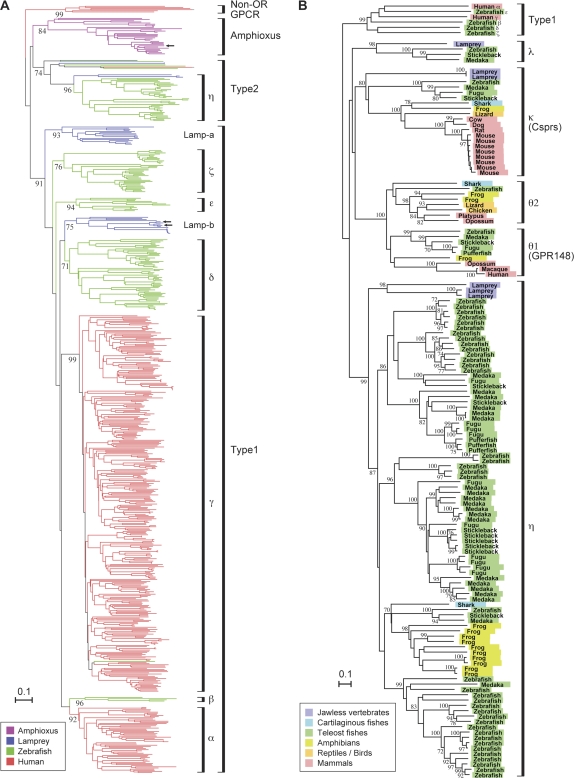
I found 31 putatively functional vertebrate-type OR-like genes from the amphioxus genome. Phylogenetic analyses showed that these genes form a monophyletic clade with all vertebrate OR genes (fig. 1A and supplementary fig. 1, Supplementary Material online). The 31 genes form an amphioxus-specific clade, suggesting that gene expansion has occurred in the amphioxus lineage. A putative OR gene identified from another species of amphioxus, Branchiostoma belcheri (Satoh 2005), is also contained in this clade (shown by the arrow in fig. 1A). Amphioxus OR genes are highly divergent from vertebrate OR genes and are characterized by long C-terminal tails. The average length of the 31 functional OR genes in amphioxus is 441 amino acids, which is much longer than the average lengths of mammalian OR genes (314 amino acids) and fish OR genes (317 amino acids). I also examined the genome sequences of ascidians C. intestinalis and C. savignyi, the larvacean O. dioica, and the sea urchin S. purpuratus. However, no OR-like sequences including pseudogenes were found from these genomes. Satoh (2005) reported that C. intestinalis gene Ci0100130320 is an OR-like gene. However, the analysis in the present study showed that this gene is closely related to β-adrenergic receptor genes (data not shown) and is clearly different from vertebrate OR genes.
FIG. 1.—
(A) Neighbor-Joining (NJ) phylogenetic tree for 615 OR-like genes and six non-OR GPCR genes as the outgroup. This tree was constructed using 31 functional OR genes in the amphioxus (magenta) and all functional Type 1 and Type 2 genes in the sea lamprey (blue), zebra fish (green), and human (red; see fig. 2). One B. belcheri OR-like gene (GenBank accession number, AB182635; Satoh 2005) and two river lamprey OR genes (AJ012708 and AJ012709; Freitag et al. 1999) were also used (indicated by arrows). Outgroup genes were randomly chosen from non-OR rhodopsin-like GPCR genes in humans (Fredriksson et al. 2003). The following genes were used as the outgroup: alpha-1B-adrenergic receptor (NP_000670.1), cholinergic receptor, muscarinic 1 (NP_000729.2), somatostatin receptor 5 (NP_001044.1), chemokine-binding protein 2 (NP_001287.2), GPCR 35 (NP_005292.2), and GPCR G2A (NP_037477.1). Bootstrap values obtained from 500 resamplings are shown only for major clades. The number of amino acid sites used was 184. The scale bar represents the estimated number of amino acid substitutions per site. (B) NJ phylogenetic tree for all (134) functional Type 2 genes identified in this study with six Type 1 genes as the outgroup. The following genes were used as the outgroup: group α, HsOR1.1.3; β, DareOR15.62; γ, HsOR11.3.2; δ, DareOR15.1; ϵ, DareOR10.29; and ζ, DareOR10.1 (Niimura and Nei 2003; this study). Bootstrap values obtained from 500 resamplings are shown for the clades with >70% bootstrap values. The species names are colored according to the color code. The number of amino acid sites used was 234.
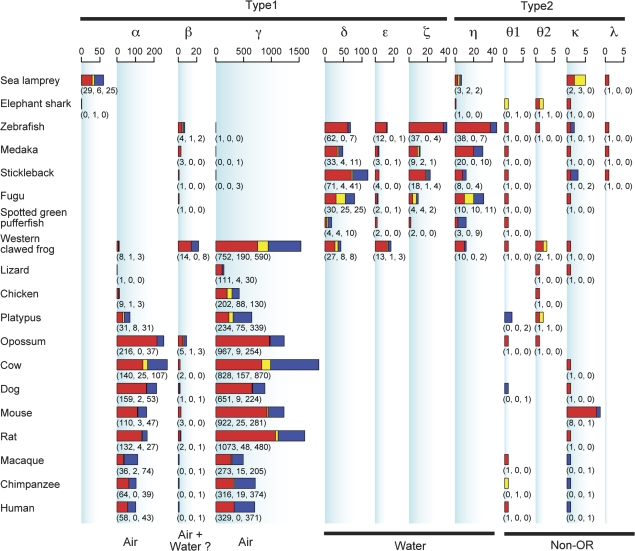
Thirty-two putatively functional OR genes were identified from the sea lamprey genome, whereas only one intact gene and one truncated gene were found from the elephant shark genome (table 1; see Discussion). It is generally said that teleost fishes have ∼100 OR genes. However, the estimated numbers of functional OR genes in this study showed an ∼10-fold difference among teleost fishes, ranging from 15 for spotted green puffer fish to 155 for zebra fish. (These numbers represent the sum of intact genes and truncated genes; using only intact genes, the difference becomes even larger.) This range is much larger than that in mammals, for which the difference is <4-fold (from ∼330 for macaques to ∼1,260 for rats). Previously, we detected 410 and 78 intact OR genes from the western clawed frog and chicken genomes (Niimura and Nei 2005b). In this study, the numbers considerably increased because of the improved qualities of the genome sequences for these species. Western clawed frogs have a surprisingly large number of OR genes (table 1), which is comparable to that in mammals.
Classification of OR Genes
Vertebrate OR genes identified in this study are separated into Type 1 and Type 2 (fig. 1A), as we reported previously (Niimura and Nei 2005b). Both Type 1 and Type 2 clades contain OR genes found from the sea lamprey genome. Therefore, it is suggested that the divergence between Type 1 and Type 2 genes was more ancient than that between jawless and jawed vertebrates (Niimura and Nei 2005b). In other words, the MRCA among all vertebrates already had Type 1 and Type 2 genes. The bootstrap support for a clade containing amphioxus OR genes and Type 2 genes is low (fig. 1A); therefore, the phylogenetic relationships among amphioxus OR genes, Type 1 genes, and Type 2 genes are unclear.
In a previous study, we classified Type 1 genes in jawed vertebrates into six groups named α–ζ, each of which corresponds to at least one ancestral gene in the MRCA between teleost fishes and tetrapods (Niimura and Nei 2005b). This classification was well supported in this study as well (fig. 1A). In addition, it was found that lamprey genes belonging to the Type 1 clade could not be classified in any of these six groups. Lamprey Type 1 genes were separated into two groups (lamp-a and lamp-b in fig. 1A). Two OR genes identified from the river lamprey Lampetra fluviatilis (Freitag et al. 1999) were included in clade lamp-b (indicated by arrows). The phylogenetic relationships among the two lamprey clades and the clades α–ζ were unresolved (fig. 1A). Therefore, under the parsimonious principle, it is likely that the divergence among clades α–ζ occurred in the jawed vertebrate lineage after the divergence from jawless vertebrates. Moreover, I found that one truncated gene in the elephant shark genome was the most closely related with lamp-a genes. This observation suggests that the divergence among the clades α–ζ probably occurred after the divergence between cartilaginous fishes and teleost fishes (see fig. 4).
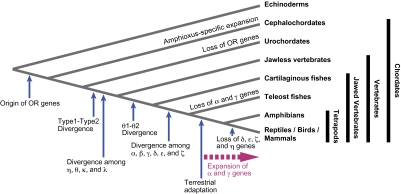
FIG. 4.—
Schematic illustration of the evolution of OR gene families in chordates. In the tetrapod lineage, the number of group α and γ genes has dramatically expanded, probably due to the importance of olfactory information in terrestrial life (see Niimura and Nei 2005b).
Type 2 genes were classified into five groups that were named η, θ1, θ2, κ, and λ (fig. 1B). Groups η, θ, and κ had been identified previously (Niimura and Nei 2005b), whereas group λ was newly identified. In this study, group θ was split into two groups, θ1 and θ2, because the phylogenetic relationships among genes within each of the groups θ1 and θ2 are consistent with the relationships among species (fig. 1B), which suggests that each group corresponds to one ancestral gene in the MRCA between teleost fishes and tetrapods.
Because groups η, κ, and λ include sea lamprey genes, the divergence among groups η, θ (θ1 and θ2), κ, and λ should be earlier than the divergence between jawless and jawed vertebrates (see fig. 4). Interestingly, the elephant shark has η, θ1, θ2, and κ genes (including truncated genes), though it has only one (truncated) Type 1 gene (see fig. 2). Therefore, it was suggested that the θ1–θ2 split occurred before the divergence between cartilaginous fishes and teleost fishes. (In fig. 4, I assumed that the θ1–θ2 split occurred in the jawed vertebrate lineage after the divergence from jawless vertebrates under the parsimonious principle because neither of the groups θ1 and θ2 contain sea lamprey genes.)
FIG. 2.—
The number of genes belonging to each group in 19 vertebrate species. Red, yellow, and blue bars represent functional genes, truncated genes, and pseudogenes, respectively. The numbers in parentheses below each bar indicate those of functional genes, truncated genes, and pseudogenes. For mammalian OR genes, groups α and β correspond to class I and group γ corresponds to class II (Glusman et al. 2000; Niimura and Nei 2005b, 2006). The numbers of class I and class II (group γ) genes in mammals were reported in Niimura and Nei (2007) and Go and Niimura (2008). The number of group α genes in a mammalian species was obtained by subtracting the number of group β genes (see fig. 3) from that of class I genes. Note that the scales of bar graphs are different among groups because of large variations in the number of genes belonging to each group. Putative functions are also shown at the bottom. “Air” and “Water” represent the detection of airborne and water-soluble odorants, respectively.
OR Genes for Water-Soluble and Airborne Odorants
Figure 2 indicates the number of OR genes belonging to each group for each species. Generally, the results are consistent with our previous study (Niimura and Nei 2005b). Group α and γ genes are present in amphibians, reptiles, birds, and mammals, but they are absent in fish with the exception of one intact gene in zebra fish and a few pseudogenes in medaka and stickleback. On the other hand, group δ, ϵ, ζ, and η genes are present in teleost fishes and amphibians, whereas they are completely absent in reptiles, birds, and mammals. These observations support our previous hypothesis that group α and γ genes are for detecting airborne odorants and group δ, ϵ, ζ, and η genes are for water-soluble odorants (Niimura and Nei 2005b).
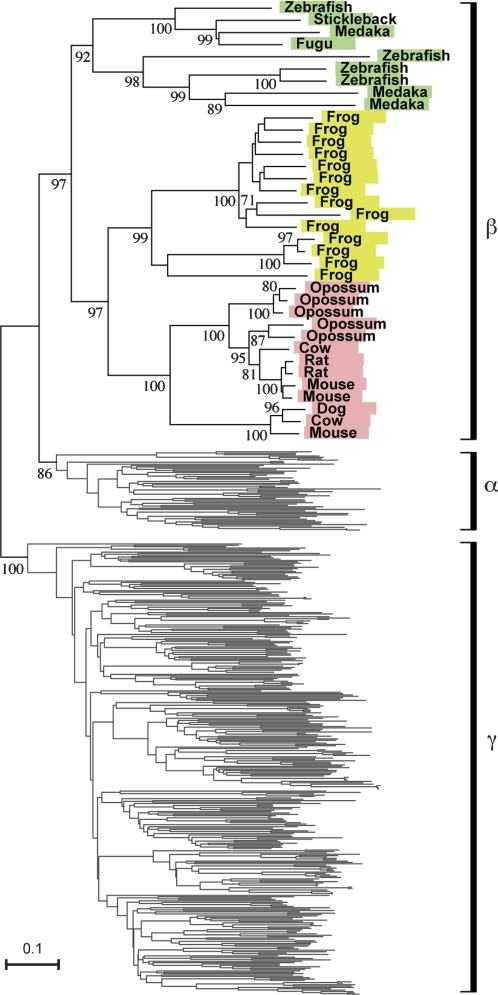
Group β genes, however, were found to be present both in aquatic and terrestrial vertebrates (figs. 2 and 3). As explained earlier, it is well known that mammalian OR genes can be classified into class I and class II (Glusman et al. 2000). Class II corresponds to group γ, whereas class I corresponds to both group α and group β (Niimura and Nei 2005b). As shown in figure 3, several early-diverging class I genes in mammals form clade β with some amphibian and fish genes. It is therefore possible that group β genes detect odorants that are both water soluble and airborne (see Discussion).
FIG. 3.—
Neighbor-Joining phylogenetic tree for 36 group β genes identified in this study with all (387) human functional OR genes. Bootstrap values were obtained from 500 replications and are shown for the clades with >70% bootstrap values (in clade β) and for major clades. Species names are colored in the same manner as figure 1B. The number of amino acid sites used was 232. Names of group β genes are provided in supplementary data set 3 (Supplementary Material online).
Type 2 Genes
By analyzing mammalian genome sequences, I found several genes belonging to groups θ1, θ2, and κ. Some of these genes are regarded as non-OR genes. A human gene belonging to group θ1 is found in databases, and its symbol, assigned by the HUGO Gene Nomenclature Committee (http://www.genenames.org), is GPR148 (RefSeq ID, NM_207364; Gloriam et al. 2005). The function of the GPR148 protein is unknown, but it was reported that this gene is expressed in the testis, brain, and spinal cord (Parmigiani et al. 2004).
Group κ genes in mice are designated as Csprs (component of Sp100-rs; Weichenhan et al. 2001). The function of the Csprs protein is unknown. It is known that chromosome 1 of the house mouse M. musculus contains a tandem cluster of Sp100-rs genes. Sp100-rs is a fusion gene of Csprs and the 5′ portion of Sp100, which encodes a nuclear dot protein (Weichenhan et al. 2001). The Sp100-rs cluster consists of about 60–2,000 repeats and encompasses 6–200 Mb of the M. musculus genome but is absent in the genome of the Asiatic mouse Mus caroli (Traut et al. 2001). It was estimated that the M. musculus B6 strain, for which the whole-genome sequences are available, has about 60 copies of an Sp100-rs gene, but this genomic portion is unassembled (Mouse Genome Sequencing Consortium 2002). Therefore, although I identified eight intact group κ (Csprs) genes from the mouse genome (fig. 2), this number does not reflect an actual number.
The evolutionary dynamics of group θ1, θ2, and κ genes are in sharp contrast to typical OR genes. They are present both in aquatic and terrestrial vertebrates, and the number of genes is usually one in each species. Moreover, the phylogenetic tree in figure 1B suggests that gene gains and losses are rare in these groups. These observations support the idea that the genes belonging to groups θ1, θ2, and κ are non-OR genes. Here, I assume that group λ genes are also non-OR genes because no gene duplications were observed in this group.
On the other hand, group η genes are likely to be OR genes that detect water-soluble odorants because they are specific to aquatic vertebrates and many lineage-specific gene expansions have occurred. In fact, it was reported that at least one group η gene (GenBank accession number, CO810666) is expressed in the olfactory epithelium of zebra fish (Alioto and Ngai 2005). Furthermore, expression analyses by reverse transcriptase–polymerase chain reaction using X. tropicalis suggested that group η genes are expressed in the olfactory epithelium in tadpoles, whereas the expression of group θ or κ genes in the X. tropicalis olfactory epithelium was detected in neither larvae nor adults (Amano T, unpublished data). Therefore, among Type 2 genes, group θ1, θ2, κ, and λ genes are likely to be non-OR genes, whereas group η genes are OR genes for water-soluble odorants (fig. 2).
Discussion
The findings in this study can be summarized in the following ways. 1) Amphioxus has vertebrate-type OR genes that were expanded in a lineage-specific manner. 2) Ascidians and larvaceans examined have lost all vertebrate-type OR genes. 3) The number of OR genes in teleost fishes is highly variable. 4) Type 1 and Type 2 genes diverged before the divergence between jawless and jawed vertebrates. 5) Group α, β, γ, δ, ϵ, and ζ genes diverged after the divergence between jawless and jawed vertebrates (and probably after the divergence between cartilaginous fishes and teleost fishes) and before the divergence between teleost fishes and tetrapods. 6) Group η, θ, κ, and λ genes diverged before the divergence between jawless and jawed vertebrates (θ1 and θ2 genes were separated before the divergence between cartilaginous fishes and teleost fishes). 7) Group α and γ genes are suggested to be for detecting airborne odorants, whereas group δ, ϵ, ζ, and η genes are for water-soluble odorants. 8) Group β genes are present in both aquatic and terrestrial vertebrates. 9) The evolutionary dynamics of θ1, θ2, κ, and λ genes are in contrast to those of typical vertebrate OR genes, suggesting that they are non-OR genes. From these observations, the evolution of OR gene families in chordates can be illustrated as in figure 4.
Amphioxus is called an “acraniate,” meaning a headless organism. It lacks an identifiable olfactory organ, and almost nothing is known of its sensitivity to chemical stimuli (Lacalli 2004). Nevertheless, many vertebrate-type OR-like genes were found in the amphioxus genome. Therefore, the origin of vertebrate-type OR genes can be traced back to the common ancestor of chordates. (Recently, Grus and Zhang [2009] suggested the same time for the origin of the OR gene family.) Satoh (2005) reported that at least one OR-like gene is broadly expressed in bipolar neurons embedded within the rostral epithelium of adult amphioxus. Further studies will be necessary to examine the cell types in which amphioxus OR genes are expressed.
I also examined the sea urchin genome but found no genes that are located within a clade containing amphioxus and vertebrate OR genes in a phylogenetic tree (data not shown). Raible et al. (2006) identified 979 rhodopsin-like GPCR genes from the sea urchin genome and argued that some of the genes are likely to be chemosensory receptors. However, their argument was not based on sequence similarities, but on the findings that these genes have specifically expanded in the sea urchin lineage and are expressed in pedicellariae and tube feet of adult sea urchins, structures that react to chemical stimuli. Therefore, my results are consistent with those of Raible et al. (2006). Rhodopsin-like GPCR genes are abundantly present in the genomes of the fruit fly Drosophila melanogaster, the malaria mosquito Anopheles gambiae, and the nematode worm Caenorhabditis elegans (Fredriksson and Schiöth 2005), as well as the sea urchin (Raible et al. 2006). It is therefore inferred that vertebrate-type OR genes emerged from one of the rhodopsin-like GPCR genes that was present in the ancestral bilaterian species.
Recent phylogenomic analyses revealed that urochordates rather than cephalochordates are the sister group to vertebrates (Delsuc et al. 2006; Putnam et al. 2008). The absence of vertebrate-type OR-like genes in the urochordate genomes examined suggests that all OR genes were lost in the lineages of these species. Larvaceans are very distant from ascidians and may be the most basal group among urochordates (Nishino and Satoh 2001). Therefore, the loss of vertebrate-type OR genes might have occurred in the common ancestor of extant urochordates. It was reported that urochordate genomes have lost many genes that are conserved between amphioxus and vertebrates (Holland et al. 2008). Ascidians are sessile filter feeders, whereas larvaceans have a floating planktonic lifestyle. Tadpole larvae of ascidians swim, but they do not feed. Reflecting their relatively inactive lifestyles, the nervous systems of urochordates are highly reduced and sensory receptors are poorly developed (Brusca and Brusca 2003). However, it is difficult to consider that they completely lack chemical senses, and thus, other gene families may function as chemosensory receptors in urochordates as in sea urchins (Raible et al. 2006).
Sea lampreys possess a well-developed olfactory system and have a relatively large olfactory bulb (Osório and Rétaux 2008). They are anadromous and migrate to shallow-water streams for spawning by utilizing odor cues. A migratory pheromone (a mixture of sulfated steroids; Sorensen et al. 2005) and a male sex pheromone (a bile acid; Li et al. 2002) in the sea lamprey were isolated, though the receptors for these chemicals are still unknown. Hashiguchi and Nishida (2007) identified at least 21 putatively functional TAAR genes from the sea lamprey genome that apparently act as odorant receptors.
In contrast to sea lampreys, only one intact OR gene and one truncated gene were found in the elephant shark genome (excluding group θ1, θ2, and κ genes). The coverage of the elephant shark genome is low (1.4×), but the estimated genome coverage is ∼75% (Venkatesh et al. 2007). It therefore appears that the number of OR genes in the elephant shark is surprisingly small. Sharks are famous for their remarkably acute sense of smell (see http://www.elasmo-research.org/). However, the elephant shark belongs to Holocephali, which is distantly related to Elasmobranchii including sharks and rays. Elephant sharks live in the deep sea (∼200 m) and their ecology is not yet well understood. At this stage, therefore, no conclusions can be made about shark olfaction.
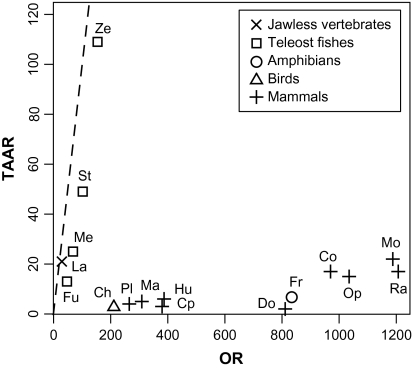
As shown in table 1, the number of OR genes is highly variable among teleost fishes, suggesting that olfactory sensitivities may be quite different among species. Interestingly, in the sea lamprey and teleost fishes, the numbers of OR genes and those of TAAR genes are similar to each other, which is in sharp contrast to the cases for tetrapods (fig. 5). It was reported that mouse TAARs are used for the detection of volatile amines in urine that function as sex pheromones (Liberles and Buck 2006). Similar repertoire sizes of OR genes and TAAR genes in fish suggest that both gene families are equally important for them and may further suggest that amines are general olfactory cues in fish. Actually, several studies have reported olfactory sensitivities to catecholamines or polyamines in goldfish (Hubbard et al. 2003; Rolen et al. 2003) and zebra fish (Michel et al. 2003). If we consider the total number of OR genes and TAAR genes, the number is not particularly small in teleost fishes compared with that in tetrapods.
FIG. 5.—
Numbers of functional OR genes and those of functional TAAR genes in 16 vertebrate species. A dashed line indicates the points in which the number of OR genes and that of TAAR genes are equal. The numbers of TAAR genes in the sea lamprey, zebra fish, stickleback, medaka, fugu, western clawed frog, and chicken were obtained from Hashiguchi and Nishida (2007). Those in the platypus, opossum, cow, and dog were taken from Grus et al. (2007), and those in the mouse, rat, chimpanzee, and human were from Lindemann et al. (2005). The number of TAAR genes in the macaque was obtained from Nei et al. (2008). La, sea lamprey; Ze, zebra fish; St, stickleback; Me, medaka; Fu, fugu; Ch, chicken; Fr, western clawed frog; Pl, platypus; Op, opossum; Co, cow; Do, dog; Mo, mouse; Ra, rat; Ma, macaque; Cp, chimpanzee; and Hu, human.
Previously, it was proposed that mammalian class I genes are “fish like” based on inaccurate phylogenetic analyses (see Niimura and Nei 2006). Later, it was revealed that, in general, fish OR genes are distantly related to both mammalian class I and class II genes (Alioto and Ngai 2005; Niimura and Nei 2005b). The functional difference between class I and class II genes is still unclear, but Zhang and Firestein (2002) hypothesized that class I genes are for detecting relatively hydrophilic compounds, whereas class II genes are for hydrophobic compounds. In this study, I found that several early-diverging class I genes (group β genes) are actually orthologous to some fish OR genes (fig. 3). Therefore, mammalian group β genes are truly fish like, and these genes may detect chemicals that are both volatile and water soluble, such as alcohol. For instance, several studies showed that fish can recognize a low concentration of β-phenylethyl alcohol, which has a pleasant rose-like smell (Neurath 1949; Teichman 1959; Nevitt et al. 1994). Moreover, I found that mouse OR genes named S6 (MmOR7.5.3) and S50 (MmOR7.5.2) in Malnic et al. (1999) belong to group β, and they were reported to respond to nonanedioic acid (azelaic acid), which is a dicarboxylic acid and is soluble in water.
As explained above, group θ1, θ2, κ, and λ genes appear not to be OR genes. They form a monophyletic (Type 2) clade with group η genes, which are bona fide OR genes (fig. 1A and B). This means that, in the vertebrate lineage, receptors for odor detection have evolved twice independently (for Type 1 genes and group η genes), or ancestral genes for groups θ1, θ2, κ, and λ acquired new functions after they diverged from group η. The ligands of group θ1, θ2, κ, and λ genes are unknown, but rare gene duplications and losses in evolution may suggest that they encode receptors for chemicals that are important to the survival of various organisms.
In this study, near-complete repertoires of OR genes identified from 23 chordate genomes were surveyed to investigate the origin and evolution of vertebrate OR genes. The results shown here should provide fundamental information for future physiological, behavioral, and evolutionary studies of olfaction.
Supplementary Materials
Supplementary data sets S1–S3 and figure S1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
I appreciate Toshikazu Amano, Takeshi Igawa, Todd Johnson, Kaoru Kubokawa, Atsushi Matsui, Masatoshi Nei, Masafumi Nozawa, and Takayuki Shoji for helpful comments and discussion. I also thank Jianzhi Zhang and Peng Shi for the use of the amphioxus genome and Byrappa Venkatesh for the elephant shark genome. In addition, I would like to extend my deep gratitude to the data producers of genome sequences, who made the sequences available before the publication of a whole-genome analysis paper. Such unpublished data were produced by the Genome Sequencing Center at Washington University School of Medicine in St Louis (sea lamprey), the Sanger Institute and the zebra fish community (zebra fish), the Broad Institute (stickleback and lizard), Department of Energy Joint Genome Institute (western clawed frog), and the Baylor College of Medicine Human Genome Sequencing Center (cow). This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan (20770192).
References
- Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Berghard A, Dryer L. A novel family of ancient vertebrate odorant receptors. J Neurobiol. 1998;37:383–392. [PubMed] [Google Scholar]
- Brusca RC, Brusca GJ. Invertebrates. 2nd ed. Sunderland (MA): Sinauer Associates; 2003. [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Schiöth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- Freitag J, Beck A, Ludwig G, von Buchholtz L, Breer H. On the origin of the olfactory receptor family: receptor genes of the jawless fish (Lampetra fluviatilis) Gene. 1999;226:165–174. doi: 10.1016/s0378-1119(98)00575-7. [DOI] [PubMed] [Google Scholar]
- Gloriam DE, Schiöth HB, Fredriksson R. Nine new human Rhodopsin family G-protein coupled receptors: identification, sequence characterisation and evolutionary relationship. Biochim Biophys Acta. 2005;1722:235–246. doi: 10.1016/j.bbagen.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Glusman G, et al. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome. 2000;11:1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Go Y, Niimura Y. Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol Biol Evol. 2008;25:1897–1907. doi: 10.1093/molbev/msn135. [DOI] [PubMed] [Google Scholar]
- Grus WE, Shi P, Zhang J. Largest vertebrate vomeronasal type 1 receptor gene repertoire in the semiaquatic platypus. Mol Biol Evol. 2007;24:2153–2157. doi: 10.1093/molbev/msm157. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 2009;26:407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Evolution of trace amine-associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard PC, Barata EN, Canário AV. Olfactory sensitivity to catecholamines and their metabolites in the goldfish. Chem Senses. 2003;28:207–218. doi: 10.1093/chemse/28.3.207. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Rev. 2001;36:46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Lacalli TC. Sensory systems in amphioxus: a window on the ancestral chordate condition. Brain Behav Evol. 2004;64:148–162. doi: 10.1159/000079744. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lindemann L, et al. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85:372–385. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Michel WC, Sanderson MJ, Olson JK, Lipschitz DL. Evidence of a novel transduction pathway mediating detection of polyamines by the zebrafish olfactory system. J Exp Biol. 2003;206:1697–1706. doi: 10.1242/jeb.00339. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of chemosensory receptor gene repertoires in vertebrates and insects: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Neurath H. Über die Leistung des Geruchssinnes bei Elritzen. Z Vgl Physiol. 1949;31:609–626. [Google Scholar]
- Nevitt GA, Dittman AH, Quinn TP, Moody WJ., Jr Evidence for a peripheral olfactory memory in imprinted salmon. Proc Natl Acad Sci USA. 1994;91:4288–4292. doi: 10.1073/pnas.91.10.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A. The family of genes encoding odorant receptors in the channel catfish. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005a;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA. 2005b;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino A, Satoh N. The simple tail of chordates: phylogenetic significance of appendicularians. Genesis. 2001;29:36–45. doi: 10.1002/1526-968x(200101)29:1<36::aid-gene1003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Olender T, et al. The canine olfactory subgenome. Genomics. 2004;83:361–372. doi: 10.1016/j.ygeno.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Osório J, Rétaux S. The lamprey in evolutionary studies. Dev Genes Evol. 2008;218:221–235. doi: 10.1007/s00427-008-0208-1. [DOI] [PubMed] [Google Scholar]
- Parmigiani RB, et al. A novel human G protein-coupled receptor is over-expressed in prostate cancer. Genet Mol Res. 2004;3:521–531. [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Quignon P, et al. Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol. 2003;4:R80. doi: 10.1186/gb-2003-4-12-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, et al. Opsins and clusters of sensory G-protein-coupled receptors in the sea urchin genome. Dev Biol. 2006;300:461–475. doi: 10.1016/j.ydbio.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in the goldfish Carassius auratus. J Exp Biol. 2003;206:1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Satoh G. Characterization of novel GPCR gene coding locus in amphioxus genome: gene structure, expression, and phylogenetic analysis with implications for its involvement in chemoreception. Genesis. 2005;41:47–57. doi: 10.1002/gene.20082. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, et al. Miniature genome in the marine chordate Oikopleura dioica. Science. 2001;294:2506. doi: 10.1126/science.294.5551.2506. [DOI] [PubMed] [Google Scholar]
- Sorensen PW, et al. Mixture of new sulfated steroids functions as a migratory pheromone in the sea lamprey. Nat Chem Biol. 2005;1:324–328. doi: 10.1038/nchembio739. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Teichman H. Über die Leistungs des Geruchssinnes beim Aal (Anguilla anguilla L. Z Vgl Physiol. 1959;42:206–254. [Google Scholar]
- Traut W, Rahn IM, Winking H, Kunze B, Weichehan D. Evolution of a 6-200 Mb long-range repeat cluster in the genus Mus. Chromosoma. 2001;110:247–252. doi: 10.1007/s004120100152. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson JP, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenhan D, et al. Source and component genes of a 6-200 Mb gene cluster in the house mouse. Mamm Genome. 2001;12:590–594. doi: 10.1007/s00335-001-3015-9. [DOI] [PubMed] [Google Scholar]
- Young JM, et al. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0018. research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.