Abstract
Dramatic examples of repeated sequences occur in the genomes of some naturally competent bacteria, which contain hundreds or thousands of copies of short motifs called DNA uptake signal sequences. Here, we analyze the evolutionary interactions between coding-region uptake sequences and the proteomes of Haemophilus influenzae, Actinobacillus pleuropneumoniae, and Neisseria meningitidis. In all three genomes, uptake sequence accumulation in coding sequences has approximately doubled the frequencies of those tripeptides specified by each species’ uptake sequence. The presence of uptake sequences in particular reading frames correlated most strongly with the use of preferred codons at degenerately coded positions, but the density of uptake sequences correlated only poorly with protein functional category. Genes lacking homologs in related genomes also lacked uptake sequences, strengthening the evidence that uptake sequences do not drive lateral gene transfer between distant relatives but instead accumulate after genes have been transferred. Comparison of the uptake sequence-encoded peptides of H. influenzae and N. meningitidis proteins with their homologs from related bacteria without uptake sequences indicated that uptake sequences were also preferentially located in poorly conserved genes and at poorly conserved amino acids. With few exceptions, amino acids at positions encoded by uptake sequences were as well conserved as other amino acids, suggesting that extant uptake sequences impose little or no constraint on coding for protein function. However, this state is likely to be achieved at a substantial cost because of the selective deaths required to eliminate maladaptive mutations that improve uptake sequences.
Keywords: competence, transformation, Haemophilus, Neisseria, Pasteurellaceae
Introduction
Small intrinsic biases in genome-wide processes can have major impacts on genome properties when compounded over millions or billions of years (Frank and Lobry 1999; Knight et al. 2001). One striking example is the DNA uptake signal sequences, a class of very abundant short dispersed repeats in the genomes of bacteria in the family Pasteurellaceae and the genus Neisseria. These bacteria are naturally competent, and their uptake sequences are thought to have accumulated because of the sequence preferences of their DNA uptake machineries. Many of these repeats occur in genes, where they may constrain protein coding and thus gene function.
Uptake sequences are called uptake signal sequences (USSs) and DNA uptake sequences (DUSs) in the Pasteurellaceae and Neisseria, respectively (Smith et al. 1995, 1999; Ambur et al. 2007); in this paper, we will use “USSs” and “DUSs” when describing the uptake sequences specific to each group and “uptake sequences” or “USs” when discussing general properties of all uptake sequences. The Pasteurellacean USS was first characterized in Haemophilus influenzae, whose 1.83-Mb genome contains 1,471 perfect-consensus copies of the 9-bp core of the motif (table 1); alignment of these cores extended the consensus to a 30-bp three-part motif (Smith et al. 1995). Analysis of other Pasteurellacean species found that five of eight sequenced species form a subclade sharing this motif, with Actinobacillus pleuropneumoniae and two other sequenced species in a separate subclade sharing a related USS motif differing at a number of positions from the H. influenzae motif (Redfield et al. 2006). The Neisserial DUS was first identified in Neisseria gonorrhoeae but has been best characterized in Neisseria meningitidis, initially as a 10-bp motif with 1,891 perfect consensus sites in the 2.18-Mb genome (table 1) (Goodman and Scocca 1988; Smith et al. 1999). This motif is also present in Neisseria lactamica and has recently been shown to extend through two additional upstream bases (Ambur et al. 2007). Table 1 gives sequences and frequencies of the core motifs of H. influenzae, A. pleuropneumoniae, and N. meningitidis.
Table 1.
Genome Sequence Information
| Species | H. influenzae | A. pleuropneumoniae | N. meningitidis |
| Genome | Rd KW20 | L20 | Z2491 |
| NCBI accession | NC_000907 | NC_009053 | NC_003116 |
| Size (Mb) | 1.83 | 2.27 | 2.18 |
| % G + C | 38% | 41% | 51% |
| % coding | 84% | 86% | 80% |
| # ORFs | 1,657 | 2,012 | 2,065 |
| Consensus-core uptake sequence | AAGTGCGGT | ACAAGCGGT | GCCGTCTGAA |
| # US in genome | 1,471 | 765 | 1,892 |
| # US in ORFs | 956 (65%a) | 377 (49%a) | 653 (34.5%a) |
Percentage of total USs in genome.
Although most other naturally competent bacteria will take up all double-stranded DNAs equally well, competent members of the Pasteurellaceae and Neisseria preferentially take up DNA fragments containing their respective uptake sequences. In fact, USSs and DUSs were originally identified by experiments seeking the molecular basis of these species’ preferential uptake of “self” DNA over foreign DNA (Danner et al. 1980; Goodman and Scocca 1988). We now know that these preferences extend to related species that share the same uptake sequence (Redfield et al. 2006). Analysis of uptake of radioactively labeled DNA has shown that uptake sequences promote the initial stages of DNA uptake (Scocca et al. 1974; Ambur et al. 2007); to date, there is no evidence that they also affect subsequent stages of DNA transport or recombination, although this has not been ruled out. They are thus thought to function by interacting at the cell surface with sequence-specific DNA-binding proteins (DNA receptors) and/or components of the type 4 pilus system responsible for DNA uptake.
The Pasteurellacean and Neisserial uptake sequences have many features in common although their DNA sequences are unrelated (Smith et al. 1999). The genomic abundances of their cores are similar—0.80 9-mer USSs/kb in H. influenzae and 0.89 10-mer DUSs/kb in N. meningitidis—and are 200- to 1,000-fold higher than expected for randomly occurring sequences. Unlike transposable elements, which spread by copying and insertion into preexisting sequences, both USSs and DUSs arise and spread by a combination of mutation and homologous recombination (Redfield et al. 2006; Treangen et al. 2008). Both are distributed more evenly around their genomes than randomly positioned repeats would be, with similar numbers in both orientations (uptake sequences are not palindromic). Both are underrepresented in coding sequences, USSs less so than DUSs. When not in coding regions, both are frequently found as dyad symmetry pairs in intergenic locations where they are predicted to function as transcriptional terminators, again USSs less so than DUSs (Goodman and Scocca 1988; Kingsford et al. 2007; Treangen et al. 2008). Although the two subtypes of Pasteurellacean USS derive from sequences in a common ancestor (Redfield et al. 2006), the Pasteurellacean and Neisserial uptake sequences are likely to have arisen independently, as uptake sequences are not known in other members of the Proteobacteria.
Because DNA fragments containing them are preferentially taken up by competent cells, uptake sequences are thought to have accumulated by recombination with homologous fragments brought into cells by the DNA uptake machinery (natural transformation) (Danner et al. 1980; Bakkali 2007; Treangen et al. 2008). Under this model, mutations that created improved matches to the uptake bias were, over evolutionary time, more often transmitted to other cells by this biased transformation. Uptake sequence accumulation would thus be a direct but unselected consequence of DNA uptake, making it a form of molecular drive. This term, originally proposed by Gabriel Dover, refers to evolutionary forces that arise from biases in population-level DNA turnover processes (e.g., gene conversion and unequal crossing over) and lead to long-term changes in genome properties (Dover 1982, 2002). The molecular drive arising from biased DNA uptake would reinforce selection for uptake sequences linked to beneficial alleles and oppose the elimination of uptake sequences associated with deleterious mutations.
In coding sequences, accumulation of uptake sequences is expected to affect the outcome of natural selection on protein function. Consider mutations that create better uptake sequences. Those that also improve gene function will be fixed more rapidly, those with neutral and nearly neutral consequences for gene function will be fixed more often, and those that are significantly deleterious will be eliminated only after more selective deaths. (The converse will be true for mutations that worsen uptake sequences.) Because beneficial mutations are rare, the latter two effects are likely to be the most significant, which is why Danner et al. (1980) proposed in 1980 that USS would be found randomly distributed wherever in the genome they are not disadvantageous. This is consistent with the observed distributions; although uptake sequences are underrepresented in coding sequences, 39% of H. influenzae genes and 22% of N. meningitidis genes contain them.
Uptake sequences are also nonrandomly distributed within coding regions. First, uptake sequences are more common in some reading frames than others, and preferential use of these reading frames has been hypothesized to reduce the impact of uptake sequences on protein function (Karlin et al. 1996; Smith et al. 1999). Second, Davidsen et al. (2004) analyzed the frequency and distribution of uptake sequences in two Neisseria species and in H. influenzae and Pasteurella multocida. They reported that uptake sequences were preferentially found in genes responsible for genome maintenance (polymerases, repair enzymes, etc.), which was hypothesized to reflect selection for preferential transfer of such genes between strains. Finally, van Passel (2008) recently reported a modest bias of USS, but not DUS, toward gene termini.
Although these gene function and reading frame preferences suggest that some uptake sequences are better tolerated than others, the actual extent of constraint imposed by protein function on uptake sequences, and by uptake sequences on protein function, has not been investigated. In this report, we analyze the reciprocal effects of uptake sequence accumulation and selection on bacterial genomes and proteomes.
Materials and Methods
Genome Sequences
The H. influenzae Rd, A. pleuropneumoniae L20, and N. meningitidis Z2491 genome sequences were obtained from GenBank (NCBI RefSeq Accession numbers NC_000907, NC_009053, and NC_003116, respectively). For the gene homologue analyses, genome sequences were obtained for the Gammaproteobacteria Escherichia coli K12, Pseudomonas aeruginosa PA01, and Vibrio cholerae O1, and for the Betaproteobacteria Bordetella pertussis Tomaha 1, Methylobacillus flagellatus KT, and Nitrosomonas europaea ATCC19718. NCBI Accession numbers: Ec: NC_000913; Pa: NC_002516; Vc: NC_002505 and NC_002506; Bp: NC_002929; Mf: NC_007947; and Ne: NC_004757.
Computational Methods
Acquisition, handling, and organization of data were automated using software written for this work in Perl (including the BioPerl modules). The BlastP program of the Blast software suite (Altschul and Lipman 1990) was used to define groups of protein homologs across species, which were then aligned using the ClustalW program (Thompson et al. 1994). Slices of sequence alignments were then selected for further analysis.
Determination of USS-Encoded Residues in Proteomes
All open reading frames (ORFs) in the genome sequences of H. influenzae, A. pleuropneumoniae, and N. meningitidis were searched for core 9- or 10-nt uptake sequences (hiUSS = AAGTGCGGT; apUSS = ACAAGCGGT; nmDUS = GCCGTCTGAA) on both DNA strands. For each genome, we created a Fasta format file of the translated sequences of all ORFs as well as files of subsets of ORFs containing defined numbers of uptake sequences. For each uptake sequence located in an ORF, we determined its orientation, its reading frame with respect to the start codon, and the sequences of the tripeptide it specified.
Analysis of Peptide Frequencies
For each proteome, we determined the number of instances of each of the tripeptides that could be encoded by its own uptake sequence (e.g., the frequency of SAV in the H. influenzae proteome). Degenerate uptake sequence/reading frame combinations (those that specified more than two tripeptides) were not analyzed further. For comparison, we also determined the frequencies of the same tripeptides in the E. coli K-12 proteome and in the other two proteomes with different uptake sequences. The statistical significance of differences between proteomes was determined using a one-way analysis of variance with repeated measures followed by Tukey’s multiple comparisons test.
Calculation of Codon Scores
Two codon scores were first calculated for each nondegenerate tripeptide encodable by an uptake sequence, first if it was encoded by the uptake sequence and second if it was encoded by the most frequent codons for those amino acids in that genome. These codon scores were calculated as the sums of the frequencies of the individual codons specifying each tripeptide using the codon frequency tables provided by the Codon Usage Database (http://www.kazusa.or.jp/codon) (Nakamura et al. 2000). The final codon score for each uptake sequence and reading frame was then calculated as the ratio of these two codon scores.
Analysis of Uptake Sequence Densities in Cluster of Orthologous Genes Groups
The A. pleuropneumoniae genes in cluster of orthologous genes (COG) group L were subdivided into groups L+ and L− as specified by the supplementary material of Davidsen et al. (2004). For other genomes, the COG assignments of Davidsen et al. were used. For each COG group in each genome, the total number of US cores in the genes was divided by the total number of nucleotides in the same genes. A Tukey–Kramer test was applied to the means of these ratios to determine whether any of the pairwise comparisons found significant differences.
Identifying Gene Homologs
For the proteins specified by each genome with uptake sequences (the “query” sequences), homologs were identified in three closely related species whose genomes have no US. For H. influenzae, the comparison genomes used were E. coli (Enterobacteraceae), V. cholerae (Vibrionaceae), and P. aeruginosa (Pseudomonadaceae). For N. meningitides, the comparison genomes used were B. pertussis (Alcaligenaceae), M. flagellatus (Methylophilales), and N. europaea (Nitrosomonadaceae). To identify homologs, we used the BlastP program from the Blast software suite with a maximum E value of 1 × 10−9, selecting the single best hit in each comparison species (Altschul and Lipman 1990). This resulted in subsets of H. influenzae and N. meningitidis protein sequences with three, two, one, or no homologues in the comparison genomes. For each protein in the H. influenzae and N. meningitidis proteomes that had homologs in all three comparison species, the pairwise percent identities between the query sequence and each of its three homologs were determined.
Alignment and Analysis of USS-Encoded Residues
For each proteome, we selected translated sequences of all ORFs that contained a single uptake sequence and had homologs in all three comparison genomes. The ClustalW program with the BLOSUM matrices (Henikoff S and Henikoff JG 1992) was then used to obtain an overall alignment of each translated query sequence with its three homologues. Sequences at the ends of each overall alignment were trimmed to give 99-aa segments centered on a tripeptide encoded by an uptake sequence. We eliminated from consideration any sequences containing gaps in the 9-aa region of the alignment centered on the US-encoded peptide.
For selected slices of the alignments (diagrammed in supplementary fig. 2 [Supplementary Material online]) we determined 1) the mean pairwise percent sequence identities of the USS-encoded amino acids in the query sequences to their comparison homologs (three query-by-homolog comparisons) and 2) the mean pairwise percent identities of the homologs to each other (three homolog-by-homolog comparisons). The BLOSUM62 matrix was then used to score the degree of query-versus-homolog and homolog-versus-homolog similarity for each slice, summing the values for all amino acids in each sequence pair. As above, we calculated the mean of the three query–homologue pairwise similarities and the mean of the three homolog–homolog pairwise similarities for each query sequence.
Results
Because individual uptake sequences are conserved over long evolutionary periods, closely related genomes cannot be used as independent samples (Bakkali et al. 2004). Thus, we restricted our analysis to the three genomes exemplifying the known types of uptake sequences: H. influenzae, A. pleuropneumoniae, and N. meningitidis. For convenient reference, the basic properties of these genomes and their uptake sequences are summarized in table 1 (the only information not previously published is the proportion of A. pleuropneumoniae perfect-core USSs in coding sequences). To allow analysis of the effect of uptake sequence accumulation on the bacterial proteomes, we restricted our study to the perfect-consensus core 9-bp USS and 10-bp DUS sequences.
Peptide Frequencies in Proteomes
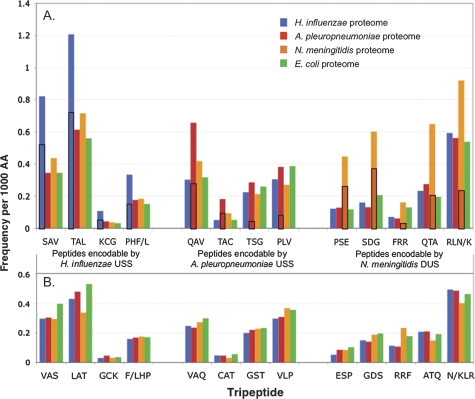
The first finding was that as uptake sequences accumulated in genomes, the peptides they can encode accumulated in the corresponding proteomes. Table 2 lists the tripeptide sequences that can be encoded by each uptake sequence in each of its six possible reading frames. We counted all of these US-encodable tripeptides in the proteomes of H. influenzae, A. pleuropneumoniae, and N. meningitidis, using the E. coli proteome as a negative control. (Combinations of uptake sequence and reading frame that encoded more than two possible tripeptides or included a stop codon were omitted from the analysis.) As a control for the differing effects of base composition and codon usage on the frequencies of the individual amino acids, we also counted reverse-order tripeptides in all of the proteomes (e.g., VAS served as a control for SAV). The results are shown in figure 1 as the frequency of each tripeptide per 1,000 amino acids for each proteome, with the reverse-order control tripeptides shown below the uptake sequence-encodable tripeptides for each uptake sequence type.
Table 2.
Distribution of Uptake Sequences in Coding Regions
| H. influenzae | Peptide | A. pleuropneumoniae | Peptide | N. meningitidis | Peptide | |
| Core uptake sequence | AAGTGCGGT | ACAAGCGGT | GCCGTCTGAA | |||
| US in ORFs | 956 | 377 | 653 | |||
| Coding strand | 386 (40.4%a) | 239 (63.4%) | 293 (44.9%) | |||
| ReadingFrame1 | 29 (3.0%) | KCG | 34 (9.0%) | TSG | 0 | AVstop |
| ReadingFrame2 | 87 (9.1%) | XVRb | 20 (5.3%) | ZKRc | 138 (21.1%) | RLN/K |
| ReadingFrame3 | 270d (28.2%) | SAV | 185 (49.1%) | QAV | 155 (23.7%) | PSE |
| Reverse strand | 570 (59.6%) | 138 (36.6%) | 360 (55.1%) | |||
| ReadingFrame1 | 375 (39.2%) | TAL | 64 (17.0%) | TAC | 16 (2.5%) | FRR |
| ReadingFrame2 | 114 (11.9%) | ZRTd | 24 (6.4%) | ZRLc | 122 (18.7%) | QTA |
| ReadingFrame3 | 81 (8.5%) | PHF/L | 50 (13.3%) | PLV | 222 (34.0%) | SDG |
All numbers in brackets are percentage of total USs in ORFs.
X = first amino acid may be any of Q/K/E/stop.
Z = first amino acid may be any of Y/H/N/D.
The most common reading frame locations for each uptake sequence are in bold.
FIG. 1.—
Frequencies of uptake sequence-encodable tripeptides in bacterial proteomes. H. influenzae proteome, blue; A. pleuropneumoniae proteome, red; N. meningitidis proteome, orange; Escherichia coli proteome, green. (A) Tripeptides that can be specified by each of the three uptake sequences: left group by H. influenzae USS; center group by A. pleuropneumoniae USS; and right group by N. meningitidis DUS. The enrichment in the H. influenzae proteome of peptides encodable by the H. influenzae USS was significant (P = 0.010), the enrichment in the A. pleuropneumoniae proteome of peptides encodable by the A. pleuropneumoniae USS was just above the significance threshold (P = 0.058), and the enrichment in the N. meningitidis proteome of peptides encodable by the N. meningitidis DUS was highly significant (P < 0.0001). Boxed areas of bars indicate the fraction of peptides specified by perfect-core uptake sequences. (B) Reversed-sequence tripeptide controls. None of the controls showed significant enrichment.
This analysis revealed that the tripeptides that can be encoded by each uptake sequence are specifically enriched in the corresponding proteome. For example, the blue bar at the far left of panel A in figure 1 indicates that the peptide SAV occurs twice as often in the H. influenzae proteome (blue) as in other proteomes (red, green, and orange). Both the E. coli control and the reverse-tripeptide controls confirm that the enrichment is specific to tripeptides that can be encoded by each genome’s own core uptake sequence. Enrichments were seen for all of the tripeptides that can be specified by uptake sequences, both those in the preferred reading frames and those in the less common reading frames. The boxed areas show the contributions of each genome’s uptake sequences, using the values in table 2. A. pleuropneumoniae has the fewest uptake sequences in its proteome and also has slightly lower peptide enrichments relative to the other genomes (mean 1.76-fold, P = 0.58), but the N. meningitidis enrichment is higher than that of H. influenzae (means 2.67-fold, P < 0.0001, and 2.24-fold, P = 0.01, respectively), even though it has fewer uptake sequences in its proteome. This tripeptide enrichment is best understood as a simple consequence of the accumulation of uptake sequences in proteomes, and it can now be added to the list of properties shared by all uptake sequences.
Reading Frame Biases
Table 2 also shows how often each uptake sequence occurs in each of the six reading frames. The numbers for H. influenzae and N. meningitidis have been previously reported; minor changes are due to improvements in the RefSeq annotation of these genomes (Smith et al. 1999; Davidsen et al. 2004). Like the other uptake sequences, A. pleuropneumoniae uptake sequences in coding regions exhibit a strong reading frame bias, with almost half in frame three in the coding orientation (table 2). Unlike DUS and H. influenzae USSs, most A. pleuropneumoniae USSs in coding regions are in the coding orientation, showing that USS orientation with respect to transcription and translation is not itself an important factor.
We considered two functional explanations for these reading frame preferences. The first is that the preferred reading frames may simply be those in which uptake sequences encode functionally versatile tripeptides that would in any case be common in the proteome, as proposed by Smith et al. (1999). Because each tripeptide occurred at similar frequencies in all genomes except where influenced by uptake sequences, the analysis shown in figure 1 suggests that generally similar forces determine tripeptide frequencies in all proteomes. The forward and reverse tripeptides also occurred at similar frequencies across these genomes, suggesting that, in the absence of uptake-sequence effects, the differing frequencies of tripeptides within and between proteomes are determined largely by the frequencies of the constituent amino acids, with only a modest effect due to interactions between neighboring amino acids (Pasamontes and Garcia-Vallve 2006).
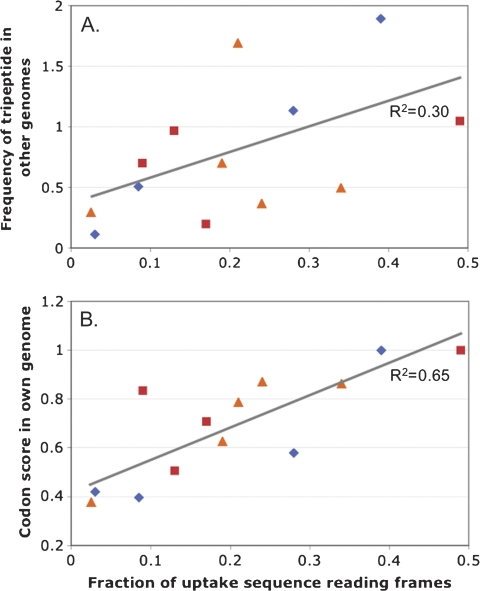
To investigate whether the use of common tripeptides causes the reading frame biases, we plotted how often each uptake sequence used each reading frame against the mean frequency of the US-encodable tripeptide in the three genomes without that uptake sequence (to exclude the peptide enrichment associated with uptake sequences in their own genomes) (fig. 2A). Although the correlation was strong for the four H. influenzae data points (R2 = 0.97), the other uptake sequences showed little correlation and the overall R2 coefficient was only 0.30.
FIG. 2.—
Analysis of tripeptides encodable by uptake sequences in the genomes of H. influenzae, A. pleuropneumoniae, N. meningitidis, and Escherichia coli. (A) Average frequencies in genomes without the specified uptake sequence of tripeptides encodable by that uptake sequence, plotted against reading-frame usage by each uptake sequence (data from table 2) (R2 = 0.30). (B) Codon score for tripeptides encodable by that uptake sequence (calculated as described in Materials and Method) plotted against reading frame usage by each uptake sequence (R2 = 0.65). H. influenzae USS, blue diamonds; A. pleuropneumoniae USS, red squares; N. meningitidis DUS, orange triangles.
The second explanation for the reading frame distribution is that the preferred reading frames may be more efficiently translated because they use relatively common codons for their amino acids. Both H. influenzae and N. meningitidis are known to exhibit moderately strong codon-bias effects (dos Reis et al. 2004; Sharp et al. 2005). To estimate the contribution of this bias to the reading frame distribution of each uptake sequence, we calculated a simple codon efficiency score for each tripeptide of interest and plotted this as a function of the percent of uptake sequences specifying each tripeptide (fig. 2B). The correlation coefficients for H. influenzae and N. meningitidis were moderately strong (0.85 and 0.83, respectively), and the overall correlation coefficient was 0.65. This suggests that uptake sequences in the less common reading frames may have been selected against because they reduce the translatability of the amino acids they encode and thus reduce fitness.
Uptake Sequence Density and Gene Function
If the genetic benefits of recombination counteract the constraints on uptake sequence locations caused by codon bias and tripeptide versatility, uptake sequences should be most frequent in those genes whose transfer is most beneficial. Davidsen et al. (2004) analyzed uptake sequence frequency in several genomes as a function of assigned COG functional category (www.ncbi.nlm-nih.gov/COG). They concluded that uptake sequences are preferentially found in “genome maintenance genes”—a subset (L+) of COG category L genes (replication, recombination, and repair functions) that 1) excludes transposases and other genes not strictly involved in genome housekeeping and 2) includes two genes from other COG categories. The calculated overrepresentation was strongest in N. meningitidis (∼2-fold) and 1.6- to 1.8-fold in the other species examined (N. gonorrhoeae, H. influenzae, and P. multocida). The authors interpreted this bias as reflecting selection for preferential uptake and recombination of genes that contribute to maintenance and variation of the genome.
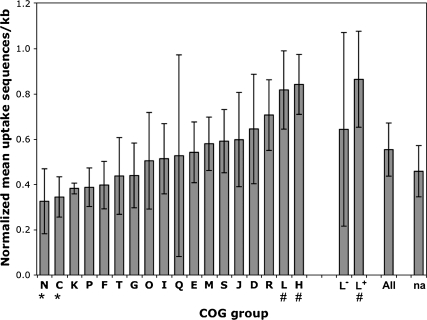
We have reanalyzed this data set (kindly provided by T. Tonjum), combining it with a new analysis of A. pleuropneumoniae COG groups and uptake sequences. Figure 3 shows the combined analysis for all five species whose COG assignments are available, now encompassing the full diversity of known uptake sequences. The standard deviations indicated by the error bars show that most of the differences are not significant (supplementary fig. 1 [Supplementary Material online] shows the analysis of each genome separately). Although the genome maintenance genes in category L+ do have uptake sequence densities higher than average, they are not the highest ranked COG category in any species other than N. meningitidis. Furthermore, N. meningitidis appears to be exceptional in lacking uptake sequences in the L− category (transposase genes) as this category contains many uptake sequences in other genomes.
FIG. 3.—
Normalized mean density of uptake sequences in genes assigned to each COG group of five bacterial genomes (N. meningitidis, N. gonorrhoeae, H. influenzae, Pasteurella multocida, and A. pleuropneumoniae). Error bars are standard deviations. A. pleuropneumoniae had no annotated genes in COG group N. na: not assigned. Groups L+ and L− are as specified by Davidsen et al. (2004). The columns marked by * differ significantly by a Tukey–Kramer test from the columns marked by #; differences between other columns are not significant.
Sequence Conservation
The final set of analyses used measures of amino acid sequence conservation to detect constraints on uptake sequence location. Our strategy to measure divergence and conservation was to compare 1) translated amino acid sequences of the H. influenzae and N. meningitidis genes containing uptake sequences to 2) the sequences of homologous proteins present in a standard set of three “comparison” genomes from species without uptake sequences. For H. influenzae, the comparison genomes were from the three closest related orders in the Gammaproteobacteria; for N. meningitidis, the analysis used sequenced genomes from three other orders of the Betaproteobacteria (the specific genomes used are listed in Materials and Methods). A. pleuropneumoniae was excluded from this analysis because of its close relationship to H. influenzae. Use of amino acid alignments allowed homologous sequences to be identified over much longer evolutionary distances than are possible with nucleotide alignments, and using standard genomes for the comparisons ensured consistency.
Relationship between BLAST Identities and Numbers of Uptake Sequences
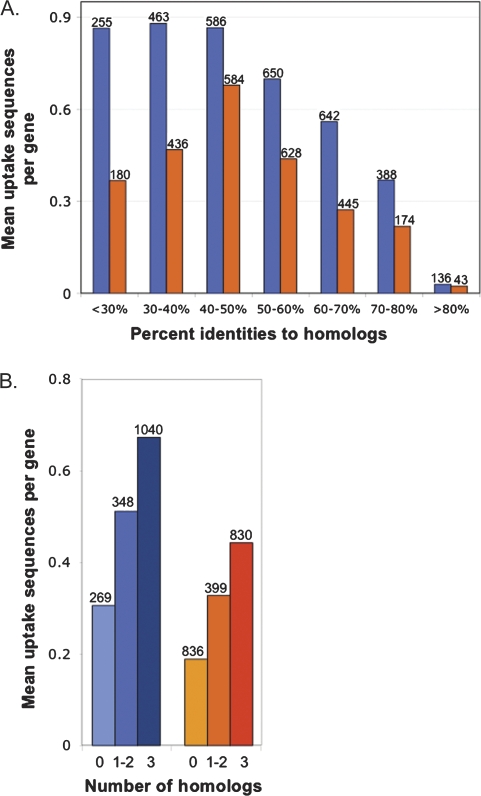
If uptake sequences preferentially accumulate where they are least harmful to gene function rather than where genetic exchange is most beneficial, we expect their density to be highest in the least conserved genes. We began by examining the numbers of uptake sequences in genes with different degrees of conservation. Figure 4A shows that highly conserved genes of both H. influenzae and N. meningitidis had fewer uptake sequences than other genes, as predicted by this “minimum harm” hypothesis.
Fig. 4.—
Relationships between the numbers of uptake sequences in genes and protein sequence similarity. Blue, H. influenzae; orange, N. meningitidis. (A) Mean number of uptake sequences per gene for genes with homologs in all three comparison genomes grouped by pairwise percent identity to each homolog. The numbers above the bars are the numbers of alignments in each category. (B) Mean number of uptake sequences per gene versus the number of comparison genomes having homologs. The numbers above the bars are the numbers of genes in each category.
Surprisingly, the least conserved genes also had fewer uptake sequences rather than more. An explanation for this effect was proposed by Treangen et al. (2008), who found that N. meningitidis DNA sequences acquired by horizontal gene transfer (HGT) have fewer DUSs than other genes. They hypothesized that these sequences entered the genome with no DUS, presumably by transduction or another DUS-independent mechanism, and gradually acquired DUS in their new genome. Uptake sequence frequency would then be a property of acquired genes that, like base composition or codon usage, only very slowly comes to resemble that of the new host genome (Lawrence and Ochman 1997).
We tested whether this explanation might also account for the low uptake sequence frequencies of the poorly conserved genes in figure 4A, by examining the frequencies of uptake sequences in those genes that had been excluded from the analysis because the proteins they encoded did not have good homologs in all three of the comparison genomes (BlastP E value ≤ 1 × 10−9). Figure 4B shows that the number of uptake sequences a gene has is directly correlated with how many strong Blast matches it had in the comparison genomes. When the overall higher numbers of uptake sequences in the H. influenzae data were taken into account, the relationship between number of matches and number of uptake sequences was nearly identical for the H. influenzae and N. meningitidis genomes despite their deep evolutionary distance.
To further test the hypothesis that uptake sequences do not help cells acquire novel genes, the 269 H. influenzae genes that had no Blast matches in comparison genomes were sorted by whether or not they had a good homolog in A. pleuropneumoniae. The 177 genes that lacked such homologs are likely to have been acquired by transfer into H. influenzae after the divergence of the two species; only 22% of these had USSs. Forty of these 177 genes were found in a database of genes predicted to have entered the genome by HGT (Garcia-Vallve et al. 2003) and only three of these (7.5%) had USSs. The other 92 genes had good A. pleuropneumoniae homologs; the subset found in the HGT database had a similar USS frequency to the others (43% and 47%, respectively), suggesting that genes transferred before the divergence of H. influenzae and A. pleuropneumoniae have had enough time to accumulate a “normal” density of USSs.
Genes horizontally transferred into the H. influenzae lineage thus appear to have acquired USSs only after transfer. Because this conclusion is based on comparisons of amino acid sequences between bacteria in different orders, our results extend and confirm those of Treangen et al. (2008), which were based on nucleotide sequence comparisons between bacteria in the same genus. We can thus add rarity in horizontally transferred genes to the list of properties shared by all uptake sequences.
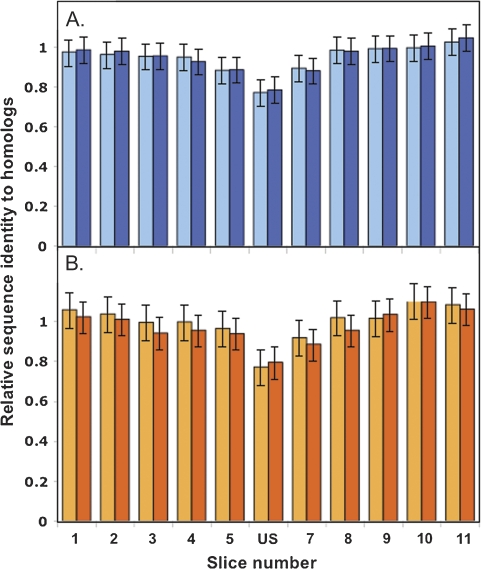
Two further analyses examined the within-gene effects of uptake sequences, asking first whether uptake sequences are more common in less conserved parts of proteins and then whether uptake sequences interfere with optimal coding and thus might reduce fitness. For simplicity, this analysis was limited to ORFs that 1) contained only a single uptake sequence, 2) had homologs in all three comparison genomes, each alignable over a 99-aa segment centered on the uptake sequence, and 3) had no gaps in the alignments of the uptake sequence itself or of two nearby 3-aa segments. The numbers of sequences meeting these criteria are given in table 3, and a diagram illustrating the analysis is provided as supplementary figure S2 (Supplementary Material online). Analysis of sequential 9-aa segments of these 99-aa alignments showed that uptake sequences preferentially occur in less-constrained parts of proteins (fig. 5A and B). The reduced conservation is not caused by the uptake sequences themselves because the same local decrease in conservation is seen for the alignments of the comparison homologs from genomes without uptake sequences. The magnitude of the effect is remarkably similar to that reported by Treangen et al. (2008) for within—N. meningitidis divergence of nucleotide sequences (10).
Table 3.
Analysis of Uptake Sequence–Encoded Peptides in Proteomes
| Set of proteins | H. influenzae Rd KW20 | N. meningitidis Z2491 |
| Proteins encoded by genome | 1657 | 2065 |
| Proteins containing UEPsa | 643 (38.8%) | 460 (22.3%) |
| Translated ORFs with a single UEPa | 425 | 335 |
| with homologs in three genomes | 282 | 173 |
| 99-aa alignment centered on UEPa | 140 | 91 |
| Number analyzed (no gaps at UEP) | 131 | 86 |
| Mean pairwise identity QHb | 54.6% | 51.0% |
| Mean pairwise identity HHc | 54.4% | 55.0% |
Note.—For all ORFs containing a single US in each genome, homologs were defined based on Blast hits (BlastP with E value < 1 × 10−9) of the protein sequence against the translated genomes of three related bacteria as described in Materials and Methods.
UEP, US-encoded peptide.
QH, query–homolog.
HH, homolog–homolog.
FIG. 5.—
Peptide sequence identity as a function of distance from the uptake sequence position for US-containing genes with three comparison homologs (see table 3). Each protein was aligned with its three homologs, and the 99-aa region centered on the US-encoded peptide was divided into 11 9-aa slices. The mean pairwise percent identity of the three query–homolog pairs and three homolog–homolog pairs was determined for each slice and was normalized by the corresponding value for the overall trimmed alignment of each protein to its 3 homologs. Error bars are 95% confidence limits from a one-way analysis of variance. (A) Average identities for slices of 131 H. influenzae sequences aligned to homologs: mean of three query–homolog comparisons (light blue); mean of three homolog–homolog comparisons (dark blue). (B) Average identities for slices of 86 N. meningitidis sequences aligned to homologs: mean of three query–homolog comparisons (light orange); mean of 3 homolog–homolog comparisons (dark orange).
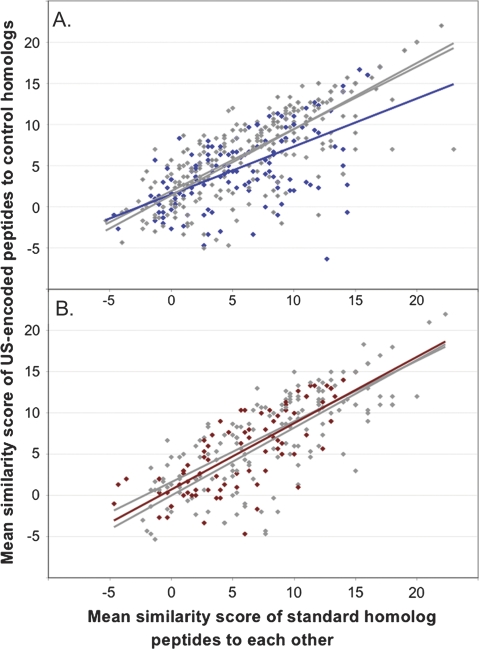
Finally, we asked whether the uptake sequences that have persisted in the proteome impose any ongoing fitness costs. Because the most likely cost is interference with coding for functionally optimal amino acids, we investigated whether amino acids specified by core uptake sequences are more divergent than other amino acids. Our strategy was to measure the deviation of these amino acids from the consensus seen for homologous peptides in the three comparison genomes. Details about the genes used and their comparison homologs are provided in Materials and Methods and in supplementary table S3 (Supplementary Material online), and figures 6A and B show the mean similarity scores for each uptake sequence analyzed. The gray symbols and lines show the scores of control 3-aa segments 9 aa upstream and downstream of the three US-encoded aa’s, and the colored points and lines show the scores of the US-encoded segments. If being encoded by uptake sequences does not constrain amino acid sequences, we expect the upstream, downstream, and US data sets to be very similar. This is seen for N. meningitidis (fig. 6B) and for the H. influenzae peptides that are not strongly conserved in the homologs (points on the left side of fig. 6A). However, for peptides whose homologs are strongly conserved, the H. influenzae USS-encoded segments are more divergent from their homologs than are segments upstream and downstream of the USSs. Although the 95% confidence intervals of the slopes overlap slightly, the difference suggests that that accumulation of USSs in highly conserved proteins may occasionally interfere with protein coding.
FIG. 6.—
Amino acid similarity of US-encoded tripeptides to aligned sequences of homologs in comparison proteomes. y axis: scores of H. influenzae or N. meningitidis peptides against comparison homolog peptides; x axis: scores of comparison peptides against each other. Colored points and lines, similarities of US-encoded amino acids to aligned tripeptides in homologs; gray points and lines, similarities of control tripeptides located 9 aa upstream and downstream of UEPs to aligned sequences in homologs. (A) H. influenzae and control peptides (blue); (B) N. meningitidis and control peptides (dark red).
How is N. meningitidis able to accommodate its uptake sequences without interference with protein coding? Its uptake sequences are not in less constrained positions; the mean similarity score of its homologs to each other is actually slightly higher than those of H. influenzae (6.17 vs. 5.88). Furthermore, the analysis in figure 1 and table 2 also shows that the amino acids specified by N. meningitidis DUSs in their preferred reading frames are actually much “less” common than those specified by H. influenzae USSs in their preferred frames, and the codon scores of the two uptake sequences are not significantly different (fig. 2B). The explanation may be that differences in population structure or transformation frequency allow N. meningitidis to more efficiently eliminate deleterious mutations.
Discussion
This study used the present distributions of uptake sequences in bacterial genes as evidence of how natural selection and molecular drive have acted on them. Our context is a simple model for the origin of uptake sequence, incorporating 1) random mutations in uptake sequences, 2) selection on the direct phenotypic consequences of these mutations, and 3) the molecular drive created by biased DNA uptake, which favors alleles that better match its bias. In this model, uptake sequences perfectly matching the core consensus originate throughout the population by mutation from imperfect uptake sequences. These variants then spread to other members of the species by natural transformation because they are favored substrates for the DNA uptake machinery. Mutations creating worse uptake sequence variants also arise, but their spread is opposed by the uptake bias. The importance of transformational recombination in uptake sequence evolution is supported by the correspondence between DUS spacing and the length distribution of N. meningitidis recombination tracts (Treangen et al. 2008). The DNA uptake and recombination advantage that uptake sequences have over other mutations (due to the bias of the uptake machinery) causes them to accumulate in genomes over evolutionary time, even if the recombination they promote has no fitness benefits. However, this molecular drive will be opposed by selection for genome functions, especially optimal protein coding.
To appreciate the interplay of forces, first consider how selection acts on deleterious mutations that do not affect uptake sequences. At the population level, all new deleterious mutations have the same ultimate cost—each will eventually be eliminated by a selective death, with the severity of the defect determining how long this is likely to take. For naturally competent cells without uptake specificity, DNA uptake and homologous recombination simply add noise to this process, as cells will sometimes acquire new alleles and lose old ones by random transformation as well as by random mutation. However, if DNA uptake is biased and some of the mutations create sequences preferred by the uptake machinery, more cells will acquire these deleterious mutations than other mutations. Elimination of each of these acquired mutations will necessitate another selective death. In the context of the mutation–selection balance usually used to predict the equilibrium frequency of deleterious mutations, we can consider biased transformation as increasing the effective mutation rate to alleles that increase uptake; by increasing how often these alleles enter the genome, biased uptake increases their equilibrium frequency. Mutations that instead directly increase fitness are rare in well-adapted organisms, but if such mutations also improve uptake sequences, they will be more strongly favored; those that worsen uptake may be lost despite their fitness benefit. On the other hand, mutations that decrease both fitness and uptake will fare especially badly.
Because uptake sequences are rare in the genes H. influenzae acquired since its divergence from A. pleuropneumoniae, their accumulation in other transferred genes is likely to have taken hundreds of millions of years. Additional evidence that the force driving uptake sequence accumulation is weak comes from the correlation between codon-bias effects and uptake sequence–reading frame use, which shows that the strength of drive favoring individual uptake sequences is comparable to the selection favoring use of optimal codons in a coding sequence (Hartl et al. 1994). But even weak forces can have large effects when they act over long periods, and accumulation of core uptake sequences has dramatically modified their associated proteomes, more than doubling the frequencies of many of the tripeptides they can encode (fig. 1).
It is difficult to evaluate how the fitness consequences of recombining linked alleles contribute to uptake sequence evolution. First, because these consequences are indirect, their effects are predicted to be weak relative to molecular drive. Second, any fitness benefits may be overwhelmed by fitness costs. The impact of maladaptive combinations of alleles is always difficult to measure because they are usually removed by natural selection, whereas beneficial combinations are preserved. However, theoretical analysis of the costs and benefits of recombination, mainly in the context of the evolution of sexual reproduction in eukaryotes, has found net benefits to be much smaller and less ubiquitous than previously thought (Otto and Gerstein 2006). Although our analysis did not confirm a strong association between uptake sequences and genome maintenance genes, it did find significant overrepresentation and underrepresentation of uptake sequences in some COG functional categories across very distantly related genomes. These differences could reflect selection on recombination of alleles linked to uptake sequences, but they might also reflect different levels of conservation and/or frequencies of horizontal transfer in these groups.
Considerations of tractability limited our analysis to sequences that perfectly matched the core consensus of each uptake sequence type. The number of sequence types needing to be considered increases dramatically as the consensus is relaxed and/or the motif is extended, but their contribution to uptake bias appears to be small (Ambur et al. 2007; Bakkali 2007). Nevertheless, both flanking sequences and mismatched cores could have substantial impacts on protein coding. Sequences with a single mismatch to the uptake sequence consensus provide the pool from which the perfect uptake sequences arise; they are overrepresented in genomes and preferred by uptake machinery over nonuptake sequences, though less so than perfect matches (Bakkali et al. 2004). Sequences with two mismatches are also overrepresented; this may reflect similar but weaker uptake biases, but could also just be produced by mutation from the better matches. Substantial coding constraints are likely to arise from the strong-consensus AT-rich tracts that flank one side of the H. influenzae and A. pleuropneumoniae USSs (Redfield et al. 2006); we do not yet know how well these are accommodated.
Although the genome and proteome properties of uptake sequences are now well characterized, we know almost nothing about how they contribute to DNA uptake. One attractive hypothesis is that uptake sequences provide sites for DNA deformation required by the uptake process—H. influenzae cells are known to be able to transport closed circular plasmids through the outer membrane, a step that is likely to require both sharp kinking of double-stranded DNA and deformation of the secretin pore (Kahn and Smith 1984; Assalkhou et al. 2007). Thus, one promising line of future investigation is characterization of the molecular interactions of uptake sequences with DNA uptake proteins.
Supplementary Material
Supplementary figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/our_journals/gbe).
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes for Health Research and the US National Institutes of Health (grant number RO1 GM60715). Substantial contributions to early stages of this work were made by Ta-Yuan Chen and H.-C. Lee and to later stages by John Nash. We also thank Tone Tonjum for COG analysis data and Heather Maughan for help with statistical analysis and dN/dS interpretation.
References
- Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambur OH, Frye SA, Tonjum T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol. 2007;189:2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assalkhou R, et al. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology. 2007;153:1593–1603. doi: 10.1099/mic.0.2006/004200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkali M. Genome dynamics of short oligonucleotides: the example of bacterial DNA uptake enhancing sequences. PLoS ONE. 2007;2:e741. doi: 10.1371/journal.pone.0000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkali M, Chen TY, Lee HC, Redfield RJ. Evolutionary stability of DNA uptake signal sequences in the Pasteurellaceae. Proc Natl Acad Sci USA. 2004;101:4513–4518. doi: 10.1073/pnas.0306366101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner DB, Deich RA, Sisco KL, Smith HO. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980;11:311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Davidsen T, et al. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 2004;32:1050–1058. doi: 10.1093/nar/gkh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive. Trends Genet. 2002;18:587–589. doi: 10.1016/s0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn ME, Smith HO. Transformation in Haemophilus: a problem in membrane biology. J Membr Biol. 1984;81:89–103. doi: 10.1007/BF01868974. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J, Campbell AM. Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res. 1996;24:4263–4272. doi: 10.1093/nar/24.21.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Gerstein AC. Why have sex? The population genetics of sex and recombination. Biochem Soc Trans. 2006;34:519–522. doi: 10.1042/BST0340519. [DOI] [PubMed] [Google Scholar]
- Pasamontes A, Garcia-Vallve S. Use of a multi-way method to analyze the amino acid composition of a conserved group of orthologous proteins in prokaryotes. BMC Bioinformatics. 2006;7:257. doi: 10.1186/1471-2105-7-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ, et al. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocca JJ, Poland RL, Zoon KC. Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J Bacteriol. 1974;118:369–373. doi: 10.1128/jb.118.2.369-373.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HO, Gwinn ML, Salzberg SL. DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol. 1999;150:603–616. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- Smith HO, Tomb J-F, Dougherty BA, Fleischmann RD, Venter JC. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ambur OH, Tonjum T, Rocha EP. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9:R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MW. An intragenic distribution bias of DNA uptake sequences in Pasteurellaceae and Neisseriae. Biol Direct. 2008;3:12. doi: 10.1186/1745-6150-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.