Abstract
Miniature inverted-repeat transposable elements (MITEs) are a particular type of defective class II transposons present in genomes as highly homogeneous populations of small elements. Their high copy number and close association to genes make their potential impact on gene evolution particularly relevant. Here, we present a detailed analysis of the MITE families directly related to grapevine “cut-and-paste” transposons. Our results show that grapevine MITEs have transduplicated and amplified genomic sequences, including gene sequences and fragments of other mobile elements. Our results also show that although some of the MITE families were already present in the ancestor of the European and American Vitis wild species, they have been amplified and have been actively transposing accompanying grapevine domestication and breeding. We show that MITEs are abundant in grapevine and some of them are frequently inserted within the untranslated regions of grapevine genes. MITE insertions are highly polymorphic among grapevine cultivars, which frequently generate transcript variability. The data presented here show that MITEs have greatly contributed to the grapevine genetic diversity which has been used for grapevine domestication and breeding.
Keywords: Vitis, transposon, MITE
Introduction
Miniature inverted-repeat transposable elements (MITEs) are a particular type of defective class II transposons. They share some features with nonautonomous class II transposons: They are characterized by their terminal inverted repeat (TIR) structure, the flanking short direct repeats formed by target site duplication (TSD), and their absence of gene-coding capacity. Most MITE families share extensive sequence similarities with class II transposons from which they are supposed to derive by internal deletion (Feschotte and Mouches 2000; Yang and Hall 2003; Jiang, Feschotte, et al. 2004; Zhang et al. 2004; Moreno-Vázquez et al. 2005) and which can mobilize them in trans (Dufresne et al. 2007; Miskey et al. 2007; Yang et al. 2007). On the other hand, MITEs are distinguished from other nonautonomous class II transposons by their high copy number, the high uniformity of their copies, and in some cases their potential to form single strand secondary structures. It has been proposed that MITEs are generated by a two-step process, in which a small number of particular class II defective elements are amplified by a still unknown replicative mechanism becoming the founder elements of new MITE families (Feschotte et al. 2002; Casacuberta and Santiago 2003). MITE families can reach very high copy numbers. For example, the Glider element is present in more than 20,000 copies in the genome of Xenopus laevis (Lepetit et al. 2000).
MITEs are often found close or within genes where they can affect gene expression by providing new splicing sites, transcription start sites, new exons, and poly(A) sites (Santiago et al. 2002; Ohmori et al. 2008; Kuang et al. 2009). Additionally, MITEs can give rise to short interfering RNA genes and regulate genes that are not necessary in their proximity (Piriyapongsa and Jordan 2007, 2008; Kuang et al. 2009). Their high copy number and frequent association with genes makes MITEs major players in the evolution of genes and the plasticity of the genomes.
Grapevine is a widely cultivated crop that has accompanied human cultures since its domestication in the Neolithic period (c. 8500–4000 BC). Cultivated grapevine (Vitis vinifera spp. sativa) is supposed to have been domesticated from wild grapevine populations (Vitis vinifera spp. sylvestris Gmelin) in the Near East and West Europe (Arroyo-Garcia et al. 2006; This et al. 2006). Although sexual crossing has been a major driver of grapevine evolution, its vegetative propagation enhanced the impact of somatic mutations and has been important for grapevine diversity. Clonal selection of superior individuals identified by growers has led to many clones with different phenotypes while maintaining the same cultivar name (Forneck 2005). Some of these mutations exist and are maintained in a chimeric state affecting only single cell layers (Franks et al. 2002), the phenotype of the plant being the result of the combination in different cells of two different genotypes.
Transposable elements (TEs) are known to be major contributors to genome variability and, in particular, to somatic mutations (Collier and Largaespada 2007; Deragon et al. 2008). Thus, TEs have probably played a major role in grapevine domestication and breeding. We recently described 51 families of class II transposons in grapevine and 15 putative families of domesticated transposons (Benjak et al. 2008). In this work, we analyze the MITE subfamilies that are related to those transposons and provide evidence for their major role in shaping the grapevine genome.
Materials and Methods
Transposon Mining
We performed our analyses using the whole-genome shotgun sequences of the two sequenced grapevine genomes made available at National Center for Biotechnology Information (NCBI; Jaillon et al. 2007; Velasco et al. 2007). We used previously described TEs (Benjak et al. 2008) as queries in Blast searches (Altschul et al. 1990) to retrieve the putative MITEs. To check for transcription of MITEs, representatives of each MITE family were used as queries in Blast searches against the grapevine expressed sequence tag (EST) collection at NCBI. The matching ESTs were then used as queries in Blast searches against the nucleotide database to determine the source sequence for each transcript. As both Velasco et al. (2007) and Jaillon et al. (2007) performed computational gene predictions, the NCBI contains a significant number of predicted (but not annotated) Vitis proteins which were useful to characterize the transduplicated sequences. For each MITE group, multiple alignments (including gapped positions) were created from which similarity matrices were calculated using BioEdit software, version 7.0.5.3 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Multiple alignments are available upon request. Average similarities were calculated from similarity matrices in Microsoft Excel software.
To look for mPifvine-3-related elements having conserved only the TIRs but containing internal sequences unrelated to mPifvine-3, we used the TRANSPO 1.0 software (http://alggen.lsi.upc.es/) (Santiago et al. 2002). The search was done looking for mPifvine-3 TIRs separated by 150–350 nt. The number of mismatches allowed for the TIR sequences (18 nt) in the TRANSPO program was two. Retrieved sequences were clustered into groups by blasting against each other. For each subfamily, a consensus sequence was created and was used as query for Blast searches in grapevine chromosomes CU462738–CU462756 (Jaillon, 2007 #21). Consensus sequences representing the MITE families were deposited at the Repbase database.
Plant Material
A list of samples and their source is given in supplementary table 1 (Supplementary Material online). DNA from all samples derived from Germany was extracted using E.Z.N.A. SP Plant DNA Mini Kit (Omega Bio-tek). DNA of other samples was obtained from different laboratories.
Polymerase Chain Reaction Analysis
Primers were designed using Primer3 (Rozen and Skaletsky 2000) and FastPCR programs (www.biocenter.helsinki.fi/bi/programs/fastpcr.htm). Each primer was blasted against the whole Vitis genomic database to check for specificity. The list of primers is given in supplementary table 2 (Supplementary Material online). Polymerase chain reactions (PCRs) were done in 20 μl reaction volumes using approximately 30 ng of DNA template, 0.5 μl of each primer (10 pmol/μl), and TaKaRa Ex Taq in the following conditions: 94°C for 2 min + 40 cycles (94°C for 25 s, 58–62°C [depending on primer] for 45 s, and 72°C for 1 min). PCR products were run in 1.2% agarose gels with ethidium bromide in a 1× Tris-acetate-EDTA buffer and visualized under UV light.
Gene and MITE Positions’ Extraction and Analysis
We developed a set of Perl scripts to extract and compare the positions of genes and MITEs in the Vitis genome. Positions of all predicted genes, including intron, exon, and untranslated region (UTR) coordinates when available, were extracted from the grapevine chromosomes CU462738–CU462756 (Jaillon, 2007 #21). The positions of MITEs were extracted from Blast results of MITE consensus sequences against the Vitis chromosomes. We considered only Blast hits that corresponded to at least 70% of the consensus length. Genes and MITE coordinates are given in the supplementary file 1 (Supplementary Material online).
Results
Grapevine Contains MITEs Related to Different Superfamilies of Class II Transposons
Although the first MITEs to be described in plants were related to elements of the PIF/Pong and Mariner families (Feschotte and Mouches 2000; Feschotte et al. 2003; Zhang et al. 2004), MITEs related to most families of class II TEs have been described in plants later on (Yang and Hall 2003; Saito et al. 2005; Kuang et al. 2009). We have recently described the “cut-and-paste” transposon landscape of the grapevine genome and found that it contains representatives of four of the five superfamilies present in plants (Benjak et al. 2008). Here, we present an analysis of the MITEs directly associated to the described grapevine transposons. We searched for elements that share TIRs and subterminal sequences with those elements are devoid of transposase coding capacity and are present in high number of copies highly homogeneous in size and sequence. This was done by visual inspection of Blast results from searches of the published grapevine genome (Velasco et al. 2007) with representatives of each of the major families of class II transposons previously described in Vitis (Benjak et al. 2008). We have not found any potential MITEs related to grapevine MULEs. Although we found defective MULEs, they were not present in multiple copies. On the contrary, we have found potential MITEs related to the other three transposon superfamilies present in grapevine, the CACTA, hAT, and PIF superfamilies, and we named these putative MITEs according to the previously given family names (m-“TE family name”.MITE subfamily number). These elements, related to 8 families of transposons, are highly homogeneous in size and sequence, which suggest that they have been amplified from a single or few founder elements, as it is usually the case for MITEs (Feschotte et al. 2002; Casacuberta and Santiago 2003; Deragon et al. 2008). Seven of these subfamilies (mCactavine-4.1, mHatvine-2.1, mHatvine-3.1, mHatvine-10.1, mPifvine-1.1, mPifvine-2.n, and mPifvine-4.1) are composed by a moderate copy number of relatively long elements, and only mPifvine-3.1 elements are present at high copy number and are of a size similar to the typical MITE families described in plants (table 1).
Table 1.
MITEs Found in Grapevine and Their Properties
| MITE Name | Average Length in bp (standard deviation) | Similarity to Autonomous TEs (%) | Approximate Copy Number | Total Coverage in kb | TIR Length | TSDs Length | Average Identity | ESTs Matching to MITEs | Representative | Coordinates | Repbase Name |
| mPifvine-1.1 | 715 (δ = 27.2) | 97 | 51 | 37.2 | 20 | 3 | 0.88 | 1 | AM485510.1 | 38610–37877 | Harbinger-1N1_VV |
| mPifvine-2.na | ∼1 kb | 93 | 65 | 91.7 | 26 | 3 | 0.77 | 2 | AM452748.2 | 1881–2798 | Harbinger-1N1_VV |
| mPifvine-3.1b | 274 (δ = 10.8) | 80 | 1,298 | 355.8 | 18 | 3 | 0.83 | ∼50 | AM468072.2 | 15068–15345 | Harbinger-3N1_VV |
| mPifvine-4.1 | 1243 (δ = 90.2) | 90 | 76 | 94.5 | 11 | 3 | 0.81 | 1 | AM450168.2 | 3569–4849 | VHARB-N4_VV |
| mHatvine-2.1 | 769 (δ = 14.4) | 94 | 22 | 16.9 | 23 | 8 | 0.92 | 2 | AM436283.2 | 16403–17173 | VIHAT2-N1_VV |
| mHatvine-3.1 | 740 (δ = 77.7) | 89 | 30 | 22.2 | 16 | 8 | 0.71 | 3 | AM458859.2 | 11690–12492 | VIHAT3-N1_VV |
| mHatvine-10.1 | 1274 (δ = 23.7) | 89 | 20 | 22.9 | 11 | 8 | 0.89 | 0 | AM432725.2 | 460–1725 | hAT-10N1_VV |
| mCactavine-4.1 | 3243 (δ = 23.8) | 94 | 30 | 97.3 | 6 | 3 | 0.84 | 31 | AM457287.1 | 34741–37976 | EnSpm-4N1_VV |
We have described 13 subfamilies of mPifvine-2 MITEs (see table 2).
Three additional subfamilies of mPifvine-3 accounting for around 2,000 copies were found using different methods. Details are given in the text.
Pack-MITEs: MITEs Transduplicating Gene Sequences
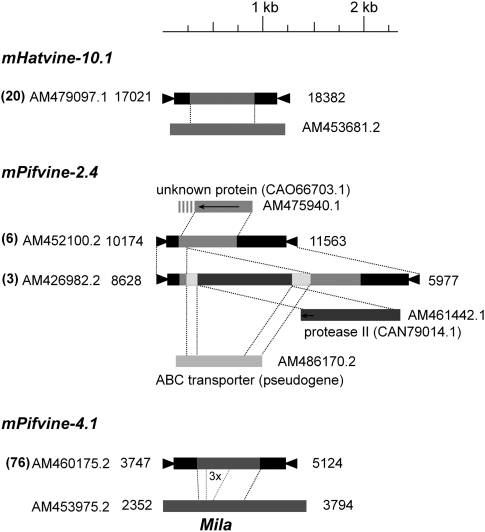
Although all the MITE families here described were found because they show extensive sequence similarity with the related TE families (table 1), some of them (mCactavine-4.1, mHatvine-10.1, mPifvine-2.n, and Pifvine-4.1) also contain sequences unrelated to their corresponding long elements. In the case of mCactavine-4.1, the internal sequence, which is 900 bp long and is highly conserved in all copies (88%), does not have similarity to any other sequence (not shown). For the other MITE subfamilies, the internal sequences are highly similar to grapevine genomic sequences. This suggests that MITEs can capture, mobilize, and amplify host genomic sequences as typical DNA transposons do (Jiang, Bao, et al. 2004; Kawasaki and Nitasaka 2004; Zabala and Vodkin 2005, 2007; Hanada et al. 2009) in a process that has been named as transduplication (Juretic et al. 2005). We have thus analyzed these possible examples of transduplication in detail. mHatvine-10.1 elements share TIRs and subterminal regions with Hatvine-10 TEs but contain a central 583-bp long region not related to Hatvine-10 but to a grapevine nongenic region (82% identity, fig. 1). This region is highly conserved in all 20 mHatvine-10.1 elements (average identity 88.5%) and suggests that an ancestral mHatvine-10.1 transduplicated a genomic region and that the composite element was later on amplified. A similar scenario could also explain the structure of mPifvine-2 elements that also contain a central region unrelated to Pifvine-2 transposons. However, this central sequence is not the same in all mPifvine-2 copies as each internal sequence is shared only by few elements (1–12). We grouped these elements in 13 subfamilies, each of which has a different internal sequence. In all the cases, the internal sequence shows high sequence similarity to a grapevine genomic sequence (table 2) suggesting that all have been transduplicated by mPifvine-2 elements (fig. 1 and table 2). Most transduplicated sequences are coding sequences corresponding to expressed grapevine sequences, and fragments from different genes can be present in a single mPifvine-2 element (fig. 1 and table 2). In most cases, the sequences found within mPifvine-2 elements have not conserved their coding capacity (exceptions are the transduplicated fragments in mPifvine-2.3, mPifvine-2.11, and mPifvine-2.12). Interestingly, in the cases when a subfamily has transduplicated several gene fragments, elements with a different number of such fragments are found, suggesting that a subset of elements containing transduplicated gene fragments have undergone additional rounds of transduplication and amplification (fig. 1).
FIG. 1.—
Transduplication of genomic sequences by MITEs. Triangles represent TIRs. Arrows represent exons. Accession numbers for all sequences are given as well as the coordinates for the TEs. Copy number is given in brackets.
Table 2.
Information on transduplicated sequences in some mPifvine-2 subfamilies.
| mPifvine-2 subfamily | Copy number | Accession n° for the representative | Coordinates | Trans-duplication in bp | Genomic source of transduplication | Identity of the original sequence to its transduplication | Predicted protein | Tentative protein annotation | Transduplicated exons |
| 1 | 6 | AM452748.2 | 1881–2798 | 500 | AM462940.1 | 88% | N.P. | Thaumatin-like | yes |
| 2 | 4 | AM442314.2 | 17623–19508 | 1300 | AM449452.1 | 94% | CAO21921.1 | BAH-AAA-containing protein | yes |
| 3 | 4 | AM477430.2 | 3327–5005 | 800 | AM441647.2 | 93% | CAN62993.1 | Glycosyl transferase | yes |
| 4 | 12 | AM426982.2 | 5977–8628 | 700 | AM486170.2 | 91% | N.P. | ABC transporter (pseudogene) | yes |
| 1000 | AM461442.1 | 86% | CAN79014.1 | Protease II | yes | ||||
| 95 | AM475940.1 | 91% | CAO66703.1 | Unknown protein | yes | ||||
| 5 | 1 | AM447383.2 | 3750–4593 | 400 | CU459360.1 | 83% | CAO66902 | DUF1296-containing protein | yes |
| 6 | 2 | AM467559.2 | 7724–9445 | 700 | AM437259.2 | 91% | CAN82620.1 | Glycosyl hydrolase | yes |
| 7 | 6 | AM436343.2 | 7645–9489 | ∼1000 | N.H. | N.D. | similar to CAO46980.1 | Unknown protein | yes |
| 8 | 6 | AM449479.2 | 9036–10309 | 80 | N.H. | N.D. | similar to CAO40038.1 | Sec15-containing protein | yes |
| 100 | N.H. | N.D. | similar to CAO49645.1 | Unknown protein | yes | ||||
| 600 | AM463124.2 | 91% | CAO39675.1 | RING finger | yes | ||||
| 9 | 12 | AM445491.2 | 6050–7622 | 830 | AM452971.1 | 82% | N.P. | N.P. | no |
| 10 | 4 | AM429887.2 | 6356–7627 | 630 | AM425582.2 | 92% | N.P. | N.P. | no |
| 11 | 2 | AM432699.2 | 8138–9741 | 700 | AM458836.2 | 92% | CAN72319.1 | Serine protease | yes |
| 12 | 4 | AM431974.2 | 47527–48887 | 860 | AM471293.1 | 92% | CAO46017.1 | Sulfate transporter like protein | yes |
| 13 | 2 | AM433436.2 | 1665–2724 | 545 | AM450890.2 | 97% | CAO42680.1 | Pectinesterase | yes |
N.H.: no hits; N.D.: not determined; N.P.: not predicted
In addition to the abovementioned examples, we have characterized another example of transduplication that has particular characteristics. mPifvine-4.1 MITEs, present in more than 70 copies that are around 1,200 bp long (table 1), have a 780-bp long central region which is not found in the full-length Pifvine-4 elements. Differently to most transduplicated sequences, this central region does not correspond to a single copy sequence found elsewhere in the genome, but to a repetitive sequence present in more than 180 copies in grapevine, which we have named Mila (its reference is given in fig. 1). Mila seems to be highly expressed as it matches more than 100 ESTs deposited in the grapevine EST databases (not shown). Mila is 1.5 Kb long and is flanked by direct repeats of 7 bp that could represent TSD generated upon insertion. All these characteristics suggest that Mila is a potentially active mobile element, which is not related to any of the known families of TEs, as its sequence and structure differ from that of the known TEs. mPifvine-4.1 could be a composite transposon containing a Mila insertion nested, but because the sequence included within mPifvine-4 elements is only an internal part of Mila (which also contains a partial tandem duplication of a central motif) and it is not flanked by TSDs, we suggest that Mila sequences have been transduplicated and further amplified by mPifvine-4.1 elements.
mPifvine-3.1 Distribution with Respect to Grapevine Genes
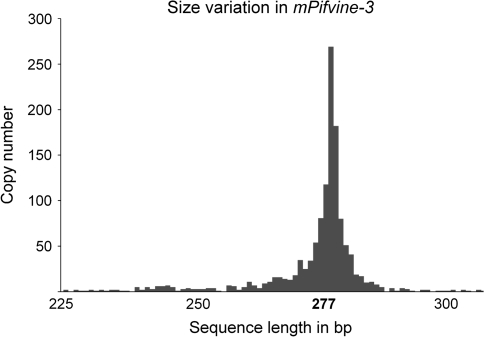
Although we have described putative MITEs belonging to eight different TE families, only the Pifvine-3–related MITE subfamily (named mPifvine-3.1) is present in high copy number (more than 1,000) and is of a size (274 bp) similar to that of typical MITEs. A phylogenetic analysis of mPifvine-3.1 elements did not allow grouping them into distinct clusters (not shown) suggesting that the mPifvine-3.1 founder elements at the origin of the bursts of amplification that generated the whole family were very similar in sequence. The fact that the 1,200 mPifvine-3.1 are extremely homogeneous in size (fig. 2) and sequence (the overall sequence similarity is on average 86% for the 90% most conserved copies) suggests that their amplification took place recently during Vitis genome evolution. This has prompted us to analyze this MITE subfamily in more detail.
FIG. 2.—
Size variation in mPifvine-3.1.
Most MITEs are supposed to originate by internal deletions of class II transposons, and consequently, they usually show high sequence similarity to them. However, for some MITEs, this is not the case as their sequence similarity to class II transposons is limited only to the transposase-recognized TIR sequences (Feschotte et al. 2003; Zhang et al. 2004; Quesneville et al. 2006). We have thus decided to look for mPifvine-3 subfamilies that could have limited sequence similarity to Pifvine-3 elements using the TRANSPO software (Santiago et al. 2002) that looks for sequences of a given range of lengths containing a specified TIR. With this approach, we could define three new subfamilies of mPifvine-3, which we have named as mPifvine-3.2, mPifvine-3.3, and mPifvine-3.4. These subfamilies contain the Pifvine-3 TIRs that flank different internal sequences not related to the autonomous Pifvine-3 element. By using the consensus sequences from each subfamily of MITEs as query in Blast searches in the Vitis shotgun sequences (Jaillon, 2007 #21), we approximated the copy number to be more than 1,000 for mPifvine-3.2, some 300 for mPifvine-3.3 and around 20 for mPifvine-3.4.
MITEs are very frequently found associated to genes in plant genomes (Casacuberta and Santiago 2003; Feschotte and Pritham 2007). The potential for MITEs to generate gene variants for evolution is thus very high. We have, therefore, decided to analyze the distribution of mPifvine-3 elements with respect to Vitis-coding sequences. To extract the positions of all predicted genes, including intron, exon, and UTR coordinates when available, we used the partially assembled grapevine chromosomes (CU462738–CU462756) (Jaillon, 2007 #21) which had a total of 303 Mb (∼70% of the whole genome). We analyzed the distribution of all mPfivine-3 subfamilies, except of the mPifvine-3.4 because of its low copy number (table 3). In cases of insertion within a predicted gene, we analyzed whether the insertion had occurred in 5′ UTR, exons, introns, or 3′ UTR.
Table 3.
Analysis of the Distribution of mPifvine-3 MITEs in the Grapevine Genome Compared with Genes
| Number of MITEs per Mb of Genomic Sequence |
|||||
| Subfamily | Nongenic DNA | Intron | Exon | 5’ UTR | 3’ UTR |
| mPifvine-3.1 | 6.24 | 3.43 | 0.14 | 10.45 | 12.76 |
| mPifvine-3.2 | 5.75 | 2.37 | 0 | 1.21 | 1.14 |
| mPifvine-3.3 | 0.56 | 0.28 | 0 | 0 | 0.19 |
NOTE.—In addition to full length elements, we also considered fragmented MITEs that contain at least 70% of the consensus full length.
As expected, the three mPifvine-3 subfamilies are excluded from coding sequences (table 3). Moreover, mPifvine-3.2 and mPifvine-3.3 elements are more frequently found in nongenic sequences than in noncoding genic sequences (table 3). This probably reflects purifying selection against elements inserted in functional regions. Note that in both the cases, the presence in introns, which are particularly long in grapevine (Jaillon et al. 2007; Velasco et al. 2007), is more frequent than in 5′ and 3′ UTRs. mPifvine-3.1 elements show a very different distribution. Although the frequency of mPifvine-3.1 elements within introns is lower that in nongenic sequences, these elements are highly abundant in 5′ UTRs and, especially, in 3′ UTRs (table 3). This makes the mPifvine-3.1 family of MITEs particularly interesting.
Recent Transposition and Amplification of mPifvine-3.1 MITEs in Vitis Species
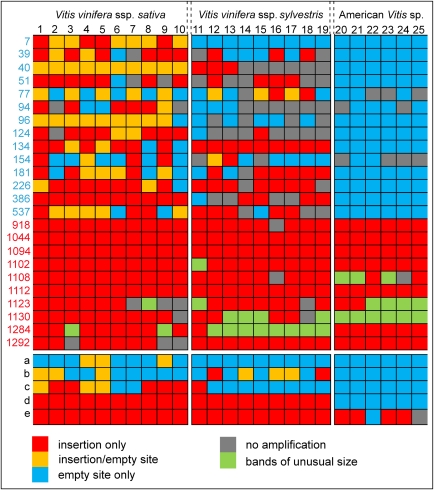
In order to look for evidences of recent transposition and amplification of mPifvine-3.1, we analyzed the presence of these elements at particular loci in 10 different cultivars of the domesticated grapevine V. vinifera ssp. sativa, 9 genotypes of the European wild species V. vinifera ssp. sylvestris, and 6 more distantly related North American Vitis species (see Materials and Methods for details). We looked for insertion polymorphisms of mPifvine-3.1 elements by amplifying by PCR 24 loci that contained an mPifvine-3.1 insertion in the published genome, that is, V. vinifera ssp. sativa cv. Pinot noir (Velasco et al. 2007). Although the mPifvine-3.1 family of MITEs is recent, pairwise comparisons of the 1298 mPifvine-3.1 elements showed a range of sequence conservation among them allowing us to identify elements that are probably more recent than others. We have thus chosen to analyze 14 loci representing more recent insertions (designated by blue numbers in fig. 4), as judged by their high degree of sequence similarity in pairwise comparisons, and 10 loci that probably represent older insertions, as judged by the same criterion (designated by red numbers in fig. 4). An example of the polymorphism analysis is shown in figure 3, and the summary of the results obtained is presented in figure 4. None of the mPifvine-3.1 recent insertions is present in the corresponding locus of any of the six American Vitis species, suggesting that these insertions occurred after the split of the European and American Vitis species. Of those, 3 of 14 insertions are not present in any of the corresponding locus of the European wild species of V. vinifera, suggesting that they occurred after V. vinifera domestication.
FIG. 4.—
Summary of the mPifvine-3.1 insertion polymorphisms analysis. Summary of the PCR amplification of different loci from DNA obtained from 25 different V. vinifera ssp. sativa cultivars, V. vinifera ssp. sylvestris genotypes, and American Vitis species. Samples are numbered on the top and details are given in supplementary table 1 (Supplementary Material online). Numbers and letters on the left indicate loci names: red numbers correspond to relatively old insertions; blue numbers correspond to relatively recent insertions (see Materials and Methods), and letters (a to e) correspond to insertions of mPifvine-3.1 elements within genes and designate the same genes shown in figure 5.
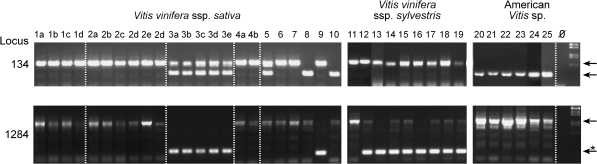
FIG. 3.—
Analysis of mPifvine-3.1 insertion polymorphisms. Examples of PCR results with primers flanking mPifvine-3.1 insertions for two chosen loci. Pictures from different gels were joined for this figure to match the order of the samples given in supplementary table 1 (Supplementary Material online). Arrows represent sizes of bands expected for insertion and empty sites. In the locus 1284, shown are only bands corresponding to the mPifvine-3.1 insertion and unusually small bands (arrow with “*”) corresponding to a larger deletion of the allele. These small bands were found in most sylvestris genotypes and only in two domesticated cultivars, Chardonnay and We 70-281-37.
Our results thus show that mPifvine-3.1 has actively transposed during the evolution of grapevine. Some mPifvine-3.1 elements were already present and transposing in the ancestor Vitis species, whereas other copies have transposed accompanying grapevine domestication and breeding. Almost all recent insertions show a high polymorphism among the 10 cultivated genotypes that were tested, which stresses the high heterozygosity of this species (Velasco et al. 2007). On the contrary, the 10 loci analyzed corresponding to older mPifvine-3.1 insertions seem to be almost fixed in the population of cultivated V. vinifera, wild European sylvestris genotypes and American Vitis species.
For a number of mPifvine-3.1 insertion loci, bands of unexpected sizes were obtained (figs. 3 and 4). The sequencing of bands deriving from loci 1102 and 1284 showed that these unexpected bands corresponded to deletions that occurred within the loci. In the case of locus 1102, there is a partial deletion of the mPifvine-3.1 and its 5′ flanking sequence, and in the case of locus 1284, there is a larger deletion of the whole mPifvine-3.1 and its flanking sequence. Whereas the latter could be the result of an abortive gap repair upon mPifvine-3.1 excision, the former does not seem to be related to the transposition of the mPifvine-3.1 element. Regardless of the origin of the deletions, these events happened after mPifvine-3.1 insertion and can be used as new markers in genotyping that could provide useful information on the origin of grapevine varieties. For example, it is interesting to note that the locus 1123 presents an unusual short band in four of six American wild Vitis species and, although its presence in European wild V. vinifera ssp. sylvestris is rare (only one of nine genotypes has the band), it is found in a domesticated grapevine variety. Similarly, the unusual short band of locus 1284 is present in all but one European wild V. vinifera ssp. sylvestris, whereas most of the V. vinifera ssp. sativa genotypes that were analyzed do not contain this allele.
The frequent association of mPifvine-3.1 elements with 5′ and 3′ UTRs suggests that these elements should be frequently present within grapevine transcripts. We thus looked for the presence of mPifvine-3.1 sequences in the grapevine ESTs collections available at NCBI by Blast search. The matching ESTs were blasted back to the genome database, and the genomic region was manually checked to confirm that a given MITE was the source of the EST.
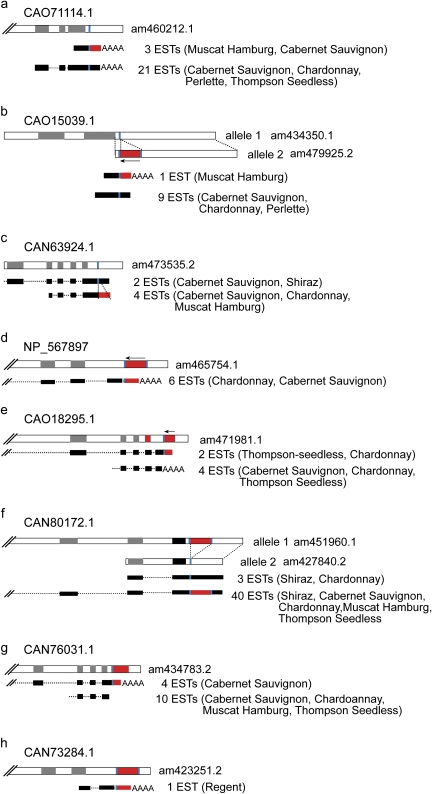
We found some 50 different ESTs that match to mPifvine-3.1 elements. In most cases, they correspond to insertions located within the predicted 3′ UTR of a gene. Interestingly, in some cases, different ESTs corresponding to the same gene are polymorphic with respect to the presence of the MITE (fig. 5). This can be the result of an insertion polymorphism among different cultivars (as the EST collections contain sequences obtained from different cultivars) or can be due to the existence of two alleles of the same gene in a particular cultivar. For example, two different alleles for a putative isoflavone reductase gene (CAN80172.1) were found in Pinot noir, one of which has an mPifvine-3 insertion at the 3′ end of the gene (fig. 5, f), suggesting that Pinot noir has two alleles of the gene, only one of them containing the mPifvine-3.1 insertion, and that both alleles are transcribed. Indeed, transcripts for both alleles were found in ESTs deriving from Shiraz and Chardonnay, but Cabernet Sauvignon, Muscat Hamburg, and Thompson Seedless only had ESTs with the mPifvine-3.1 insertion. Similarly, we found different ESTs corresponding to a gene coding for a putative saccharopine dehydrogenase (CAO15039.1) with or without an mPifvine-3.1 element inserted in the 3′ UTR.
FIG. 5.—
Presence of the m-Pifvine-3.1 MITEs (in red) in different genes and their transcripts. For each gene (designated by a letter from a to h), genomic sequence is given on the top (white boxes represent introns and gray boxes represent exons) and the corresponding ESTs are shown below as black boxes. The inserted m-Pifvine-3.1 is sown as a red box. The total number of ESTs found and their origin (cultivar name) are given on the right. All genes are 5′–3′ oriented. Arrows indicate cases where MITEs are in reverse orientation. Accession numbers of genomic sequences are given on the right and the accessions of predicted proteins corresponding to the genes are shown on top of each gene. Target sequences and TSDs are shown in blue, and the polyadenylation tails are shown by “AAAA.”
We have selected five mPifvine-3.1 insertions present in grapevine EST collections to further analyze their insertion polymorphisms among 25 different Vitis genotypes. All these insertions generated new transcription termination sites for the genes where they were inserted in, as deduced from the analysis of ESTs collections. One of these insertions (designated as “e” in figs. 4 and 5) is probably an old insertion that occurred prior to the split between European and American Vitis species and is almost fixed in the species analyzed (only one American species does not contain it). Still, the grapevine EST collections contain two different transcripts corresponding to the gene where this mPifvine-3.1 is inserted, one of them stopping just before the MITE sequence. This suggests that the insertion of the mPifvine-3.1 element provided the gene with an alternative transcriptional terminator and that this new allele was maintained and become fixed in the population during evolution. The second insertion analyzed, designated with a “d” in figures 4 and 5, is absent from all the American wild species analyzed, whereas it seems to be fixed in the European wild and domesticated species. We have only found ESTs terminating at the mPifvine-3.1 element, suggesting that the new termination site provided by the MITE substituted the terminator of the gene. The rest of the insertions analyzed are highly polymorphic among domesticated cultivars and Vitis species. Two of them (“b” and “c” in figs. 4 and 5) are also present in some European wild species suggesting that the insertion occurred before grapevine domestication, whereas the third (“a” in figs. 4 and 5) seems to be specific of the domesticated genotypes which is compatible with an insertion occurring after grapevine domestication. In all these cases, the grapevine EST collections contain two different transcripts corresponding to the two alleles found in grapevine genotypes, showing that the insertion of the mPifvine-3.1 element has generated transcript variability.
Discussion
Although the first MITEs described were related to the PIF/Pong and Mariner families, it has been shown later on that most class II families of TEs can generate MITEs. Here, we show that grapevine also contains MITEs related to most of class II TEs families present in this species. It has been proposed that MITEs are generated by a two-step process in which a subset of defective class II elements with special characteristics (e.g., small size) would be amplified to high copy numbers by a replicative-related, and still to be described, mechanism (Feschotte et al. 2002; Casacuberta and Santiago 2003). Such a mechanism implies that MITEs are amplified from typical class II elements, and thus, both types of elements should coexist in a particular genome. This is what has been found for the impala/mimp1 element of Fusarium oxysporum (Dufresne et al. 2007), but in other cases, such as that of the Arabidopsis and Medicago truncatula Emigrant/Lemi1 elements, MITEs and typical defective elements are restricted to different genomes (Guermonprez et al. 2008). The work presented here shows that defective elements and MITEs do coexist in grapevine and, more significantly, that grapevine also contains elements that could represent an intermediate type of defective elements. Indeed, seven of the eight families here described contain elements that, although being highly homogeneous in size and sequence, are relatively long and are present at moderate copy number. This new type of defective class II elements could be the result of an incomplete amplification due to suboptimal characteristics of the family founder element. In this respect, it is interesting to note the inverse relationship between the size of the elements and their copy number, suggesting that the size could be an important constraint for the high amplification of a defective element. The results presented here thus support a model for MITEs amplification from particular defective class II elements and point to a small size as one of the important characteristic for a defective element to become the founder of a new MITE family.
Although the elements described here were found because they show extensive sequence similarity with the related grapevine TEs families, some of the longest elements also contain internal sequences not related to them but to grapevine genomic sequences. Mobile genetic elements duplicate and mobilize cellular gene sequences, potentially contributing to creative mutagenic processes like exon shuffling and gene duplication. Cellular genes flanking the 3′ termini of retrotransposons can be duplicated by read-through transcription (Moran et al. 1999) and it has been shown that Helitron-mediated movement of cellular genes has massively changed the maize genome and caused a lack of gene colinearity between different maize inbred lines (Lai et al. 2005; Morgante et al. 2005). DNA transposons can also capture and mobilize genome sequences in a process that has been named transduplication (Jiang, Bao, et al. 2004; Kawasaki and Nitasaka 2004; Juretic et al. 2005; Zabala and Vodkin 2005, 2007; Hanada et al. 2009), and we have recently shown that this phenomenon also occurred in grapevine (Benjak et al. 2008). But, to our knowledge, the capacity of MITEs to transduplicate genomic sequences has not been reported to date. Transposons are usually present at low or moderate copy numbers, and with the exception of the Arabidopsis KI-MULE which is present in some 97 copies (Hoen et al. 2006), the transduplicated sequences are unique or are present at very low copy number. It has been proposed that transduplicated gene fragment may regulate paralogous gene expression through siRNA-related mechanisms or they may provide sequence reservoirs for gene conversion (Hoen et al. 2006). The capacity of MITEs to transduplicate genome sequences greatly increases the possibility of amplification and mobilization of transduplicated gene fragments and may have important implications for the evolution and regulation of the related genes. On the other hand, the capacity of MITEs that have transduplicated genomic sequences to continue to transpose and amplify suggests a mechanism to generate new MITE families with limited similarity to autonomous class II elements.
In addition to genic sequences, grapevine MITEs have also transduplicated a fragment of a previously uncharacterized transposon that we have named Mila. The amplification within a MITE of a transduplicated transposon fragment will increase the possibilities for a siRNA control of the transposon and may represent a new mechanism to control transposon activity.
One of the MITE families described here, mPifvine-3.1, has attained more than 1,000 copies in grapevine. Our results show that, although mPifvine-3.1 were already present in the ancestor of the wild Vitis species found in both Europe and America, they have transposed and amplified after their split accompanying grapevine domestication and breeding. In sharp contrast to the other related Pifvine-3 MITEs, mPifvine-3.1 elements seem to concentrate in the UTRs of grapevine genes, and especially in the 3′ UTR. Assuming that different MITE subfamilies sharing the same TIRs are mobilized by the same transposition machinery, the preferential distribution of mPifvine-3.1 within UTRs should be the result of selection rather than of a difference in insertion specificity. This preferential retention suggests a positive impact of mPifvine-3.1 element insertions, which could modify the mRNA fate in many ways, including its stability and processing or its degradation through posttranscriptional gene silencing mechanisms. A possible function for the mPifvine-3.1 insertions within gene UTRs could explain why old mPifvine-3.1 elements tend to be fixed in the population in spite of the high level of heterozygosity of grapevine species. It is interesting to note that, whereas older mPifvine-3.1 insertions are fixed in the population, the recent ones are highly polymorphic among cultivars. This polymorphism, which can be detected also at the transcriptional level, may be linked to phenotypic variability.
Although sexual crossing has been a major driver of grapevine evolution, its vegetative propagation enhanced the impact of somatic mutations and has been important for grapevine diversity. TEs are known to be the major contributors to genome variability and, in particular, to somatic mutations. Among them, MITEs seem particularly well suited to influence gene evolution. Their smaller size may allow MITEs to introduce more subtle changes in gene expression or regulation, and their high copy number makes their potential impact higher.
In addition to the fundamental interest for genome evolution studies, MITE insertions can be also used as molecular markers. The work presented here shows that mPifvine-3.1 “insertion/empty site” bands represent codominant alleles at a single locus that can be used for fingerprinting. The high copy number of mPifvine-3.1 elements as well as their frequent association to genes make them a very useful potential source for new markers to assist selection programs as well as for varietal and clone identification.
In summary, the work presented here shows that MITEs have contributed to gene evolution in grapevine, in particular during its domestication and breeding, by capturing and amplifying gene sequences as well as by inserting in a high number of grapevine genes. The work presented will allow the development of new molecular markers for grapevine selection and breeding, and the examples of MITE polymorphic insertions within genes reported here will allow in the future to experimentally test the direct impact of MITE insertions on gene expression.
Supplementary Material
Supplementary tables 1 and 2 and supplementary file are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by Ministerio de Educación y Ciencia [grant BFU2006-04005] and the Xarxa de Referència en Biotecnologia from the Generalitat de Catalunya [to J.M.C]; the Federal Ministry of Food, Agriculture and Consumer Protection of Germany through the Agency for Agriculture and Food [grants 04HS021/04HS022/04HS023 to A.F]; partially funded by an European Molecular Biology Organization Short Term Fellowship [ASTF 115-07 to A.B.].
Acknowledgments
We are grateful to Paloma Más and José-Luís Riechman for their critical reading of the manuscript. For providing the plant material, we thank Ernst Rühl (Institute of Grapevine Breeding Geisenheim, Germany), Bernd Hill (Staatliche Lehr- und Versuchsanstalt für Wein- und Obstbau Weinsberg, Germany), and Reinhard Antes (Nursery Antes, Heppenheim, Germany). For providing DNA material, we thank Prof. Andrew Walker and Summeira Riaz (Department of Viticulture and Enology, UC Davis), Patrice This (Institut National de la Recherche Agronomique, Montpellier, France), Jose-Miguel Martínez-Zapater and Rosa Arroyo-García (Instituto Nacional de Investigación Agraria, Madrid, Spain), and Dario Copetti (Applied Genomics Institute, University of Udine, Italy).
References
- Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arroyo-Garcia R, et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol Ecol. 2006;15:3707–3714. doi: 10.1111/j.1365-294X.2006.03049.x. [DOI] [PubMed] [Google Scholar]
- Benjak A, Forneck A, Casacuberta JM. Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PLoS ONE. 2008;3:e3107. doi: 10.1371/journal.pone.0003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Santiago N. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene. 2003;311:1–11. doi: 10.1016/s0378-1119(03)00557-2. [DOI] [PubMed] [Google Scholar]
- Collier L, Largaespada D. Transposable elements and the dynamic somatic genome. Genome Biol. 2007;8:S5. doi: 10.1186/gb-2007-8-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deragon J, Casacuberta JM, Panaud O. Plant transposable elements. In: Volff JN, editor. Genome Dyn. Basel: Karger; 2008. pp. 69–82. [DOI] [PubMed] [Google Scholar]
- Dufresne M, et al. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics. 2007;175:441–452. doi: 10.1534/genetics.106.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Mouches C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Ann Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneck A. Plant breeding: clonality—a concept for stability and variability during vegetative propagation. In: Esser ULK, Beyschlag W, Murata J, editors. Progress in botany. Berlin (Germany): Springer; 2005. pp. 165–183. [Google Scholar]
- Franks T, Botta R, Thomas MR, Franks J. Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet. 2002;104:192–199. doi: 10.1007/s001220100683. [DOI] [PubMed] [Google Scholar]
- Guermonprez H, Loot C, Casacuberta JM. Different strategies to persist: the pogo-like lemi1 transposon produces miniature inverted-repeat transposable elements or typical defective elements in different plant genomes. Genetics. 2008;180:83–92. doi: 10.1534/genetics.108.089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, et al. The functional role of pack-MULEs in rice inferred from purifying selection and expression profile. Plant Cell. 2009;21:25–38. doi: 10.1105/tpc.108.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen DR, et al. Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol. 2006;23:1254–1268. doi: 10.1093/molbev/msk015. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004a;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Jiang N, Feschotte C, Zhang X, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr Opin Plant Biol. 2004b;7:115–119. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Nitasaka E. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 2004;45:933–944. doi: 10.1093/pcp/pch109. [DOI] [PubMed] [Google Scholar]
- Kuang H, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Li Y, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit D, Pasquet S, Olive M, Thézé N, Thiébaud P. Glider and Vision: two new families of miniature inverted-repeat transposable elements in Xenopus Laevis genome. Genetica. 2000;108:163–169. doi: 10.1023/a:1004173315419. [DOI] [PubMed] [Google Scholar]
- Miskey C, et al. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol Cell Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Moreno-Vázquez S, Ning J, Meyers B. hATpin, a family of MITE-like hAT mobile elements conserved in diverse plant species that forms highly stable secondary structures. Plant Mol Biol. 2005;58:869–886. doi: 10.1007/s11103-005-8271-8. [DOI] [PubMed] [Google Scholar]
- Morgante M, et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Ohmori Y, Abiko M, Horibata A, Hirano H-Y. A transposon, Ping, is integrated into Intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant Cell Physiol. 2008;49:1176–1184. doi: 10.1093/pcp/pcn093. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Quesneville H, Nouaud D, Anxolabehere D. P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC Genomics. 2006;7:214. doi: 10.1186/1471-2164-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Yonemaru J, Ishikawa G, Nakamura T. A candidate autonomous version of the wheat MITE Hikkoshi is present in the rice genome. Mol Genet Genomics. 2005;273:404–414. doi: 10.1007/s00438-005-1144-7. [DOI] [PubMed] [Google Scholar]
- Santiago N, Herraiz C, Goni JR, Messeguer X, Casacuberta JM. Genome-wide analysis of the emigrant family of MITEs of Arabidopsis thaliana. Mol Biol Evol. 2002;19:2285–2293. doi: 10.1093/oxfordjournals.molbev.a004052. [DOI] [PubMed] [Google Scholar]
- This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–519. doi: 10.1016/j.tig.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Hall TC. MDM-1 and MDM-2: two mutator-derived MITE families in rice. J Mol Evol. 2003;56:255–264. doi: 10.1007/s00239-002-2397-y. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala G, Vodkin L. Novel exon combinations generatedby alternative splicing of gene fragments mobilized by a CACTA transposon in Glycine max. BMC Plant Biol. 2007;7:38. doi: 10.1186/1471-2229-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala G, Vodkin LO. The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell. 2005;17:2619–2632. doi: 10.1105/tpc.105.033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jiang N, Feschotte C, Wessler SR. PIF- and Pong-like transposable elements: distribution, evolution and relationship with Tourist-like miniature inverted-repeat transposable elements. Genetics. 2004;166:971–986. doi: 10.1093/genetics/166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]