Abstract
The deep phylogeny of eukaryotes is an important but extremely difficult problem of evolutionary biology. Five eukaryotic supergroups are relatively well established but the relationship between these supergroups remains elusive, and their divergence seems to best fit a “Big Bang” model. Attempts were made to root the tree of eukaryotes by using potential derived shared characters such as unique fusions of conserved genes. One popular model of eukaryotic evolution that emerged from this type of analysis is the unikont–bikont phylogeny: The unikont branch consists of Metazoa, Choanozoa, Fungi, and Amoebozoa, whereas bikonts include the rest of eukaryotes, namely, Plantae (green plants, Chlorophyta, and Rhodophyta), Chromalveolata, excavates, and Rhizaria. We reexamine the relationships between the eukaryotic supergroups using a genome-wide analysis of rare genomic changes (RGCs) associated with multiple, conserved amino acids (RGC_CAMs and RGC_CAs), to resolve trifurcations of major eukaryotic lineages. The results do not support the basal position of Chromalveolata with respect to Plantae and unikonts or the monophyly of the bikont group and appear to be best compatible with the monophyly of unikonts and Chromalveolata. Chromalveolata show a distinct, additional signal of affinity with Plantae, conceivably, owing to genes transferred from the secondary, red algal symbiont. Excavates are derived forms, with extremely long branches that complicate phylogenetic inference; nevertheless, the RGC analysis suggests that they are significantly more likely to cluster with the unikont–Chromalveolata assemblage than with the Plantae. Thus, the first split in eukaryotic evolution might lie between photosynthetic and nonphotosynthetic forms and so could have been triggered by the endosymbiosis between an ancestral unicellular eukaryote and a cyanobacterium that gave rise to the chloroplast.
Keywords: eukaryotic phylogeny, rare genomic changes, parsimony, substitutions, insertions, deletions
Introduction
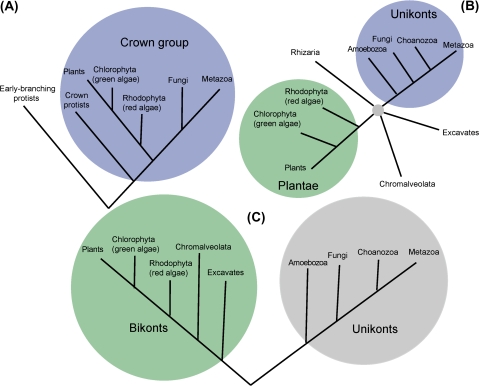
The deep phylogeny of eukaryotes is an extremely difficult and controversial problem. In the early days of molecular phylogeny, up to mid-1990s, the consensus appeared to be the crown-group phylogeny, that is, a tree that consisted of the crown including animals, fungi, plants, and some groups of unicellular eukaryotes (protists) and a number of “early branching” groups of protists (Sogin 1991; Sogin et al. 1993; Sogin and Silberman 1998). The crown-group phylogeny, in other words, the basal position of many, although not all, protist groups (fig. 1A), was supported by numerous phylogenetic analyses of rRNA as well as various conserved proteins. Even more importantly, the dominant evolutionary hypothesis at the time was the so-called archezoan scenario under which different amitochondrial protists (such as diplomonads or microsporidia) were thought to represent primitive eukaryotic forms, archezoa, one of which would become the host of the (proto)mitochondrial, α-proteobacterial endosymbiont (Cavalier-Smith 1993, 1998; Patterson 1999; Roger 1999).
FIG. 1.—
Competing topologies of the evolutionary tree of eukaryotes. (A) Crown-group topology (B) The Big Bang radiation of the five supergroups (C) The unikont–bikont topology. The trees are rendered in a simplified form, with only well-characterized groups for which complete genome sequences are available and that were included in the present analysis denoted explicitly. The branch lengths are arbitrary.
Subsequently, however, it was shown that all protists that were studied in sufficient detail carried organelles related to mitochondria (mitosomes, hydrogenosomes, and others) and possessed genes of apparent protomitochondrial (α-proteobacterial) descent (Dyall and Johnson 2000; Roger and Silberman 2002; Embley, van der Giezen, Horner, Dyal, Bell, and Foster 2003; Embley, van der Giezen, Horner, Dyal, and Foster 2003; van der Giezen and Tovar 2005; Embley and Martin 2006; Minge et al. 2008). Thus, the apparent indications from cell biology that protists lacking typical mitochondria were evolutionarily primitive were, effectively, invalidated. In parallel, the early branching of protists was repeatedly challenged once it became clear that many of these organisms, especially, parasites, evolve at a high rate, so that their basal position in trees could be a long-branch attraction artifact (Baldauf et al. 2000, 2003). Specifically, it was shown beyond reasonable doubt, by using phylogenetic methods that are relatively robust to long-branch effects, that microsporidia (one of the groups that appeared to best fit the definition of Archaezoa considering their simple cellular organization) are not a basal group, but rather, a highly derived, rapidly evolving sister group of fungi (Keeling and McFadden 1998; Keeling and Fast 2002; Fischer and Palmer 2005). Definitive phylogenetic affinities turned out to be hard to obtain for other former “archezoa,” in part, probably, owing to their rapid evolution. Nevertheless, the two major developments, the demonstration of the nonexistence of primitive amitochondrial forms among the rapidly increasing variety of well-characterized eukaryotes and of the unreliability of the basal position of protists together led to the effective collapse of the crown-group phylogeny of eukaryotes.
The concept of eukaryotic phylogeny that comes closest to being the current consensus maintains that there are five or, possibly, six distinct major branches, or supergroups, in the eukaryotic domain of cellular life, namely, unikonts (an assemblage that includes opishtokonts (Metazoa, Fungi, and related protists and Amoebozoa with the latter considered a distinct supergroup in some studies), Plantae, Chromalveolata, excavates, and Rhizaria (fig. 1B) (Adl et al. 2005; Keeling et al. 2005; Keeling 2007). The “higher” eukaryotes that comprise the core of the former crown group are thus split between two supergroups, unikonts (opisthokonts) and Plantae, whereas the remaining three supergroups consist of diverse protists. The monophyly of each of the supergroups is still questioned as exemplified by recent multigene phylogenetic analyses that employed broad taxonomic sampling and diverse methods (Philip et al. 2005; Parfrey et al. 2006; Yoon et al. 2008).
Regardless of the exact status and composition of each individual supergroup, it appears that several major branches of eukaryotes diverged in a “Big Bang”-type event, where the internal branches in the tree are extremely short, so much so that the “true” tree topology might be undecipherable (Philippe et al. 2000; Rokas et al. 2005; Rokas and Carroll 2006; Koonin 2007). Nevertheless, attempts have been made to root the tree of eukaryotes by using apparent derived shared characters (synapomorphies) along with phylogenies of highly conserved proteins. These studies led to the conclusion that the root lies between the opisthokonts (Metazoa, Choanozoa, and Fungi) and the bikonts (all groups of eukaryotes that ancestrally possess two cilia, namely, plants and most of the protists), with the position of the Amoebozoa remaining uncertain (Stechmann and Cavalier-Smith 2002) but leaning toward an affiliation with opisthokonts (Stechmann and Cavalier-Smith 2003a). The conclusion on the monophyly of the bikonts rests, primarily, on the fusion of a single pair of essential genes, those for dihydrofolate reductase (DHFR) and thymidylate synthase, purportedly, buttressed by the analysis of domain architectures and sequence-based phylogenies of some highly conserved proteins, such as myosins (Richards and Cavalier-Smith 2005).
Considering the crucial importance of the sequence of events at the earliest stages of eukaryotic evolution for understanding the emergence of the key biological features of the major groups of eukaryotes, the inference of the root position on the strength of only one or two characters; however, fundamental ones, seem unsatisfactory, given that parallel emergence of the purported derived character, such as a gene fusion, is difficult to rule out. Indeed, independent fusions of the same pairs of genes in diverse groups of eukaryotes as well as in eukaryotes and bacteria have been demonstrated in case studies (Yanai et al. 2002; Makiuchi et al. 2007). Furthermore, reversion of an ancestral fusion via the split of the fused genes in unikonts cannot be ruled out either.
We sought to reexamine the root position in the eukaryotic tree by means of a genome-wide analysis of rare genomic changes (RGCs). Lately, the analysis of RGCs that can be exemplified by diagnostic gene fusions, domain architectures of proteins, or features of genome architecture such as gene overlaps became an increasingly popular approach to the study of deep evolutionary relationship, given that these characters appear to be less prone to various artifacts than standard methods of molecular phylogeny (Rokas and Holland 2000; Iyer et al. 2004; Luo et al. 2006). Although it can be argued that RGC-based methods effectively employ parsimony and so would be prone to the same artifacts as maximum parsimony methods in sequenced-based phylogenetic analysis, this would not be the case if the RGCs were free of homoplasy (parallel changes and reversals), which is the primary problem for the maximum parsimony methods. Conceivably, if the analyzed changes are indeed rare and their number is sufficiently large, the effect of homoplasy would be minimized. It should be noticed that molecular phylogeny methods that employ sophisticated models of sequence evolution, usually within the maximum likelihood framework, are not without their own serious problems that are related, mostly, to model overspecification and misspecification (proverbial attempts to “fit an elephant”) (Kolaczkowski and Thornton 2004; Steel 2005; Thornton and Kolaczkowski 2005; Stefankovic and Vigoda 2007). Application of sequence-based phylogenetic methods within the phylogenomic approach not only has the potential to substantially increase the resolution power but also poses challenges owing to horizontal gene transfer as well as different optimal models of evolution for different genes (Phillips et al. 2004; Bucknam et al. 2006; Dagan and Martin 2006; Bapteste et al. 2008). The pitfalls that are inherent in even the most advanced maximum likelihood and Bayesian methods, in particular, in the phylogenomic setting, stimulate the search for RGCs that are most suitable for phylogenetic analysis.
Recently, we introduced a new class of RGCs designated RGC_CAMs (after conserved amino acids-multiple substitutions), which are inferred from genome-scale analysis of alignments of orthologous proteins and underlying nucleotide sequence alignments (Rogozin et al. 2007a, 2007b). The RGC_CAM approach utilizes amino acid residues that are conserved through long evolutionary spans and in major organismal lineages, with the exception of a few taxa that together comprise a candidate clade. In order to minimize homoplasy, only those amino acid replacements that require 2 or 3 nt substitutions are employed for phylogenetic inference. The RGC_CAM method, combined with a procedure for rigorous statistical testing of competing phylogenetic affinities, is specifically designed for testing (rejecting) evolutionary hypotheses that are presented as unresolved trifurcations of clades. A direct estimation of the level of homoplasy among RGC_CAMs revealed a nonnegligible number of parallel changes but nevertheless showed that the method is robust for a wide range of phylogenetic problems (Rogozin et al. 2008).
We were interested in applying the RGC_CAM approach to the relationship between the eukaryotic supergroups, a fundamental problem with an obvious bearing on the rooting of the evolutionary tree of eukaryotes. The problem with using RG_CAMs for resolving such deep evolutionary relationships is that the number of characters that support a particular clade can be quite small. Therefore, we additionally employed a relaxed version of the RGC_CAMs denoted RGC_CAs where the requirement for multiple substitutions is lifted, of course, at the price of increased homoplasy (Rogozin et al. 2008). The combined results of these RGC analyses seem to, effectively, refute the bikont–unikont split as the first bifurcation in the evolution of eukaryotes and instead suggest the affiliation of the major protist groups with the animal–fungi (opisthokont) clade. This result is compatible with the scenario where the acquisition of the cyanobacterial symbiont (the future chloroplast) by an ancestor of Plantae triggered the first divergence of major clades in the evolution of eukaryotes.
Materials and Methods
Amino Acid Alignments
Each of the 716 protein alignments (488,157 sites altogether) constructed from a previously delineated set of highly conserved clusters of eukaryotic orthologous genes or eukaryotic orthologous groups (KOGs) (Koonin et al. 2004) analyzed here included orthologs from eight eukaryotic species with completely sequenced genomes: Homo sapiens (Hs), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Arabidopsis thaliana (At), Anopheles gambiae (Ag), and Plasmodium falciparum (Pf) (Rogozin et al. 2003). To these KOGs, probable orthologs from 66 prokaryotic genomes from the COG database (Tatusov et al. 2003) were added using a modification of the COGNITOR method (Tatusov et al. 1997). Briefly, all protein sequences from the prokaryotic genomes are compared with the protein sequences previously included in the KOGs; a protein is assigned to a KOG when two genome-specific best hits to members of the given KOG are detected. We added five prokaryotic orthologs (denoted P1, P2, P3, P4, and P5) to each KOG and required these prokaryotic orthologs to belong to three or more major prokaryotic clades (see supplementary table S1, Supplementary Material online) (Basu, Rogozin, and Koonin 2008). The requirement for the availability of five diverse prokaryotic orthologs was satisfied for 396 of the initially selected 716 KOGs. To the resulting mixed COG/KOGs, probable orthologs from 25 other eukaryotic genomes, namely, those of Oryza sativa (Os), Physcomitrella patens (Ppat), Chlamydomonas reinhardtii (Crei), Ostreococcus lucimarinus (Oluc), Volvox carteri (Vcar), Monosiga brevicollis (Mb), Dictyostelium discoideum (Ddis), Entamoeba histolytica (Ehis), Giardia lamblia (Glam), Leishmania braziliensis (Lbra), Leishmania infantum (Linf), Leishmania major (Lmaj), Trypanosoma brucei (Tbru), Trypanosoma cruzi (Tcru), Babesia bovis (Bbov), Cryptosporidium hominis (Chom), Cryptosporidium parvum (Cpar), Phaeodactylum tricornutum (PhTri), Phytophthora infestans (Pinf), Phytophthora ramorum (Pram), Phytophthora sojae (Psoj), Paramecium tetraurelia (Ptet), Tetrahymena thermophila (Tthe), Theileria parva (Tpar), and Trichomonas vaginalis (Tvag) were added using COGNITOR. Amino acid sequence alignments are available at the authors’ Web site at ftp://ftp.ncbi.nlm.nih.gov/pub/koonin/RGC_CAM/eukaryotic_evolution/. To minimize misalignment problems, only conserved, unambiguously aligned regions of the alignments were subject to further analysis. Specifically, we only analyzed positions surrounded by segments of protein alignments containing no insertions or deletions with a 5-amino acid window from each side.
Rare Genomic Changes
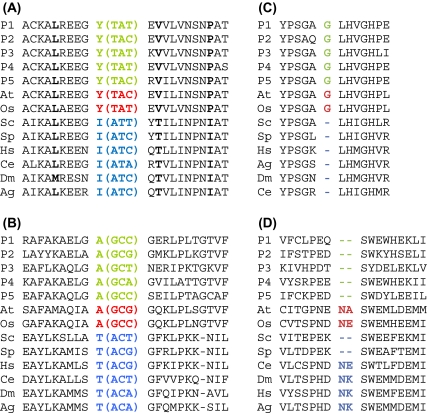
For the purpose of phylogenetic analysis using the RGC_CAM method (Rogozin et al. 2007b), we analyzed amino acid residues that are conserved in most of the included eukaryotes, with the exception of a few species and the prokaryotic outgroups. The assumption is that any character shared by the included five diverse prokaryotic outgroup species and the majority of eukaryotes is the ancestral state, whereas the deviating species possess a derived state (fig. 2A). To reduce the level of homoplasy, only amino acid replacements that require 2 or 3 nt substitutions (Rogozin et al. 2007b). Given the rarity of multiple substitutions, these double replacements are plausible RCGs (RGC_CAMs). To simplify further presentation, we use the following notation: S1 ≠ S2 = S3 means that, for a conserved amino acid position in an alignment, species S2 and S3 share the same amino acid that is different from the amino acid in the species S1. Under this notation, for example, a plasmodium-specific RGC_CAM is denoted by Pf ≠ At = Os = Sc = Sp = Hs = Dm = Ag = Ce = P1 = P2 = P3 = P4 = P5, whereas an RGC_CAM shared by the fungi and animals is denoted by Sc = Sp = Hs = Dm = Ag = Ce ≠ Pf = At = Os = P1 = P2 = P3 = P4 = P5.
FIG. 2.—
Examples of the RGCs used in this work (A) RGC_CAM: KOG0370 (B) RGC_CA: KOG0370 (C) RGC_DELL: KOG0435 (D) RGC_INS: KOG2509. For RGC_CAM (A) and RGC_CA (B), the corresponding codons extracted from the underlying nucleotide sequence alignments are shown in parentheses. The RGC positions are shown in green (five prokaryotic species used as the outgroup), red (plants), and blue (fungi, animals). H. sapiens (Hs), A. gambiae (Ag), C. elegans (Ce), D. melanogaster (Dm), S. cerevisiae (Sc), S. pombe (Sp), A. thaliana (At), O. sativa (Os), and five outgroup prokaryotic species (P1–P5).
First, we estimated the branch length for each analyzed taxon in RGC_CAM units (fig. 3). For each species or group of species, we calculated the number of amino acid residues that are different from all other species (e.g., Sc = Sp ≠ At = Os = Dm = Ag = Hs = Ce = P1 = P2 = P3 = P4 = P5 for fungi).
FIG. 3.—
The analyzed trifurcations of major eukaryotic lineages. For each analyzed trifurcation, the lengths of branches in the number of RGC_CAMs are indicated. Balanced trifurcation indicates that all three analyzed branches are of approximately equal lengths (the lengths are not significantly different as suggested by the χ2 test with 2 degrees of freedom); otherwise, that is, when there is a statistically significant difference in branch lengths, a trifurcation is considered to be unbalanced.
The next step of the RGC_CAM analysis is statistical testing of phylogenetic hypotheses. We developed a test designed to resolve ambiguous phylogenetic relationships by analyzing all possible evolutionary scenarios for three lineages. In this test, the number of RGC_CAMs shared by two lineages (e.g., Sc = Sp = Hs = Dm = Ag = Ce ≠ Pf = At = Os = P1 = P2 = P3 = P4 = P5; fungi and animals—these shared RGC_CAMs are consistent with the accepted phylogeny) is used as a variable. The values of this variable for two compared alternative topologies, along with the respective branch lengths (excluding the branch that is common to both alternatives), are put in a 2 × 2 contingency table. The test is based on a null model under which, in a comparison of two alternative hypotheses, for example, H1 = ((X − Y),Z) versus H2 = ((X − Z),Y), the number of RGC_CAMs that are shared by two lineages due to chance (NXY and NXZ) is proportional to the length of the branch, the position of which differs between the compared hypotheses, that is, Y and Z, respectively, in the above example. Specifically, we examined all three pairwise comparisons for each analyzed trifurcation, that is, hypothesis H1 = ((X − Y),Z) versus hypothesis H2 = ((X − Z),Y); H1 = ((X − Y),Z) versus H3 = ((Y − Z),X); and H2 = ((X − Z),Y) versus H3 = ((Y − Z),X), using the right-tail Fisher's exact test. In this work, P12, P23, and P13 denote the P values associated with the comparison of the respective hypotheses. It should be emphasized that all numbers in the contingency tables are independent, that is, each RGC_CAM is counted only once (Rogozin et al. 2007b).
The same approach was employed for analyses of a relaxed version of RGC_CAMs by allowing all possible amino acid replacements (as opposed to only those that require 2 or 3 nt substations in RGC_CAMs). We denote these characters RGC_CAs (fig. 2B). In addition, we analyzed deletions (RGC_DEL, fig. 2C) and insertions (RGC_INS, fig. 2D) surrounded by conserved fragments of protein alignments.
Results
Rare Genomic Changes Employed in This Analysis
Four classes of RGCs were employed in this work (see Materials and Methods for details).
RGC_CAMs. In the context of the present work, we used this method to analyze amino acid residues that are conserved in the majority of the included eukaryotes and five prokaryotes comprising the outgroup (for the list of employed prokaryotic species; see supplementary table S1, Supplementary Material online), with the exception of several eukaryotic species. The underlying assumption is that any character shared by the majority of eukaryotes and five diverse prokaryotic species is the ancestral state, whereas the deviating species possess a derived state (fig. 2A). In order to reduce the level of homoplasy, that is, the same amino acid replacements in different lineages that do not reflect common ancestry but rather represent parallel, reverse, or convergent changes (Telford and Budd 2003), the RGC_CAM method analyzes only those amino acid replacements that require two or three nucleotide substitutions (fig. 2A). Because multiple, adjacent nucleotide substitutions are rare, the level of homoplasy, in this case, is much lower than it is for amino acid changes caused by single nucleotide substitutions (Averof et al. 2000; Silva and Kondrashov 2002; Kondrashov 2003).
RGC_CAs. The same as RGC_CAMs but without the requirement for multiple nucleotide substitutions (fig. 2B). This relaxation of the requirements to the analyzed characters leads not only to a substantial increase in the number of available characters but also, inevitably, to increased homoplasy.
RGC_DELs. Deletions flanked by conserved regions of protein alignments (fig. 2C).
RGC_INSs. Insertions flanked by conserved regions of protein alignments (fig. 2D).
Reality Checks: The Plants-Animals-Fungi Trifurcation and the Animal–Choanoflagellate Clade
We first applied the RGC_CAM approach to a well-characterized case of ancient divergence of major eukaryotic lineages, namely, plants, animals, and fungi. Numerous molecular phylogenetic studies indicate that animals and fungi form a clade to the exclusion of plants (Baldauf 1999), so the existence of that clade (opisthokonts) is not seriously contested (Parfrey et al. 2006; Yoon et al. 2008).
In this case, the analyzed branches are of approximately equal lengths, that is, form a balanced tree (table 1 and fig. 3A), a situation in which the RGC analyses are most reliable (Rogozin et al. 2008). The raw number of shared RGC_CAMs was by far the greatest for the animal–fungi clade, and this excess was highly statistically significant for all combinations of plant species included in the analysis (table 1). The statistical test yielded significant P values both for the basal position of plants, that is, the animal–fungi clade (P13 and P23, table 1) and for the basal position of fungi that implies the plants–animals clade (P12, table 1). However, the support for the animals–fungi clade in most cases was much stronger (P13 and P23 < 0.0001, table 1) compared with the support for the plants–animals clade (e.g., P12 = 0.013 for the first test in the table 1). The RGC_CAs yielded qualitatively similar results, with an even stronger statistical significance owing to the larger number of characters (table 1). The raw numbers of shared RGC_DEL and RGC_INS also were the largest for the animal–fungi clade (table 1). However, there were few unique insertions and deletions, and the relative level of homoplasy appeared to be much higher compared with RGC_CAMs and RGC_CAs, so that neither hypothesis received significant statistical support (table 1).
Table 1.
Analysis of the Trifurcation Plants (P)–Animals (A)–Fungi (F)
| RGC | Hypothesis |
Branch Length |
Relative Probabilities of Hypotheses |
|||||||
| P + A | P + F | A + F | P | A | F | Stem | P12 | P13 | P23 | |
| H1 | H2 | H3 | ||||||||
| A. thaliana and O. sativa | ||||||||||
| CAM | 12 | 4 | 45 | 45 | 25 | 37 | 211 | 0.013 (H1) | <0.001 (H3) | <0.001 (H3) |
| CA | 50 | 23 | 151 | 193 | 106 | 138 | 696 | <0.001 (H1) | <0.001 (H3) | <0.001 (H3) |
| DEL | 4 | 4 | 8 | 8 | 14 | 6 | 53 | 0.283 | 0.206 | 0.828 |
| INS | 3 | 2 | 4 | 10 | 10 | 21 | 59 | 0.238 | 0.451 | 0.404 |
| A. thaliana and P. patens | ||||||||||
| CAM | 6 | 3 | 52 | 45 | 25 | 33 | 225 | 0.168 | <0.001 (H3) | <0.001 (H3) |
| CA | 38 | 20 | 156 | 173 | 107 | 137 | 718 | 0.002 (H1) | <0.001 (H3) | <0.001 (H3) |
| DEL | 3 | 4 | 10 | 8 | 15 | 3 | 65 | 0.066 | 0.0207 (H3) | 0.757 |
| INS | 2 | 3 | 4 | 10 | 7 | 20 | 69 | 0.437 | 0.930 | 0.395 |
| A. thaliana and C. reinhardtii | ||||||||||
| CAM | 12 | 3 | 36 | 24 | 24 | 29 | 175 | 0.016 (H1) | 0.027 (H3) | <0.001 (H3) |
| CA | 46 | 20 | 118 | 97 | 86 | 109 | 593 | <0.001 (H1) | <0.001 (H3) | <0.001 (H3) |
| DEL | 3 | 2 | 4 | 6 | 9 | 4 | 44 | 0.907 | 0.419 | 0.885 |
| INS | 2 | 0 | 2 | 5 | 5 | 14 | 53 | 0.100 | 0.351 | 0.318 |
| A. thaliana and O. lucimarinus | ||||||||||
| CAM | 5 | 3 | 38 | 30 | 19 | 30 | 203 | 0.190 | <0.001(H3) | <0.001(H3) |
| CA | 32 | 21 | 121 | 104 | 88 | 123 | 660 | 0.011 (H1) | <0.001 (H3) | <0.001 (H3) |
| DEL | 3 | 3 | 4 | 4 | 12 | 5 | 52 | 0.333 | 0.928 | 0.350 |
| INS | 2 | 2 | 4 | 6 | 6 | 20 | 55 | 0.283 | 0.476 | 0.436 |
| A. thaliana and V. carteri | ||||||||||
| CAM | 9 | 3 | 41 | 26 | 25 | 31 | 191 | 0.054 | 0.002 (H3) | <0.001 (H3) |
| CA | 45 | 21 | 132 | 97 | 93 | 122 | 635 | <0.001 (H1) | <0.001 (H3) | <0.001 (H3) |
| DEL | 5 | 1 | 6 | 5 | 11 | 3 | 50 | 0.939 | 0.394 | 0.382 |
| INS | 2 | 0 | 3 | 9 | 5 | 17 | 56 | 0.076 | 0.908 | 0.082 |
| All unicellular plants | ||||||||||
| CAM | 3 | 4 | 30 | 30 | 19 | 27 | 145 | 0.877 | <0.001 (H3) | <0.001 (H3) |
| CA | 28 | 21 | 96 | 86 | 75 | 100 | 476 | 0.053 | <0.001 (H3) | <0.001 (H3) |
| DEL | 2 | 3 | 3 | 5 | 9 | 3 | 39 | 0.205 | 0.412 | 0.455 |
| INS | 2 | 0 | 1 | 3 | 3 | 10 | 36 | 0.095 | 0.214 | 0.999 |
| All plants | ||||||||||
| CAM | 3 | 3 | 24 | 17 | 14 | 24 | 126 | 0.425 | 0.006 (H3) | <0.001 (H3) |
| CA | 23 | 14 | 79 | 47 | 61 | 86 | 429 | 0.019 (H1) | 0.023 (H3) | <0.001 (H3) |
| DEL | 2 | 3 | 3 | 4 | 9 | 3 | 37 | 0.205 | 0.878 | 0.378 |
| INS | 2 | 0 | 1 | 3 | 3 | 10 | 34 | 0.095 | 0.214 | 0.999 |
NOTE.—The results are given for different combinations of plant species. H1, H2, and H3 denote the three possible phylogenetic hypotheses regarding the resolution of the given trifurcation. P12, P23, and P13 denote the P values associated with the comparison of the respective hypotheses (see Materials and Methods for details). (H1) and (H2) denote the polarity of the comparison; for instance, (H1) after a P12 value indicates that, in the given comparison, H1 is significantly more likely than H2, conversely, (H2) indicates that H2 is significantly more likely than H1.
Thus, the results of this analysis of a well-established deep evolutionary relationship between major groups of eukaryotes confirm that RGC_CAMs and RGC_CAs are, in general, reliable indicators of phylogenetic affinity. Somewhat unexpectedly, we found that these characters were much more informative than indels which are more traditional markers used for deep phylogenetic analysis. Given this observation, we employed only RGC_CAMs and RGC_CAs for all analyses of uncertain phylogenetic relationships between eukaryotes.
Choanoflagellates are a group of unicellular eukaryotes that show a marked similarity to the choanocytes (feeding cells) of sponges, an observation suggesting the possibility that this group of protists, along with several apparently related groups, includes the closest living relatives of metazoans. This hypothesis is supported both by several phylogenetic analyses (Cavalier-Smith and Chao 2003; Rokas et al. 2005; Steenkamp et al. 2006) and by the analysis of the first sequenced genome of a choanoflagellate, M. brevicollis, which is remarkably complex and encodes orthologs of many signature animal proteins (King et al. 2008). We analyzed the trifurcation M. brevicollis–animals–fungi using RGC_CAMs and RGC_CAs (fig. 3B and supplementary table S2, Supplementary Material online). Clear support for the M. brevicollis–animals clade was obtained from both statistical tests and raw numbers of RGCs (supplementary table S2, Supplementary Material online). In this case, the relatively long M. brevicollis branch (unbalanced tree) did not cause problems for the RGC_CAM and RGC_CA analyses (fig. 3B and supplementary table S2, Supplementary Material online), possibly, owing to the relatively short stem branch (the branch that leads from the outgroup to the analyzed trifurcation; fig. 3B), which minimizes artifacts caused by reversals (Irimia et al. 2007; Rogozin et al. 2008).
Dictyostelium discoideum and E. histolytica: Testing the Monophyly of Amoebozoa and their Relationship with Opisthokonts
We applied the RGC_CAM approach to a well-known case of problematic phylogeny, namely, the evolutionary positions of the slime mold D. discoideum (member of the phylum Mycetozoa or social amoebae) and E. histolytica, member of the phylum Archamoebae. Several phylogenetic analyses suggested that these distantly related amoebae formed a clade that is a sister group to the opisthokont clade although the split between the two lineages of Amoebozoa is deep and is thought to have occurred shortly after the divergence of Amoebozoa from the opisthokonts (Bapteste et al. 2002; Song et al. 2005). We analyzed the trifurcation D. discoideum–plants–opisthokont (fig. 3C) using 278 KOG alignments (supplementary table S3, Supplementary Material online). A clear support for the D. discoideum–opisthokont clade was obtained from both the raw numbers of RGCs and statistical tests (supplementary table S3, Supplementary Material online). The analysis of the E. histolytica–plants–opisthokont trifurcation did not reveal such a clear picture, probably, due to the substantial decrease in the number of analyzed genes compared with the case of D. discoideum (only 191 KOGs) and the extremely long E. histolytica branch (fig. 3D and supplementary table S3, Supplementary Material online) which could result in an excess of parallel changes and reversals (Rogozin et al. 2008). Nevertheless, despite some ambiguity in the results, both the raw numbers and the statistical tests tend to support the E. histolytica–opisthokont clade (supplementary table S4, Supplementary Material online). Thus, the results of RGC analyses with both available genome of amoebas were consistent with the monophyly of Amoebozoa and opisthokonts (together comprising the unikont supergroup), in agreement with some phylogenetic tree analyses (Baldauf et al. 2000; Stechmann and Cavalier-Smith 2003b) but not others (Parfrey et al. 2006; Yoon et al. 2008).
Given the distant and uncertain relationship between the two amoebas themselves, we examined the trifurcation opisthokonts–D. discoideum–E. histolytica (fig. 3E). The raw number of shared RGC_CAMs was the largest for the D. discoideum–E. histolytica clade (table 2). The interpretation of this result requires caution because the E. histolytica branch was extremely long, so that the resulting unbalanced tree might contain an excessive number of parallel changes and reversals (Rogozin et al. 2008). However, reversals cannot have a substantial effect because of the extremely short stem branch (Irimia et al. 2007; Rogozin et al. 2007a) (table 2), whereas the effect of parallel changes is taken into account by the employed statistical test. The results of this test indicate that the most likely tree topology is ((D. discoideum + Metazoa/Fungi) E. histolytica), that is, an Opisthokonta–Mycetozoa clade, to the exclusion of E. histolytica (Archamoebae) (table 2). We employed three different settings for this analysis whereby either animals together with fungi, or four animals, or two fungi were chosen to represent the opisthokont clade, and the results were similar for all three experiments (table 2). Thus, the RGC_CAM and RGC_CA analyses suggest that D. discoideum forms a clade with the opisthokonts, to the exclusion of E. histolytica, that is, the two amoebas, according to these results, represent distinct clades within the unikont supergroup. This conclusion contradicts the results of some of the previous phylogenetic studies (Bapteste et al. 2002; Song et al. 2005) but is compatible with the topology of the trees obtained by the analysis of domain compositions of multidomain proteins (Basu et al. 2008). It seems possible that the apparent monophyly of Mycetozoa and Archamoebae that was observed in phylogenetic analyses is a long-branch attraction artifact.
Table 2.
Phylogeny of Amoebozoa: Analysis of the Trifurcation D. discoideum (DDIS)–E. histolytica (EHIS)–Opisthokonta (Animals–Fungi, AF)
| RGC | Hypothesis |
Branch Length |
Relative Probabilities of Hypotheses |
|||||||
| DDIS + EHIS | DDIS + AF | EHIS + AF | DDIS | EHIS | AF | Stem | P12 | P13 | P23 | |
| H1 | H2 | H3 | ||||||||
| Animals and fungi | ||||||||||
| CAM | 7 | 2 | 1 | 18 | 130 | 1 | 2 | 0.010 (H2) | 0.512 | 0.046 (H2) |
| CA | 26 | 9 | 8 | 84 | 393 | 6 | 14 | <0.001 (H2) | 0.012 (H3) | 0.001 (H2) |
| Animals | ||||||||||
| CAM | 12 | 3 | 1 | 24 | 157 | 13 | 3 | 0.126 | 0.055 | 0.010 (H2) |
| CA | 44 | 17 | 10 | 111 | 480 | 52 | 19 | <0.001 (H2) | 0.040 (H1) | <0.001 (H2) |
| Fungi | ||||||||||
| CAM | 9 | 4 | 3 | 30 | 160 | 17 | 3 | 0.041 (H2) | 0.570 | 0.017 (H2) |
| CA | 36 | 20 | 21 | 147 | 508 | 69 | 33 | <0.001 (H2) | 0.291 | <0.001 (H2) |
NOTE.—The results are given for different combinations of animal and fungal species. H1, H2, and H3 denote the three possible phylogenetic hypotheses regarding the resolution of the given trifurcation. P12, P23, and P13 denote the P values associated with the comparison of the respective hypotheses (see Materials and Methods for details). (H1) and (H2) denote the polarity of the comparison; for instance, (H1) after a P12 value indicates that, in the given comparison, H1 is significantly more likely than H2, conversely, (H2) indicates that H2 is significantly more likely than H1.
The Phylogenetic Position of the Chromalveolata
The Chromalveolata is an assemblage of numerous, diverse groups of protists that was proposed as a monophyletic supergroup by Cavalier-Smith as a refinement of the previously described kingdom Chromista (Cavalier-Smith 2002). The monophyly of Chromalveolata is not considered to be unequivocally established but is supported by several phylogenetic analyses (Baldauf et al. 2000; Harper et al. 2005). Most of the chromalveolates possess a chloroplast-related organelle (such as the apicoplast of the Apicomplexa) that is surrounded by a complex, multilayer membrane. Accordingly, it has been proposed that Chromalveolata is an ancient bikont branch that evolved via a secondary endosymbiosis with a red alga (Archibald and Keeling 2002; Cavalier-Smith 2003; Lane and Archibald 2008).
Taking advantage of the large number of sequenced genomes from diverse chromalveolates, we performed a detailed analysis of the relationship between Chromalveolata, Plantae, and opisthokonts (fig. 3F). The raw number of shared RGC_CAMs in the majority of comparisons (68 cases) was the greatest for the Chromalveolata–animals/fungi clade (supplementary table S5, Supplementary Material online), and overall, this clade received the strongest statistical support (table 3). However, in 20 comparisons, the raw number of shared RGC_CAMs was the greatest for the Chromalveolata–Plantae clade (supplementary table S5, Supplementary Material online), and there was some, albeit weaker, statistical support for this clade (table 3). The third topology, with the basal position of the Chromalveolates and a Plantae–opisthokont clade was poorly supported (table 3 and supplementary table S5, Supplementary Material online) and could be effectively ruled out.
Table 3.
Phylogenetic Position of Chromalveolata: Analysis of the Trifurcation Chromalveolates (CHR)–Plants (PLAN)–Opisthokonta (Animals–Fungi, AF)
| RGC | The Number of Tests Supporting a Hypothesis |
|||||
| CHR + PLAN versus CHR + AF |
CHR + PLAN versus PLAN + AF |
CHR + AF versus PLAN + AF |
||||
| > | < | > | < | > | < | |
| A. thaliana and O. sativa | ||||||
| CAM | 1 | 8 | 1 | 0 | 10 | 1 |
| CA | 0 | 20 | 11 | 7 | 14 | 2 |
| A. thaliana and P. patens | ||||||
| CAM | 0 | 7 | 6 | 0 | 12 | 0 |
| CA | 1 | 20 | 9 | 11 | 13 | 2 |
| Unicellular Plantae | ||||||
| CAM | 9 | 5 | 3 | 0 | 11 | 0 |
| CA | 5 | 8 | 10 | 8 | 14 | 4 |
| All plants | ||||||
| CAM | 10 | 0 | 1 | 8 | 2 | 0 |
| CA | 14 | 5 | 11 | 6 | 10 | 14 |
NOTE.—The results are presented for the indicated combinations of plant species. The signs ‘>’ and ‘<’ denote the polarity of the comparison; for instance, ‘>’ below CHR + PLAN indicates that, in the given comparison, the hypothesis that Chromalveolates and plants are sister taxa is significantly more likely than the hypothesis that Chromalveolates and opisthokonts are sister taxa, conversely, ‘<’ indicates that the second hypothesis is significantly more likely than the first hypothesis.
The raw numbers of shared RGCs can be an useful addition to the statistical test (see the analysis of the plants–animals–fungi trifurcation above). However, the utility of raw numbers is hampered by large differences in branch lengths (Rogozin et al. 2008). To minimize this effect, we compared the numbers of RGC_CA(M)s that supported the Chromalveolata–opisthokonts clade or the Chromalveolata–Plantae clade for cases where the branches leading to opisthokonts and plants were of approximately equal lengths (table 4). In the substantial majority of tests, the number of RGC_CA(M)s supporting the Chromalveolata–opisthokonts clade was greater than that supporting the affiliation of chromalveolates with plants (table 4).
Table 4.
Support of the Affiliation of Protist Taxa with Opisthokonta (Animals–Fungi) or with Plantae from the Comparison of Raw Numbers of RGC_CA(M)s
| Clade/RGC | Protists–Opisthokonta | Protists–Plants | Pbinom |
| Number of tests in support |
|||
| Chromalveolates | |||
| RGC_CAM | 25 | 15 | |
| RGC_CA | 30 | 17 | |
| Total | 55 | 32 | 0.009 |
| Kinetoplastids | |||
| RGC_CAM | 9 | 0 | |
| RGC_CA | 21 | 0 | |
| Total | 30 | 0 | <0.001 |
| T. vaginalis | |||
| RGC_CAM | 4 | 0 | |
| RGC_CA | 7 | 0 | |
| Total | 11 | 0 | <0.001 |
| G. lamblia | |||
| RGC_CAM | 3 | 4 | |
| RGC_CA | 8 | 1 | |
| Total | 11 | 5 | 0.105 |
NOTE.—The tests involved different combinations of plant or opisthokont species as shown in table 3. Only tests with approximately equal lengths (±5 for RGC_CAMs and ±15 for RGA_CAs) of plant and animal-fungi branches were taken into account.
In this analysis, many comparisons failed to produce a statistically significant outcome (supplementary table S5, Supplementary Material online). Moreover, some Chromalveolate species, such as C. hominis and P. infestans, but not others, possess multiple RGCs supporting the monophyly of Chromalveolates and plants (supplementary table S5, Supplementary Material online). These observations might indicate that Chromalveolates have a genuine mixed heritage, with the majority of the genes sharing common ancestry with orthologs from opisthokonts but some genes being of plant origin. To test this hypothesis, we examined the affinities of multiple RGC_CAs (RGC_CAMs were not conducive to this type of analysis because there were too few genes with multiple RGC_CAMs) within the same gene, under the reasoning that, if the apparent mixed phylogenetic signal is indeed due to distinct origins of different genes of Chromalveolates and not to noise, all RGC_CAs from the same gene should point in the same direction. Altogether, 21 KOGs contained two or more RGC_CAs, and in each case, multiple RGC_CAs within the same gene supported either the Chromalveolata–Opisthokonta clade or the Chromalveolata–Plantae clade, with the sole exception of KOG100 (supplementary table S6, Supplementary Material online). A striking example is KOG2446 (Glucose-6-phosphate isomerase) that carries up to 12 RGC_CAs (depending on the combination of species) all of which support the Chromalveolata–Plantae clade. Although apparently affected by homoplasy, their results indicate that the gene complement of Chromalveolata indeed could be heterogeneous, with the majority of the genes sharing a common ancestry with orthologs from opisthokonts but some genes derived from Plantae. The presence of multiple genes of apparent red algal origin in genomes of chromalveolates has been reported (Li et al. 2006). Taken together, these findings are compatible with the scenario under which the common ancestor of the Chromalveolata emerged as a result of engulfment of a red alga by a unikont host.
The Phylogenetic Position of Excavate Taxa: Diplomonads (Giardia), Kinetoplastids, and Parabasalia (Trichomonas)
The excavates comprise a vast assemblage of diverse organisms some of which, in particular, diplomonads and parabasalids, lack typical mitochondria and accordingly were long considered “primitive” forms and promising candidates for the archezoan status (Roger 1999; Simpson 2003). Although the discovery of mitochondria-related organelles and genes of apparent mitochondrial origin invalidates the hypothesis that some of the excavates are primary amitochondrial forms, the possibility that they are “basal” eukaryotes remains attractive given that some of these organisms are among the eukaryotic forms with the simplest cellular and genomic organization. Among the 5 eukaryotic supergroups, the monophyly of excavates is, probably, most dubious, and the phylogenetic position of many excavate taxa remains uncertain (Simpson, Inagaki, and Roger 2006; Rodriguez-Ezpeleta et al. 2007). However, a recent phylogenomic analysis of 148 genes from a broad variety of eukaryotic taxa seems to provide substantial support for an excavate clade (Hampl et al. 2009).
We applied the RGC approaches to assess the phylogenetic positions of three highly diverse excavates. Giardia lamblia, a flagellated, amitochondrial protozoan parasite, is the only representative of diplomonads for which the complete genome sequence is currently available. The genome of this organism lacks many genes that are present in all other eukaryotes (Morrison et al. 2007). Accordingly, Giardia was traditionally considered one of the best candidates for a basal position in the eukaryotic tree. However, the bikont–unikont phylogeny rejects this view in favor of the affiliation of diplomonads and associated excavate taxa with the bikont branch of eukaryotes (Stechmann and Cavalier-Smith 2002; Stechmann and Cavalier-Smith 2003a; Rodriguez-Ezpeleta et al. 2007).
We analyzed the trifurcation G. lamblia–plants–opisthokonts (fig. 3G). The raw number of shared RGC_CAMs was the greatest for the plants–animals–fungi clade as expected given the extremely long Giardia branch (fig. 3G and table 5). In this case, reversals are expected to have a substantial effect because of the long stem branch (Irimia et al. 2007; Rogozin et al. 2007a) (table 5). Thus, the trifurcation G. lamblia–plants–opisthokonts could not be unambiguously resolved using RGCs. Nevertheless, assuming that the basal position of G. lamblia is a long-branch artifact, the results of the present analysis are best compatible with the Giardia–opisthokont clade (tables 4 and 5).
Table 5.
Phylogenetic Position of the Diplomonads: Analysis of the Trifurcation G. lamblia (GLAM)–Plants (PLAN)–Opisthokonta (Animals–Fungi, AF)
| RGC | Hypothesis |
Branch Length |
Relative Probabilities of Hypotheses |
|||||||
| GLAM + PLAN | GLAM + AF | PLAN + AF | GLAM | PLAN | AF | Stem | P12 | P13 | P23 | |
| H1 | H2 | H3 | ||||||||
| Animals and fungi, A. thaliana and O. sativa | ||||||||||
| CAM | 3 | 3 | 8 | 208 | 12 | 4 | 74 | 0.267 | <0.001 (H3) | <0.001 (H3) |
| CA | 14 | 21 | 47 | 600 | 64 | 15 | 249 | <0.001 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, A. thaliana and O. sativa | ||||||||||
| CAM | 9 | 3 | 18 | 256 | 19 | 14 | 91 | 0.239 | <0.001 (H3) | <0.001 (H3) |
| CA | 29 | 26 | 95 | 731 | 93 | 66 | 300 | 0.277 | <0.001 (H3) | <0.001 (H3) |
| Fungi, A. thaliana and O. sativa | ||||||||||
| CAM | 9 | 10 | 17 | 265 | 44 | 16 | 92 | 0.036 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 36 | 46 | 89 | 764 | 149 | 78 | 311 | <0.001 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, A. thaliana and P. patens | ||||||||||
| CAM | 2 | 8 | 5 | 175 | 8 | 5 | 52 | 0.057 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 8 | 27 | 36 | 489 | 37 | 12 | 180 | <0.001 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, A. thaliana and P. patens | ||||||||||
| CAM | 9 | 9 | 7 | 212 | 10 | 13 | 63 | 0.459 | <0.001 (H3) | <0.001 (H3) |
| CA | 24 | 34 | 54 | 595 | 54 | 50 | 219 | 0.130 | <0.001 (H3) | <0.001 (H3) |
| Fungi, A. thaliana and P. patens | ||||||||||
| CAM | 5 | 14 | 8 | 213 | 15 | 13 | 70 | 0.059 | <0.001 (H3) | <0.001 (H3) |
| CA | 23 | 49 | 65 | 610 | 64 | 65 | 230 | 0.011 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, unicellular plants | ||||||||||
| CAM | 1 | 2 | 6 | 180 | 6 | 2 | 59 | 0.278 | <0.001 (H3) | <0.001 (H3) |
| CA | 5 | 12 | 27 | 464 | 23 | 9 | 176 | 0.005 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, unicellular plants | ||||||||||
| CAM | 7 | 4 | 7 | 216 | 7 | 11 | 69 | 0.181 | <0.001 (H3) | <0.001 (H3) |
| CA | 23 | 20 | 42 | 555 | 33 | 42 | 213 | 0.211 | <0.001 (H3) | <0.001 (H3) |
| Fungi, unicellular plants | ||||||||||
| CAM | 6 | 7 | 8 | 214 | 10 | 7 | 69 | 0.374 | <0.001 (H3) | <0.001 (H3) |
| CA | 17 | 25 | 46 | 571 | 39 | 54 | 224 | 0.659 | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, all plants | ||||||||||
| CAM | 1 | 2 | 4 | 164 | 3 | 1 | 50 | 0.371 | <0.001 (H3) | <0.001 (H3) |
| CA | 4 | 12 | 20 | 430 | 13 | 4 | 161 | 0.004 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, all plants | ||||||||||
| CAM | 7 | 2 | 5 | 191 | 3 | 7 | 58 | 0.051 | <0.001 (H3) | <0.001 (H3) |
| CA | 18 | 14 | 30 | 508 | 19 | 28 | 190 | 0.124 | <0.001 (H3) | <0.001 (H3) |
| Fungi, all plants | ||||||||||
| CAM | 4 | 7 | 6 | 190 | 4 | 6 | 60 | 0.880 | <0.001 (H3) | <0.001 (H3) |
| CA | 11 | 24 | 34 | 509 | 21 | 43 | 197 | 0.704 | <0.001 (H3) | <0.001 (H3) |
NOTE.—The results are given for different combinations of plant and animal–fungal species. H1, H2, and H3 denote the three possible phylogenetic hypotheses regarding the resolution of the given trifurcation. P12, P23, and P13 denote the P values associated with the comparison of the respective hypotheses (see Materials and Methods for details). (H1) and (H2) denote the polarity of the comparison; for instance, (H1) after a P12 value indicates that, in the given comparison, H1 is significantly more likely than H2, conversely, (H2) indicates that H2 is significantly more likely than H1.
The kinetoplastids, a distinct group of mitochondriate protists that includes such major parasites as trypanosomes and Leishmania, comprise another branch in the putative excavate supergroup (Simpson, Stevens, and Lukes 2006; Stevens 2008). We took advantage of the availability of five complete genomes from this group to examine the phylogenetic affinities of kinetoplastids using RGCs (fig. 3H). In the majority of the comparisons (30 cases), the greatest raw number of shared RGC_CAMs was seen for the Plantae–opisthokont clade, that is, the basal position of kinetoplastids (supplementary table S7, Supplementary Material online) that also received a strong statistical support. However, in 25 comparisons, the raw number of shared RGC_CAMs was the largest for the kinetoplastid–opisthokont clade (supplementary table S7, Supplementary Material online), and this excess was statistically supported as well (table 6). Similarly to the case of Giardia, the kinetoplastid branch was extremely long (fig. 3H and supplementary table S7, Supplementary Material online) because of which the basal position of this group, most likely, is an artifact. Under this assumption, the present results support the kinetoplastid–opisthokont clade (tables 4 and 6).
Table 6.
Phylogenetic Position of the Kinetoplastids: Analysis of the Trifurcation Kinetoplastids (KIN)–plants (PLAN)–Opisthokonta (Animals–Fungi, AF)
| RGC | The Number of Tests Supporting a Hypothesis |
|||||
| KIN + PLAN versus KIN + AF |
KIN + PLAN versus PLAN + AF |
KIN + AF versus PLAN + AF |
||||
| > | < | > | < | > | < | |
| A. thaliana and O. sativa | ||||||
| CAM | 0 | 5 | 0 | 15 | 0 | 4 |
| CA | 0 | 11 | 0 | 17 | 0 | 17 |
| A. thaliana and P. patens | ||||||
| CAM | 0 | 6 | 0 | 0 | 0 | 0 |
| CA | 0 | 12 | 0 | 10 | 0 | 18 |
| Unicellular Plantae | ||||||
| CAM | 0 | 1 | 0 | 0 | 0 | 0 |
| CA | 0 | 6 | 0 | 18 | 0 | 18 |
| All Plantae | ||||||
| CAM | 0 | 0 | 0 | 0 | 0 | 0 |
| CA | 0 | 6 | 0 | 18 | 0 | 18 |
NOTE.—The results are included for the indicated combinations of plant species. The signs ‘>’ and ‘<’ denote the polarity of the comparison; for instance, ‘>’ below KIN + PLAN indicates that, in the given comparison, the hypothesis that kinetoplastids and plants are sister taxa is significantly more likely than the hypothesis that kinetoplastids and opisthokonts are sister taxa, conversely, ‘<’ indicates that the second hypothesis is significantly more likely than the first hypothesis.
Trichomonas vaginalis is a flagellated, amitochondrial parasitic protist that represents the parabasalids, another excavate group with an uncertain phylogenetic position (Edgcomb et al. 2001; Carlton et al. 2007). We analyzed the trifurcation T. vaginalis–plants–opisthokonta (table 7). As with the other excavates, the results, at face value, seemed to support a basal position for T. vaginalis (fig. 3I and table 7). However, assuming that this position is a long-branch artifact, the T. vaginalis–opisthokonta clade was strongly supported by both raw numbers and statistical tests (tables 4 and 7).
Table 7.
Phylogenetic Position of Parabasalids: Analysis of the Trifurcation T. vaginalis (TVAG)–Plants (PLAN)–Opisthokonta (Animals–Fungi, AF)
| RGC | Hypothesis |
Branch Length |
Relative Probabilities of Hypotheses |
|||||||
| TVAG + PLAN | TVAG + AF | PLAN + AF | TVAG | PLAN | AF | Stem | P12 | P13 | P23 | |
| H1 | H2 | H3 | ||||||||
| Animals and fungi, A. thaliana and O. sativa | ||||||||||
| CAM | 0 | 3 | 12 | 163 | 17 | 6 | 70 | 0.032 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 8 | 17 | 52 | 470 | 73 | 16 | 277 | <0.001 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, A. thaliana and O. sativa | ||||||||||
| CAM | 1 | 7 | 18 | 211 | 22 | 21 | 87 | 0.047 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 22 | 33 | 88 | 597 | 98 | 68 | 348 | 0.010 (H2) | <0.001 (H3) | <0.001 (H3) |
| Fungi, A. thaliana and O. sativa | ||||||||||
| CAM | 3 | 8 | 19 | 206 | 29 | 33 | 92 | 0.192 | <0.001 (H3) | <0.001 (H3) |
| CA | 30 | 43 | 81 | 598 | 123 | 98 | 377 | 0.021 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, A. thaliana and P. patens | ||||||||||
| CAM | 0 | 5 | 15 | 169 | 15 | 7 | 69 | 0.009 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 7 | 19 | 55 | 482 | 59 | 15 | 276 | <0.001 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, A. thaliana and P. patens | ||||||||||
| CAM | 1 | 10 | 22 | 214 | 18 | 23 | 85 | 0.032 (H2) | <0.001 (H3) | <0.001 (H3) |
| CA | 18 | 36 | 91 | 603 | 79 | 68 | 341 | 0.007 (H2) | <0.001 (H3) | <0.001 (H3) |
| Fungi, A. thaliana and P. patens | ||||||||||
| CAM | 3 | 10 | 21 | 216 | 29 | 34 | 93 | 0.110 | <0.001 (H3) | <0.001 (H3) |
| CA | 31 | 46 | 81 | 609 | 106 | 91 | 375 | 0.029 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, unicellular plants | ||||||||||
| CAM | 0 | 3 | 14 | 145 | 6 | 4 | 72 | 0.122 | <0.001 (H3) | <0.001 (H3) |
| CA | 5 | 13 | 45 | 431 | 22 | 10 | 254 | 0.002 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, unicellular plants | ||||||||||
| CAM | 1 | 8 | 18 | 184 | 8 | 19 | 84 | 0.262 | <0.001 (H3) | <0.001 (H3) |
| CA | 13 | 30 | 75 | 532 | 36 | 59 | 307 | 0.250 | <0.001 (H3) | <0.001 (H3) |
| Fungi, unicellular plants | ||||||||||
| CAM | 3 | 7 | 22 | 179 | 11 | 27 | 90 | 0.867 | <0.001 (H3) | <0.001 (H3) |
| CA | 27 | 35 | 69 | 534 | 48 | 71 | 326 | 0.397 | <0.001 (H3) | <0.001 (H3) |
| Animals and fungi, all plants | ||||||||||
| CAM | 0 | 2 | 9 | 133 | 4 | 3 | 65 | 0.277 | <0.001 (H3) | <0.001 (H3) |
| CA | 4 | 12 | 38 | 384 | 13 | 7 | 234 | 0.019 (H2) | <0.001 (H3) | <0.001 (H3) |
| Animals, all plants | ||||||||||
| CAM | 1 | 5 | 13 | 167 | 5 | 15 | 76 | 0.888 | <0.001 (H3) | <0.001 (H3) |
| CA | 10 | 20 | 62 | 468 | 19 | 45 | 279 | 0.449 | <0.001 (H3) | <0.001 (H3) |
| Fungi, all plants | ||||||||||
| CAM | 2 | 5 | 14 | 158 | 6 | 25 | 81 | 0.462 | <0.001 (H3) | <0.001 (H3) |
| CA | 18 | 26 | 53 | 465 | 29 | 63 | 295 | 0.188 | <0.001 (H3) | <0.001 (H3) |
NOTE.—The results are given for different combinations of plant and animal–fungal species. H1, H2, and H3 denote the three possible phylogenetic hypotheses regarding the resolution of the given trifurcation. P12, P23, and P13 denote the P values associated with the comparison of the respective hypotheses (see Materials and Methods for details). (H1) and (H2) denote the polarity of the comparison; for instance, (H1) after a P12 value indicates that, in the given comparison, H1 is significantly more likely than H2, conversely, (H2) indicates that H2 is significantly more likely than H1.
Discussion
We employed RGCs to analyze one of the most difficult problems in the evolution of eukaryotes, the relationship between the five supergroups. At present, the best description of the radiation of the supergroups seems to be a Big Bang, a pattern that might indeed reflect rapid divergence or condensed cladogenesis, in part, driven by major events such as endosymbiosis (Philippe et al. 2000; Keeling et al. 2005; Rokas and Carroll 2006; Keeling 2007; Koonin 2007). Thus, attempts to decipher the relationships between the supergroups are important not only (and, perhaps, not so much) for establishing the true tree topology for its own sake but also for reconstructing the most likely scenario of the actual events that occurred during the early, formative stages of eukaryotic evolution.
Given the presumed rapidity of the pivotal evolutionary events at this early stage in the evolution of eukaryotes, combined with the dramatic differences in the evolutionary rates among the supergroups, definitive elucidation of the true tree topology is extremely challenging (Philippe et al. 2000). Not surprisingly, so far, despite substantial effort, traditional methods of phylogenetic analysis failed to yield a solution.
In this difficult situation, shared derived characters might offer the best chance to shed light on the early radiation of the supergroups. Attempts to implement this approach include the influential analyses of gene fusions, such as the DHFR–ThyK fusion and domain architectures, such as those of myosins (Stechmann and Cavalier-Smith 2002, 2003a; Richards and Cavalier-Smith 2005). The caveat of this type of analysis is that, with a small number of characters, ruling out homoplasy is difficult, if feasible at all. The RGCs could have an advantage because multiple, if not necessarily numerous (for deep evolutionary relationships), characters are available for analysis. In this work, we attempted both the rather traditional analysis of indels and the more recently developed classes of characters, RGC_CAMs and RGC_CAs. Somewhat unexpectedly, considering the long history of the use of indels for cladistic-type analysis (Rivera and Lake 1992; Gupta 1998; Gupta and Griffiths 2002), indels turned out to be, largely, uninformative for the elucidation of the relationships between the supergroups, whereas the RGC_CAMs and RGC_CAs seemed to carry considerable information (of course, this is not to imply that indels are not helpful in elucidating more recent evolutionary events).
Even with the use of RGCs, resolving the relationship between the supergroups remains an enormously difficult task, so perhaps, the most tangible outcome of this analysis is the rejection of certain evolutionary hypotheses. Thus, the analysis of RGC_CA(M)s allowed us to effectively rule out the basal position of Chromalveolata vis-a-vis Plantae and opisthokonts and produced evidence in favor of a Chromalveolata–opisthokonts clade as opposed to the Plantae–Chromalveolata clade that is predicted by the bikont–unikont topology of the eukaryotic tree. Notably, however, there was also a nonnegligible signal for the plant–Chromalveolata affinity that is most parsimoniously explained by the contribution of the secondary, red algal endosymbiont to the gene complement of the Chromalveolata.
For the three analyzed excavate taxa (diplomonads, kinetoplastids, and parabasalids), the basal position could not be rejected. However, in agreement with the previous conclusions based on the analysis of slowly evolving positions in conserved proteins (Philippe et al. 2000), it seems most likely that this tree topology is an artifact caused by the extremely long branches characteristic of these groups that imply large numbers of parallel changes and reversals since the divergence from other supergroups. Under the assumption that the basal position of these groups is a long-branch artifact, they all show affiliation with opisthokonts and not with Plantae.
A recent, extensive phylogenomic study suggested the existence of a “megagroup” of eukaryotes that consists of Plantae (there denoted Archaeplastida), Chromalveolata, and Rhizaria (Hampl et al. 2009). However, in addition to the usual complications that emerge in the maximum likelihood analysis of concatenated protein sequence alignments and the problems caused by the potential signal from horizontally transferred genes in Chromalveolata, the tree of Hampl et al. (2009) is unrooted, so the conclusion on the existence of the megagroup is conditioned on the root position between unikonts and bikonts (Stechmann and Cavalier-Smith 2003a). Unlike the standard phylogenetic methods, RGC approaches including RGC_CAM, their own limitations notwithstanding, are specifically geared toward the inference of the root position.
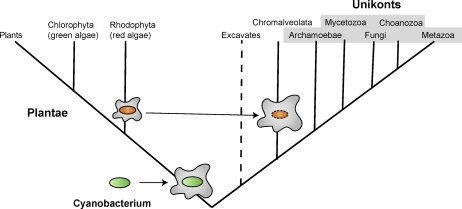
Thus, the results of the present analysis of RGCs seem to be best compatible with an unexpected phylogeny in which the first split is between Plantae, that is, primary chloroplast-containing forms and the rest of the eukaryotes (fig. 4). Although surprising in view of some of the previous inferences, this putative topology of the eukaryotic tree appears biologically plausible in that the acquisition of the cyanobacterial endosymbiont would trigger the divergence of the ancestors of Plantae from the common ancestor with the rest of the eukaryotes. Subsequently, the emergence of the Chromalveolata could have been similarly precipitated by the secondary endosymbiosis, the engulfment of a red alga.
FIG. 4.—
The scenario of evolution of eukaryotic supergroups that is best compatible with the results of the RGC analysis. The primary (postmitochondrial) endosymbiosis of a cyanobacterium with an ancient, heterotrophic, unicellular eukaryote that is thought to have precipitated the first split in the evolution of eukaryotes, that between photosynthesis and nonphotosynthetic organisms, and the secondary endosymbiosis of a red alga and a nonphotosynthetic unicellular form, which would trigger the divergence of chromalveolates from the unikont lineage, are schematically shown. The oval shape encases the traditional unikont supergroup. The excavates are shown as a single branch, although their monophyly remains uncertain as well as their position in the tree; to emphasize this uncertainty, the excavate branch is shown with a dashed line. The branch lengths are arbitrary.
Conclusions
The present results are far from being the final word on the relationship between the eukaryotic supergroups but they are at odds with some popular hypotheses, in particular, the bikont–unikont split as the primary radiation in the history of eukaryotes. Extreme caution is necessary in drawing positive conclusions from deep phylogenetic reconstructions like this one. Nevertheless, the present findings are best compatible with the monophyly of unikonts and Chromalveolata, with excavates, possibly, joining the same major assemblage of eukaryotic taxa. Under this, biologically plausible scenario, the first major split in eukaryotic evolution is between photosynthetic and nonphotosynthetic forms and would have been triggered by the endosymbiosis between an ancient heterotrophic, unicellular eukaryote and a cyanobacterium that gave rise to the chloroplast. Methodologically, the present analysis reveals the apparent advantage of RGCs based on (preferably, multiple) substitutions in otherwise highly conserved positions over indels as phylogenetic markers. Apparently, shared indels are too rare and too prone to homoplasy to be informative for resolving deep multifurcations. In addition, the results emphasize the importance of taxon sampling for RGC analysis: the availability of a diverse collection of complete genomes representing Chromalveolata provided for much more conclusive results for this supergroup than for excavates where such sampling is currently impossible. Thus, further progress of genomics of poorly characterized eukaryotic groups is expected to provide additional material for more conclusive reconstruction of the key events of the deep evolutionary past.
Supplementary Material
Supplementary tables S1–S7 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS; research grant from the National Sciences and Engineering Research Council of Canada.
Supplementary Material
Acknowledgments
We thank Laura Katz, Masatoshi Nei, Fyodor Kondrashov, Liran Carmel, and Yuri Wolf for useful discussions. The authors declare that they have no competing interests. I.B.R. and E.V.K. incepted the study; I.B.R., M.K.B., and M.C. implemented the tests and performed data analysis; I.B.R. and E.V.K. wrote the manuscript which was read, edited, and approved by all authors.
References
- Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- Averof M, Rokas A, Wolfe KH, Sharp PM. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science. 2000;287:1283–1286. doi: 10.1126/science.287.5456.1283. [DOI] [PubMed] [Google Scholar]
- Baldauf SL. A search for the origins of animals and fungi: comparing and combining molecular data. Am Nat. 1999;154:S178–S188. doi: 10.1086/303292. [DOI] [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci USA. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, et al. Alternative methods for concatenation of core genes indicate a lack of resolution in deep nodes of the prokaryotic phylogeny. Mol Biol Evol. 2008;25:83–91. doi: 10.1093/molbev/msm229. [DOI] [PubMed] [Google Scholar]
- Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu MK, Rogozin IB, Koonin EV. Primordial spliceosomal introns were probably U2-type. Trends Genet. 2008;24:525–528. doi: 10.1016/j.tig.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucknam J, Boucher Y, Bapteste E. Refuting phylogenetic relationships. Biol Direct. 2006;1:26. doi: 10.1186/1745-6150-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol Rev Camb Philos Soc. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Philos Trans R Soc Lond B Biol Sci. 2003;358:109–133. doi: 10.1098/rstb.2002.1194. discussion 133–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Johnson PJ. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr Opin Microbiol. 2000;3:404–411. doi: 10.1016/s1369-5274(00)00112-0. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Roger AJ, Simpson AG, Kysela DT, Sogin ML. Evolutionary relationships among “jakobid” flagellates as indicated by alpha- and beta-tubulin phylogenies. Mol Biol Evol. 2001;18:514–522. doi: 10.1093/oxfordjournals.molbev.a003830. [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Embley TM, et al. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- Embley TM, van der Giezen M, Horner DS, Dyal PL, Foster P. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci. 2003;358:191–201. doi: 10.1098/rstb.2002.1190. discussion 201–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WM, Palmer JD. Evidence from small-subunit ribosomal RNA sequences for a fungal origin of Microsporidia. Mol Phylogenet Evol. 2005;36:606–622. doi: 10.1016/j.ympev.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Griffiths E. Critical issues in bacterial phylogeny. Theor Popul Biol. 2002;61:423–434. doi: 10.1006/tpbi.2002.1589. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Penny D, Garcia-Fernandez J, Roy SW. Rare coding sequence changes are consistent with Ecdysozoa, not Coelomata. Mol Biol Evol. 2007;24:1604–1607. doi: 10.1093/molbev/msm105. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Genomics. Deep questions in the tree of life. Science. 2007;317:1875–1876. doi: 10.1126/science.1149593. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, McFadden GI. Origins of microsporidia. Trends Microbiol. 1998;6:19–23. doi: 10.1016/S0966-842X(97)01185-2. [DOI] [PubMed] [Google Scholar]
- King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The Biological Big Bang model for the major transitions in evolution. Biol Direct. 2007;2:21. doi: 10.1186/1745-6150-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Li S, Nosenko T, Hackett JD, Bhattacharya D. Phylogenomic analysis identifies red algal genes of endosymbiotic origin in the chromalveolates. Mol Biol Evol. 2006;23:663–674. doi: 10.1093/molbev/msj075. [DOI] [PubMed] [Google Scholar]
- Luo Y, Fu C, Zhang DY, Lin K. Overlapping genes as rare genomic markers: the phylogeny of gamma-Proteobacteria as a case study. Trends Genet. 2006;22:593–596. doi: 10.1016/j.tig.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Makiuchi T, Nara T, Annoura T, Hashimoto T, Aoki T. Occurrence of multiple, independent gene fusion events for the fifth and sixth enzymes of pyrimidine biosynthesis in different eukaryotic groups. Gene. 2007;394:78–86. doi: 10.1016/j.gene.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Minge MA, et al. Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc Biol Sci. 2008;276:597–604. doi: 10.1098/rspb.2008.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, et al. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. Am Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Philip GK, Creevey CJ, McInerney JO. The Opisthokonta and the Ecdysozoa may not be clades: stronger support for the grouping of plant and animal than for animal and fungi and stronger support for the Coelomata than Ecdysozoa. Mol Biol Evol. 2005;22:1175–1184. doi: 10.1093/molbev/msi102. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992;257:74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Roger AJ. Reconstructing early events in eukaryotic evolution. Am Nat. 1999;154:S146–S163. doi: 10.1086/303290. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Silberman JD. Cell evolution: mitochondria in hiding. Nature. 2002;418:827–829. doi: 10.1038/418827a. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Thomson K, Csuros M, Carmel L, Koonin EV. Homoplasy in genome-wide analysis of rare amino acid replacements: the molecular-evolutionary basis for Vavilov's law of homologous series. Biol Direct. 2008;3:7. doi: 10.1186/1745-6150-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Carmel L, Koonin EV. Analysis of rare amino acid replacements supports the Coelomata clade. Mol Biol Evol. 2007a;24:2594–2597. doi: 10.1093/molbev/msm218. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Carmel L, Koonin EV. Ecdysozoan clade rejected by genome-wide analysis of rare amino acid replacements. Mol Biol Evol. 2007b;24:1080–1090. doi: 10.1093/molbev/msm029. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- Rokas A, Kruger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- Silva JC, Kondrashov AS. Patterns in spontaneous mutation revealed by human-baboon sequence comparison. Trends Genet. 2002;18:544–547. doi: 10.1016/s0168-9525(02)02757-9. [DOI] [PubMed] [Google Scholar]
- Simpson AG. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota) Int J Syst Evol Microbiol. 2003;53:1759–1777. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Hinkle G, Leipe DD. Universal tree of life. Nature. 1993;362:795. doi: 10.1038/362795a0. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Silberman JD. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int J Parasitol. 1998;28:11–20. doi: 10.1016/s0020-7519(97)00181-1. [DOI] [PubMed] [Google Scholar]
- Song J, et al. Comparing the Dictyostelium and Entamoeba genomes reveals an ancient split in the Conosa lineage. PLoS Comput Biol. 2005;1:e71. doi: 10.1371/journal.pcbi.0010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003a;13:R665–666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol. 2003b;57:408–419. doi: 10.1007/s00239-003-2490-x. [DOI] [PubMed] [Google Scholar]
- Steel M. Should phylogenetic models be trying to “fit an elephant”? Trends Genet. 2005;21:307–309. doi: 10.1016/j.tig.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Stefankovic D, Vigoda E. Pitfalls of heterogeneous processes for phylogenetic reconstruction. Syst Biol. 2007;56:113–124. doi: 10.1080/10635150701245388. [DOI] [PubMed] [Google Scholar]
- Stevens JR. Kinetoplastid phylogenetics, with special reference to the evolution of parasitic trypanosomes. Parasite. 2008;15:226–232. doi: 10.1051/parasite/2008153226. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Budd GE. The place of phylogeny and cladistics in Evo-Devo research. Int J Dev Biol. 2003;47:479–490. [PubMed] [Google Scholar]
- Thornton JW, Kolaczkowski B. No magic pill for phylogenetic error. Trends Genet. 2005;21:310–311. doi: 10.1016/j.tig.2005.04.002. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6:525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Wolf YI, Koonin EV. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0024. research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, et al. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.