Abstract
Elevated levels of genetic drift are hypothesized to be a dominant factor that influences genome size evolution across all life-forms. However, increased levels of drift appear to be correlated with genome expansion in eukaryotes but with genome contraction in bacteria, suggesting that these two groups of organisms experience vastly different mutational inputs and selective constraints. To determine the contribution of small insertion and deletion events to the differences in genome organization between eukaryotes and prokaryotes, we systematically surveyed 17 taxonomic groups across the three domains of life. Based on over 5,000 indel events in noncoding regions, we found that deletional events outnumbered insertions in all groups examined. The extent of deletional bias, when measured by the total length of insertions to deletions, revealed a marked disparity between eukaryotes and prokaryotes, whereas the ratio was close to one in the three eukaryotic groups examined, deletions outweighed insertions by at least a factor of 10 in most prokaryotes. Moreover, the strength of deletional bias is associated with the proportion of coding regions in prokaryotic genomes. Considering that genetic drift is a stochastic process and does not discriminate the exact nature of mutations, the degree of bias toward deletions provides an explanation to the differential responses of eukaryotes and prokaryotes to elevated levels of drift. Furthermore, deletional bias, rather than natural selection, is the primary mechanism by which the compact gene packing within most prokaryotic genomes is maintained.
Keywords: genome evolution, genome size, mutational spectra, organismal complexity, indels
Introduction
The genome sizes of cellular organisms span at least six orders of magnitude (Gregory 2005), but the evolutionary and functional basis of this variation remains unclear. Early studies detected relationships between genome size and several phenotypic traits, such as generation time (Bennett 1972), cell and nuclear volume (Cavalier-Smith 1982), duration of mitosis and meiosis (Bennett 1987), embryonic developmental time (Jockusch 1997), and plant seed or leaf size (Chung et al. 1998). Based on such correlations, genome size was hypothesized to be under selective constraints (Gregory 2002); however, comparative studies in bacteria have failed to support such adaptive view as a general explanation. For example, although the streamlined genomes of bacteria are often regarded as an adaptation for rapid cell growth, bacterial replication rates are not correlated with genome size either within (Mikkola and Kurland 1991; Bergthorsson and Ochman 1998) or among species (Mira et al. 2001; Froula and Francino 2007). The only exception appears to be from nutrient-limited marine bacteria (Dufresne et al. 2005; Giovannoni et al. 2005), which may be under selection for reduced cell volume.
It has recently been posited that the overall size and structure of genomes are determined mainly by a nonadaptive, population-level process, namely random genetic drift (Lynch and Conery 2003). Because the accumulation of slightly deleterious mutations is facilitated by an increase in drift, lineages with relatively small effective population sizes (e.g., mammals) tend to have large genomes due to the proliferation of transposable elements and the lengthening of introns (Lynch and Conery 2003; Lynch 2006a). In contrast, lineages with relatively large population sizes (e.g., most free-living bacteria) would be expected to have more streamlined genomes on account of more effective selection against unnecessary or slightly deleterious sequences, which limits the accumulation of selfish and noncoding DNA (Lynch 2006b). Although this model provides a straightforward and seemingly unifying explanation for the evolution of genome size across all life forms, it does not explain the variation in the most genetically diverse group of organisms on the planet. Contrary to the predictions of this model, the strength of drift is “negatively” correlated with genome size in Bacteria (Kuo et al. 2009; Novichkov et al. 2009), with those bacteria the lifestyles of which cause the most dramatic reductions in effective population size having the most reduced genomes (Moran and Plague 2004; Nakabachi et al. 2006).
Because genetic drift facilitates the fixation of slightly deleterious mutations, the difference between the effects of drift on the size of eukaryotic and bacterial genomes is most likely rooted in the mutational input. Previous studies that examined small-scale indels (ranging from single to several hundred nucleotides) in pseudogenes or other nonfunctional elements revealed that deletions prevail over insertions across a wide range of taxonomic groups, including Archaea (von Passel et al. 2007), Bacteria (Andersson JO and Andersson SGE 2001; Mira et al. 2001), nematodes (Robertson 2000), insects (Petrov et al. 1996, 2000; Petrov and Hartl 1998; Bensasson et al. 2001), and mammals (Graur et al. 1989). Mutation accumulation experiments on a few model organisms offer a slightly different view: A preponderance of deletions has been observed in the bacterium Salmonella enterica (Nilsson et al. 2005), whereas insertions outnumbered deletions in the nematode Caenorhabditis elegans (Denver et al. 2004). Based on these observations, a general mutational bias toward deletions coupled with the genome-wide effects of genetic drift have been hypothesized as the major factors contributing to genome size evolution (Petrov et al. 2000; Petrov 2002; but see Gregory 2003, 2004; Vinogradov 2004).
Unfortunately, the extent to which eukaryotes and prokaryotes differ with respect to their deletional bias is unclear, mainly because the methods used to identify indels vary widely across studies and the taxon sampling in individual studies was rather limited. By taking advantage of the large collection of genome sequences available, we examined a diverse set of lineages to directly compare the impact of mutational input on genome evolution across the domains of life.
Materials and Methods
Prokaryotes: Archaea and Bacteria
To assemble data sets for examining deletional bias in these microbial taxa, we selected sets of three genomes (representing separate strains or species depending on the particular taxonomic group) from each group. In order to infer neutral indels in highly degraded pseudogenes, we required the divergence level among the three lineages to be low enough to achieve unambiguous alignments but to have incurred an ample number of indels. All of the archaeal and bacterial genomes used in this study were downloaded from NCBI GenBank (Benson et al. 2008) on 4 December 2008. The Genome Project IDs are listed in table 1. Data parsing and processing were performed with a set of custom Perl scripts written with Bioperl modules (Stajich et al. 2002).
Table 1.
Summary of Taxon Sampling
| Domaina | Phylum | Genus | Sampled Genomesb | NCBI Genome ID |
| A | Crenarchaeota | Sulfolobus | ((S. tokodaii, S. acidocaldarius), S. solfataricus) | ((246, 13935), 108) |
| A | Euryarchaeota | Methanococcus | ((M. maripaludis S2, M. maripaludis C6), M. vannielii); | ((10632, 19639), 17889) |
| B | Actinobacteria | Mycobacterium | ((M. tuberculosis, M. marinum), M. avium) | ((15642, 16725), 88) |
| B | Chlorobi | Chlorobium | ((C. limicola, C. phaeobacteroides), C. phaeovibrioides) | ((12606, 12609), 12607) |
| B | Cyanobacteria | Synechococcus | ((S. sp. CC9605, S. sp. CC9902), S. sp. WH 8102) | ((13643, 13655), 230) |
| B | Firmicutes | Bacillus | ((B. subtilis, B. amyloliquefaciens), B. licheniformis) | ((76, 13403), 12388) |
| B | Proteobacteria | Bartonella | ((B. quintana, B. henselae), B. bacilliformis) | ((44, 196), 16249) |
| B | Proteobacteria | Rickettsia | ((R. prowazekii, R. typhi), R. canadensis) | ((43, 10679), 12952) |
| B | Proteobacteria | Wolbachia | ((W. wMel, W. wPip), W. wBm) | ((272, 30313), 12475) |
| B | Proteobacteria | Neisseria | ((N. meningitidis FAM18, N. meningitidis 053442), N. gonorrhoeae) | ((255, 16393), 23) |
| B | Proteobacteria | Geobacter | ((G. metallireducens, G. sulfurreducens), G. uraniireducens) | ((177, 192), 15768) |
| B | Proteobacteria | Buchnera | ((B. aphidicola APS, B. aphidicola Sg), B. aphidicola Bp) | ((245, 312), 256) |
| B | Proteobacteria | Escherichia | ((E. coli K12, E. coli EDL933), E. coli CFT073) | ((225, 259), 313) |
| B | Spirochetes | Borrelia | ((B. turicatae, B. recurrentis), B. burgdorferi) | ((13597, 18233), 3) |
| E | Chordata | Homo/Pan | ((Homo sapiens, Pan troglodytes), Pongo pygmaeus) | NA |
| E | Arthropoda | Drosophila | ((D. sechellia, D. simulans), D. melanogaster) | NA |
| E | Ascomycota | Saccharomyces | ((S. cerevisiae, S. paradoxus), S. mikatae) | NA |
NA, Not applicable.
A: Archaea; B: Bacteria; E: Eukaryota.
Parentheses denote the phylogenetic grouping of taxa in standard Newick tree format.
For each group, we began by identifying single-copy orthologs that are shared among all three genomes. These conserved single-copy genes served as anchors to delineate orthologous noncoding regions from which indels could be identified. Sets of orthologous genes were recovered with OrthoMCL (Li et al. 2003), which is a clustering algorithm largely based on all-against-all BlastP (Altschul et al. 1990) hits and has been shown to perform well by a benchmarking study (Hulsen et al. 2006). As a conservative inference of orthology, the BlastP e-value cutoffs were set at 1 × 10−15.
After identifying conserved single-copy genes, we screened the genome for lineage-specific pseudogenes, recognized as protein-coding regions that are disrupted or truncated in only one of the three taxa and are flanked by two conserved single-copy genes. To ensure the quality of alignments, we also required the pseudogene regions between the two conserved flanking genes to be at least 50 bp in length.
The rationale for focusing on pseudogenes in Archaea and Bacteria is based on the fact that the ancestral state of such regions can be confidently inferred from the conserved, single-copy homologs that are uninterrupted in the other two genomes; thus, even a high rate of recombination, as observed among some closely related bacteria (Touchon et al. 2009), would not affect our classification of each event as either an insertion or a deletion. Considering that the first indel to disrupt an open reading frame might not be neutral, we examined only those pseudogenes that contained at least three indels. Pseudogenes that have incurred this number of indels are often unrecognizable by sequence-similarity searches but can be readily identified using our synteny-based approach. Importantly, this method also eliminates the latent bias toward detecting deletions when using full-length open reading frames to search for fragmented pseudogenes.
Orthologous regions that were identified by the described approach were aligned in Muscle (Edgar 2004) using default parameters. To improve alignment quality, sequence alignments incorporated the entire region including the adjacent flanking genes, which were not subjected to indel analysis. Indels specific to one taxon were identified by a custom Perl script; all indels were then manually curated by visual inspection in Jalview (Waterhouse et al. 2009), and poorly aligned regions were excluded.
Eukaryotes: Primates, Flies, and Yeasts
Data sets of indels for the three groups of eukaryotes were constructed and analyzed using approaches similar to those used for Archaea and Bacteria, with the differences noted below:
(1) Due to the lack of robust (or any) gene annotations in several of the eukaryotic genomes available from GenBank, we obtained each of the three eukaryote data sets from alternate databases. Data on primate genomes, including human (Homo sapiens), chimpanzee (Pan troglodytes), and orangutan (Pongo pygmaeus), were retrieved from Ensembl (Hubbard et al. 2009) release 52; Drosophila genomes, including Drosophila melanogaster, Drosophila sechellia, and Drosophila simulans, were downloaded from FlyBase (Tweedie et al. 2009) version FB2009_01; the Saccharomyces data set, including Saccharomyces cerevisiae, Saccharomyces mikatae, and Saccharomyces paradoxus, was extracted from the Saccharomyces Genome Database (Christie et al. 2004) on 27 January, 2009.
(2) To minimize the effects of paralogs in the identification of single-copy orthologs, we applied a more stringent e-value cutoff of 1 × 10−25 in the BlastP step.
(3) The organization of most eukaryotic genomes makes it problematic to identify pseudogenes and their corresponding orthologs, so we focused instead on other classes of noncoding regions, that is, introns or intergenic regions that can be readily aligned among species. Because of the low level of recombination among species and the availability of well-established phylogenies, we utilized an outgroup to infer ancestral states and to establish the polarity of all indels that are specific to only one of the two ingroup lineages. For primates and Drosophila, we selected single-copy genes with exactly one intron in all three species because the orthology among such introns can be established unequivocally. We imposed lower and upper limits on intron lengths because indels in extremely short introns may not be neutral and extremely long introns might prove difficult to align. For primates, we examined introns that were 1–20 kb in length in all three species; for Drosophila, we set the range to 0.2–10 kb. When examining introns, we included the two flanking exons (instead of genes) to ensure quality of the alignments.
(4) Due to the paucity of introns in the Saccharomyces genomes, we examined the intergenic regions that are flanked by two conserved single-copy genes. Because regulatory elements might constitute a significant fraction of short intergenic regions (and thus the indels are more likely to have a fitness effect and not represent neutral events), we excluded intergenic regions shorter than 600 base pairs (bp) in any of the three species considered.
Results
We sampled 17 broadly divergent taxonomic groups, each containing an extensive collection of genome sequences (table 1), and for each group, we selected three lineages that are closely related such that orthologous noncoding regions can be unambiguously aligned. The alignments allowed us to infer the exact boundaries and ancestral state of indels within these noncoding regions, which together provide robust estimates of the mutational input of base pair- to kilobase-sized insertions and deletions to these genomes. Note that because we focused on pseudogenes that had accumulated multiple indels in archaea and bacteria and on long noncoding regions in eukaryotes, the overwhelming majority of indel events can be considered neutral and therefore represent the background pattern of mutations in these genomes.
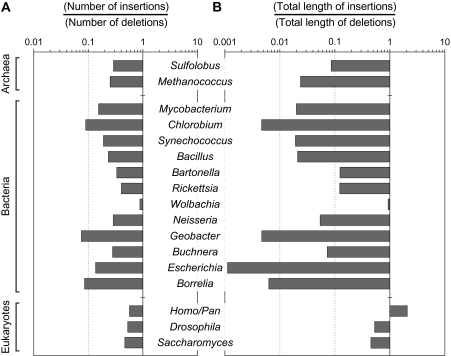
Our results revealed a pervasive bias toward deletions in all taxonomic groups examined, although the extent of bias was substantially lower in the eukaryotic lineages considered (fig. 1). Deletions outnumber insertions in all groups examined, with the extremes observed in Bacteria: The ratio of insertions to deletions ranges from a low of 0.07 in Geobacter to nearly 0.9 in Wolbachia (fig. 1 and table 2). With the exception of Primates, all sampled taxonomic groups experienced a net loss of DNA through small indels. For each bp removed from a genome through deletions, prokaryotes gained from 0.001 bp in Escherichia coli to 0.93 bp in Wolbachia through insertions; in contrast, Saccharomyces and Drosophila gained 0.45 bp and 0.53 bp, respectively, whereas primates gained 2.08 bp for each bp removed by deletions (fig. 1 and table 2). In fact, the observed biases toward deletions are likely to be underestimates: Several deletions were excluded because we required inference of the exact ancestral state, and in the majority of cases, the focal lineage possessed a deletion that was >50 bp, but the exact length of this deletion could not be established because a shorter indel was present in the corresponding region in the other two lineages.
FIG. 1.—
Extent of indel bias in cellular genomes. (A) Ratios of deletion to insertion events. A ratio of less than one indicates a bias toward deletions. (B) Indel bias based on the total length of DNA gained and lost. A ratio of less than 1 indicates a bias toward DNA loss.
Table 2.
Summary of Indel Statistics
| Noncoding Regions Examined |
Insertional Events |
Deletional Events |
|||||
| Domaina | Genus | Number | Total Length (bp)b | Number | Total Length (bp) | Number | Total Length (bp) |
| A | Sulfolobus | 14 | 9,210 | 20 | 173 | 69 | 2,015 |
| A | Methanococcus | 21 | 12,925 | 45 | 178 | 180 | 7,514 |
| B | Mycobacterium | 28 | 17,017 | 21 | 121 | 136 | 6,089 |
| B | Chlorobium | 19 | 6,440 | 11 | 33 | 123 | 7,090 |
| B | Synechococcus | 14 | 4,761 | 15 | 64 | 79 | 3,334 |
| B | Bacillus | 50 | 15,682 | 79 | 318 | 339 | 15,195 |
| B | Bartonella | 34 | 30,775 | 123 | 1,433 | 370 | 11,582 |
| B | Rickettsia | 16 | 17,606 | 64 | 345 | 158 | 2,823 |
| B | Wolbachia | 10 | 10,915 | 31 | 471 | 35 | 507 |
| B | Neisseria | 13 | 9,728 | 24 | 216 | 84 | 4,016 |
| B | Geobacter | 22 | 3,401 | 9 | 35 | 121 | 7,516 |
| B | Buchnera | 29 | 23,111 | 105 | 676 | 377 | 9,334 |
| B | Escherichia | 12 | 4,909 | 8 | 9 | 59 | 8,223 |
| B | Borrelia | 18 | 6,3532 | 17 | 59 | 199 | 9,335 |
| E | Homo/Pan | 136 | 1,182,162 | 235 | 3,343 | 412 | 1,610 |
| E | Drosophila | 170 | 235,213 | 204 | 1,372 | 385 | 2,582 |
| E | Saccharomyces | 99 | 167,335 | 374 | 980 | 801 | 2,168 |
A: Archaea; B: Bacteria; E: Eukaryota.
Sequence length in focal lineages before alignment.
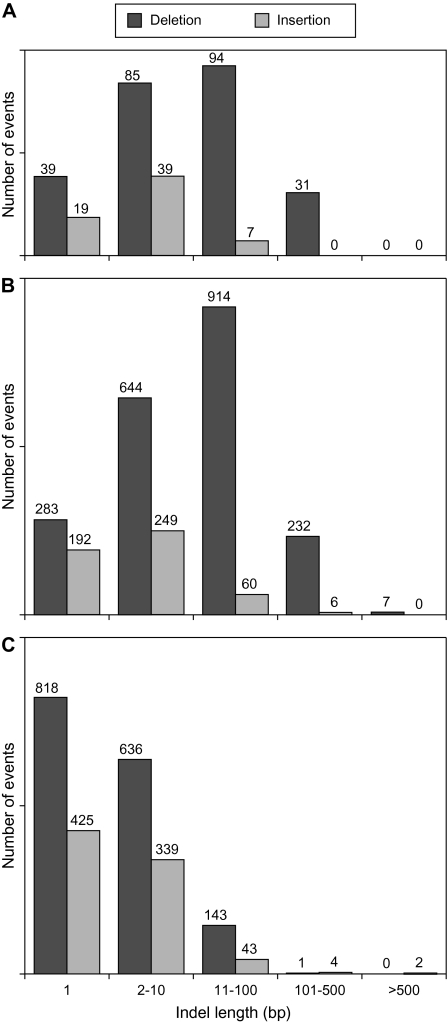
There is a clear difference in the length distribution of indels among three domains, which contributes to the disparity in genomes sizes between prokaryotes and eukaryotes (fig. 2). In Archaea and Bacteria, deletions are more frequent, and on average longer, than insertions, which results in the strong bias toward DNA loss (fig. 2A and B). In contrast, the length distributions of insertions and deletions in eukaryotes are not markedly different, with the majority of observed indels in the 1–10 bp range (fig. 2C). Of the three eukaryotic groups, single bp indels account for 46% of the observed indels in primates, 40% in Drosophila, and over 60% in Saccharomyces (supplementary fig. S1, Supplementary Material online).
FIG. 2.—
Length distribution of small indels across the three domains of life. (A) Archaea, (B) Bacteria, and (C) Eukaryotes.
Archaea and Bacteria
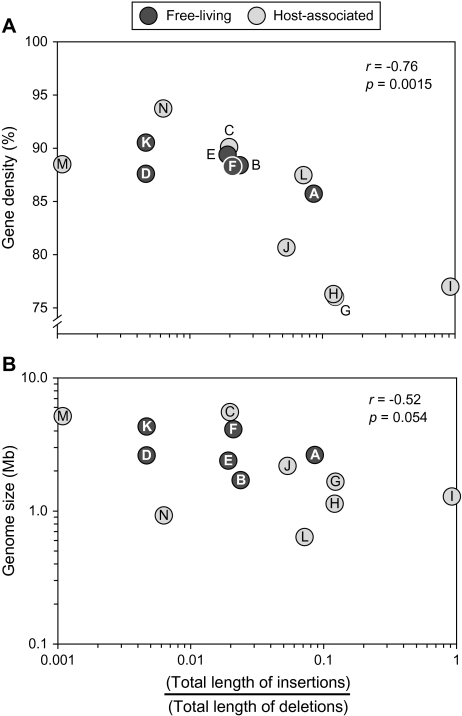
The level of bias toward deletions varies considerably among the prokaryotic genomes examined (fig. 1), allowing us to test two hypotheses concerning the role of deletional bias in genome evolution. First, as the bias toward deletions increases, one expects a more rapid deterioration of nonfunctional regions, resulting in the more compact packing of genes within a genome. Consistent with this hypothesis, gene density (i.e., the proportion of a genome that consists of annotated genes) among prokaryotes is significantly correlated with strength of deletional bias (fig. 3A r= −0.76, P = 0.0015). Second, in that overall genome size in prokaryotes is largely a function of the number of genes in the genome (Mira et al. 2001; Giovannoni et al. 2005; Kuo et al. 2009), we expect little association between the extent of deletional bias in noncoding regions and overall genome size. Because this association borders the conventional significance threshold (fig. 3B r = −0.52, P = 0.054), a more extensive taxon sampling would be necessary to further test this hypothesis.
FIG. 3.—
Correlation between indel bias and genomic features in prokaryotes. (A) Gene density. (B) Genome size. Data points are labeled as follows: A, Sulfolobus; B, Methanococcus; C, Mycobacterium; D, Chlorobium; E, Synechococcus; F, Bacillus; G, Bartonella; H, Rickettsia; I, Wolbachia; J, Neisseria; K, Geobacter; L, Buchnera; M, Escherichia; and N, Borrelia.
We note that the genera with the weakest biases toward deletions are members of the alphaproteobacteria (i.e., Bartonella, Rickettsia, and Wolbachia). Although each of these groups forms obligate associations with eukaryotic hosts, it is unlikely that this lifestyle alone or the age of the association with their respective hosts can explain the observed pattern. The extent of deletional bias in other obligate pathogens (e.g., Borrelia and Neisseria) and endosymbionts (i.e., Buchnera) span much of the observed range. Therefore, diminished biases toward deletions are probably taxonomic characteristic of this bacterial group.
Eukaryotes
The mutational input in Saccharomyces is dominated by small indels. Among 1,175 indels recognized in 99 intergenic regions, the longest insertion was only 65 bp and the longest deletion was 73 bp. Although the length distributions of insertions and deletions do not differ in Saccharomyces (supplementary fig. S1, Supplementary Material online), deletions outnumbered insertions, resulting in a net loss of DNA.
In contrast to prokaryotes and Saccharomyces (both of which lack long insertions), transposable elements provide a major source of DNA gains in primates and Drosophila. Although our analyses in these two eukaryotes were restricted to orthologous introns, which favor the identification of shorter indels, we detected one 1,102-bp insertion in P. troglodytes genome (containing two LINE and one SINE) and one 703-bp insertion in D. sechellia (containing a FB4 element). Despite their rare occurrences, these insertions of transposable elements offset the loss of DNA through frequent small deletions; and in fact, in the case of the primates, such rare long insertions are sufficient to result in a net gain of DNA in introns.
Discussion
The mutational input of insertions and deletions to a genome, as measured either by the number of events or the total length of DNA segments, is inherently biased toward deletions across a wide range of taxonomic groups representing the three domains of life. With the exception of alphaproteobacteria, the deletional biases in prokaryotes were at least one order of magnitude higher than those observed in eukaryotes (fig. 1). Although the prevalence of transposable elements in primates and Drosophila contribute to this difference, the indel pattern in Saccharomyces suggests that eukaryotic genomes have lower intrinsic rates of DNA loss through small indels. In spite of the limitation on taxon sampling imposed by the current availability of eukaryotic genome sequences, the strong differences in mutational input observed between prokaryotes and eukaryotes have played the major role in shaping the genome size and organization within these two groups.
Limitation on Taxon Sampling
Despite recent increases in sequence databases, the availability of genome sequences from closely related lineages remains the limiting factor in making reliable comparisons among divergent taxa. In addition to requiring a set of three genomes for each group, our analyses also demanded that their divergence levels be within a fairly narrow range, low enough to allow confident alignments of noncoding regions, yet sufficiently high to allow for the accumulation of indels. Such requirements limited the number of lineages that could be sampled, and therefore, we are presently unable to extend the generality of our findings to plants or protozoans.
Mutational Input at Larger Scales
To ascertain the mutational input to a genome, the present study focused on small indels occurring in noncoding regions; however, there are several classes of large-scale mutations that can contribute to genome size evolution. For example, whole-genome duplications are a major evolutionary force in many eukaryotic groups (Kellis et al. 2004; Adams and Wendel 2005; Dehal and Boore 2005; Aury et al. 2006), and alternatively, in prokaryotes, large-scale deletions have been detected in both experimental and comparative analyses (Moran and Mira 2001; Nilsson et al. 2005). Because such changes are accompanied by large changes in gene content, they often have a substantial effect on organismal fitness and highly variable fixation probabilities, and therefore, their incidence cannot fully portray the underlying pattern of mutational events.
Despite the strong bias toward deletions in most prokaryotic genomes, the constant influx of novel genes though lateral gene transfer (Garcia-Vallvé et al. 2000; Gogarten et al. 2002; Lerat et al. 2005) will offset the frequent deletions in noncoding regions and can even lead to large increases in genome size. These newly acquired genes seem to represent the most fluid portion of prokaryotic genomes and are the primary contributor to the observed differences in genome size and gene contents among closely related taxa (von Passel et al. 2008; Kuo and Ochman 2009; Touchon et al. 2009).
Transposable elements represent a special class of mutations that can greatly influence the genome size. For example, data from available genome sequences indicate that the quantity of transposable elements is the major determinant of genome size in eukaryotes (Gregory 2005). Because the proliferation of transposable elements is generally viewed as deleterious, the number of transposable elements within a genome is hypothesized to be under the control of purifying selection and a decrease in effective population size would inevitably lead to genome expansion (Lynch and Conery 2003). However, bacteria appear to be an exception to this rule (Kuo et al. 2009; Novichkov et al. 2009), possibly due to strong deletional biases, as observed in this study. Whereas transposable elements and insertion sequences are observed to proliferate during the initial stage of drift-associated genome reduction in bacteria, these elements are eventually eliminated and are virtually absent from the highly reduced genomes of bacterial symbionts (Moran and Plague 2004).
Evolution of Genome Organization
Among the starkest differences in genome architecture between prokaryotes and eukaryotes is the variation in gene density. The lower bound of gene density in prokaryotic genomes appears to be ∼50%, and the vast majority of lineages having a gene density of well over 80% (Kuo et al. 2009). In contrast, the eukaryotic genomes that have been sequenced to date encompass a very wide distribution (Gregory 2005), ranging from about 90% in the microsporidian Encephalitozoon cuniculi (Katinka et al. 2001) to less than 2% in humans (International Human Genome Sequencing Consortium 2004). Much of the variation in gene density in eukaryotes is due to the prevalence of transposable elements and introns, whose fixation probability is in turn controlled by the balance between selection and drift (Lynch and Conery 2003; Gregory 2005).
The association between genome size and effective population size among eukaryotes has lead to the hypothesis that elevated levels of drift are the main cause of genome expansion in eukaryotes (Lynch and Conery 2003). Intriguingly, bacteria exhibit the opposite trend, such that genome reduction usually coincides with an increase level of genetic drift (Kuo et al. 2009; Novichkov et al. 2009). Our results suggest that this difference between prokaryotes and eukaryotes is due in large part to the mutational input of insertions and deletions to a genome. With a strong bias toward deletions, DNA segments that do not contribute to organismal fitness in prokaryotic genomes are likely to be purged, even in the absence of selection. And because drift promotes the fixation of slightly deleterious mutations, which are likely to instigate gene inactivation in gene-rich prokaryotic genomes, a reduction in effective population size (e.g., by switching from a free-living to an obligate endosymbiotic lifestyle) can lead to the loss of function in many nonessential genes. Subsequently, these newly formed noncoding regions are removed through the mutational bias toward deletions, thereby maintaining high gene densities.
Although nonadaptive processes, such as biases in mutational input and genetic drift, appear to be dominant forces that influence the evolution of genome size, natural selection will also govern the overall size of some genomes. The genome reduction observed in certain marine bacteria has been attributed to selection for decreased cell volume and energetic efficiency in light of limiting nutrients (Dufresne et al. 2005; Giovannoni et al. 2005). And aside from the selective constraints imposed on the proliferation of transposable elements (Lynch and Conery 2003) and introns (Lynch 2006a), other mutations with extremely small effects on organismal fitness will be influenced by selection when population sizes are sufficiently large. For example, E. coli is thought to have a large effective population size when compared with other bacteria (Kuo et al. 2009), and this species also exhibits the strongest deletional bias among the prokaryotes examined in this study (fig. 1 and table 2). These observations, along with the relatively rapid removal of pseudogenes in E. coli (Lerat and Ochman 2004), might be taken to indicate that positive selection is operating on small-scale deletions to foster the elimination of pseudogenes. Although pseudogenes are generally considered to be selectively neutral, this suggests the possibility that the presence of pseudogenes incurs some detrimental effects, such as the energetic costs associated with their transcription and translation or the potential hazard of synthesizing anomalous proteins.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by the National Institutes of Health grant [GM56120 to H.O.].
Supplementary Material
Acknowledgments
We thank B. Nankivell for administrative assistance and preparation of the figures.
References
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Andersson SGE. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proc R Soc Lond B Biol Sci. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Variation in genomic form in plants and its ecological implications. New Phytol. 1987;106:177–200. [Google Scholar]
- Bensasson D, Petrov DA, Zhang DX, Hartl DL, Hewitt GM. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol Biol Evol. 2001;18:246–253. doi: 10.1093/oxfordjournals.molbev.a003798. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Skeletal DNA and the evolution of genome size. Annu Rev Biophys Bioeng. 1982;11:273–302. doi: 10.1146/annurev.bb.11.060182.001421. [DOI] [PubMed] [Google Scholar]
- Christie KR, et al. Saccharomyces Genome Database. SGD. provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Lee JH, Arumuganathan K, Graef GL, Specht JE. Relationships between nuclear DNA content and seed and leaf size in soybean. Theor Appl Genet. 1998;96:1064–1068. [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froula JL, Francino MP. Selection against spurious promoter motifs correlates with translational efficiency across bacteria. PLoS One. 2007;2:e745. doi: 10.1371/journal.pone.0000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallvé S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Graur D, Shuali Y, Li WH. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J Mol Evol. 1989;28:279–285. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size and developmental complexity. Genetica. 2002;115:131–146. doi: 10.1023/a:1016032400147. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Is small indel bias a determinant of genome size? Trends Genet. 2003;19:485–488. doi: 10.1016/S0168-9525(03)00192-6. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, Huynen M, de Vlieg J, Groenen P. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jockusch EL. An evolutionary correlate of genome size change in plethodontid salamanders. Proc R Soc Lond B Biol Sci. 1997;264:597–604. [Google Scholar]
- Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kuo CH, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009 doi: 10.1101/gr.091785.109. DOI: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. The fate of new bacterial genes. FEMS Microbiol Rev. 2009;33:38–43. doi: 10.1111/j.1574-6976.2008.00140.x. [DOI] [PubMed] [Google Scholar]
- Lerat E, Ochman H. Psi-Phi: exploring the outer limits of bacterial pseudogenes. Genome Res. 2004;14:2273–2278. doi: 10.1101/gr.2925604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 2006a;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006b;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Mikkola R, Kurland CG. Is there a unique ribosome phenotype for naturally occurring Escherichia coli? Biochimie. 1991;73:1061–1066. doi: 10.1016/0300-9084(91)90148-t. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:research0054.1–research0054.12. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nilsson A, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 2009;191:65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. Mutational equilibrium model of genome size evolution. Theor Pop Biol. 2002;61:531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL. Evidence for DNA loss as a determinant of genome size. Science. 2000;287:1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- Robertson HM. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 2000;10:192–203. doi: 10.1101/gr.10.2.192. [DOI] [PubMed] [Google Scholar]
- Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Evolution of genome size: multilevel selection, mutation bias or dynamical chaos? Curr Opin Genet Dev. 2004;14:620–626. doi: 10.1016/j.gde.2004.09.007. [DOI] [PubMed] [Google Scholar]
- von Passel MWJ, Marri PR, Ochman H. The emergence and fate of horizontally acquired genes in Escherichia coli. PLoS Comput Biol. 2008;4:e1000059. doi: 10.1371/journal.pcbi.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Passel MWJ, Smillie CS, Ochman H. Gene decay in archaea. Archaea. 2007;2:137–143. doi: 10.1155/2007/165723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.