Abstract
Theoretical studies predict X chromosomes and autosomes should be under different selection pressures, and there should therefore be differences in sex-specific and sexually antagonistic gene content between the X and the autosomes. Previous analyses have identified an excess of genes duplicated by retrotransposition from the X chromosome in Drosophila melanogaster. A number of hypotheses may explain this pattern, including mutational bias, escape from X-inactivation during spermatogenesis, and the movement of male-favored (sexually antagonistic) genes from a chromosome that is predominantly carried by females. To distinguish among these processes and to examine the generality of these patterns, we identified duplicated genes in nine sequenced Drosophila genomes. We find that, as in D. melanogaster, there is an excess of genes duplicated from the X chromosome across the genus Drosophila. This excess duplication is due almost completely to genes duplicated by retrotransposition, with little to no excess from the X among genes duplicated via DNA intermediates. The only exception to this pattern appears within the burst of duplication that followed the creation of the Drosophila pseudoobscura neo-X chromosome. Additionally, we examined genes relocated among chromosomal arms (i.e., genes duplicated to new locations coupled with the loss of the copy in the ancestral locus) and found an excess of genes relocated off the ancestral X and neo-X chromosomes. Interestingly, many of the same genes were duplicated or relocated from the independently derived neo-X chromosomes of D. pseudoobscura and Drosophila willistoni, suggesting that natural selection favors the traffic of genes from X chromosomes. Overall, we find that the forces driving gene duplication from X chromosomes are dependent on the lineage in question, the molecular mechanism of duplication considered, the preservation of the ancestral copy, and the age of the X chromosome.
Keywords: gene duplication, retrotransposition, sex chromosomes, neo-X chromosomes, X-inactivation
Introduction
Sex is determined in many animals by heteromorphic chromosomes (Charlesworth 1996). Generally, heteromorphic sex chromosomes come in two varieties: XY and ZW systems. In XY systems, females are homogametic (XX) and males are heterogametic (XY). Therefore, the X is found in females 2/3 of the time and in males 1/3 of the time (the autosomes, on the other hand, spend equal time in males and females), and X chromosomes are hemizygous in males and homozygous in females. Because they are under different selection pressures, sex-specific and sexually antagonistic gene content should differ between X chromosomes and the autosomes (Rice 1984; Vicoso and Charlesworth 2006). Male-favorable genes are expected to be located on the X chromosome if beneficial mutations in these genes are recessive as recessive alleles on the X are exposed to selection in hemizygous males. However, if beneficial mutations in male-favored genes are dominant or if selection acts on standing genetic variation, these genes should be located on the autosomes because they will be exposed to selection more often on the autosomes than the X. Differences in sex-biased gene content between the X chromosome and the autosomes can evolve either by the migration of sex-biased genes between the autosomes and X chromosomes or by the loss/gain of sex-biased functions of genes in particular chromosomal contexts (Vicoso and Charlesworth 2006).
Additionally, X chromosomes are silenced in the germ line of a diverse array of animal taxa (Kelly et al. 2002; Hense et al. 2007; Turner 2007). In Drosophila, X-inactivation is limited to spermatogenesis (Hense et al. 2007). Spermatogenic X-inactivation is thought to be the selective force driving the excess retrotransposition of genes from the X to the autosomes in both Drosophila melanogaster and humans (Betrán et al. 2002; Emerson et al. 2004; Potrzebowski et al. 2008). The autosomal copies of these paralogs tend to be testis expressed, suggesting that these new genes are preferentially retained because they allow for escape from X-inactivation. However, there is still a deficiency of genes with male-biased expression on Drosophila X chromosomes after testis-expressed genes are removed from the comparison (Parisi et al. 2003; Sturgill et al. 2007). Therefore, it is possible that sexually antagonistic selection—and not simply selection for testis-expressed derived copies on the autosomes—drives the duplication of male-favorable genes from the X to the autosomes (Wu and Xu 2003; Vicoso and Charlesworth 2006).
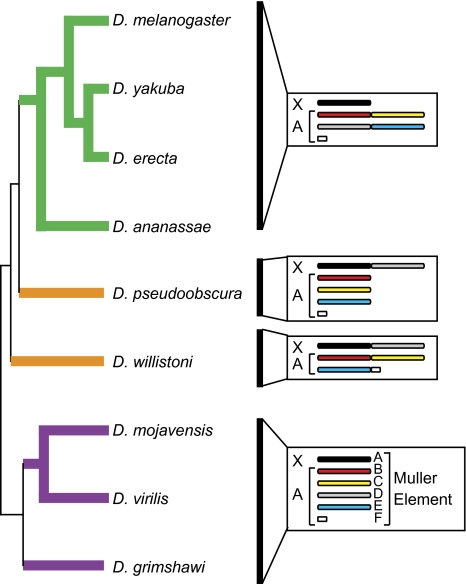
Because previously published analyses of gene duplication from the X chromosome to the autosomes in Drosophila have been limited to only retroposed genes and to only the D. melanogaster genome (Betrán et al. 2002; Dai et al. 2006; Bai et al. 2007), it is unclear whether these patterns of movement hold for all types of duplications and for the entire genus. We identified duplicated and relocated genes in multiple sequenced Drosophila genomes to examine the evolutionary dynamics of gene traffic between X chromosomes and autosomes. In our analysis, we focused on gene duplication events that occurred along multiple different evolutionary lineages, allowing for both retroposed and DNA-based duplications. The species sampled in this genus represent a large swath of evolutionary time, with species from two major subgenera: Drosophila and Sophophora. Furthermore, the sequenced genomes contain two independent X-autosome fusions (fig. 1) (Drosophila 12 Genomes Consortium 2007), allowing us to examine what happens when autosomes become X linked. We find that most lineages are biased for X-to-autosome duplications and that this bias is driven by retroposition in almost all lineages. Our results suggest that the X-to-autosome retroposition is driven by selection for escape from spermatogenic X-inactivation. We also find that an excess of genes was relocated from the X to the autosomes (i.e., duplication followed by loss of the ancestral copies) along multiple lineages, and we conclude that this pattern is not driven by mutational biases. We cannot, however, identify the specific selective force responsible for the excess relocation off the X chromosome.
FIG. 1.—
Phylogeny of some Drosophila species with sequenced genomes and their karyotypes. Lineages upon which duplicated and relocated genes were identified are highlighted in bold. Homologous chromosomes arms (Muller elements) are the same color for all species. X, X chromosome; A, autosome.
Materials and Methods
Identification of Gene Families
Coding sequence annotations from the Drosophila 12 Genomes Consortium (2007) were used in this analysis. Three species were excluded (Drosophila simulans, Drosophila sechellia, and Drosophila persimilis) because they are very closely related to other sampled species and had their genomes sequenced to low coverage. We therefore analyzed genes in the remaining nine species (fig. 1). Briefly, genes in non-D. melanogaster species were identified via a variety of methods, and the gene models were reconciled using GLEAN (Honeybee Genome Sequencing Consortium 2006; Elsik et al. 2007). The GLEAN predictions, along with D. melanogaster gene models (release 4), were assigned to homologous gene families using fuzzy reciprocal Blast (FRB). The FRB gene families were then filtered for known transposable elements and aligned using MUSCLE (Edgar 2004), as described previously (Hahn et al. 2007). Neighbor-Joining trees were built using these alignments, with distances calculated using the amino acid sequences of the genes. The gene trees and species trees were reconciled using NOTUNG (Durand et al. 2006).
Assignment of Scaffolds to Chromosome Arms
Drosophila genomes are organized into five major chromosome arms and a dot chromosome (fig. 1). Each arm is referred to as a Muller element (Muller 1940), and the ancestral karyotype consists of an acrocentric X chromosome (Muller element A), four acrocentric major autosomes (Muller elements B–E), and a small autosome (Muller element F). All the largest scaffolds created in the 12 Genomes Sequencing projects have been assigned to both a Muller element and a chromosome arm (Drosophila 12 Genomes Consortium 2007; Schaeffer et al. 2008). The coordinates of the genes within the scaffolds were used to assign genes to Muller elements A–E for the following species: D. melanogaster, Drosophila yakuba, Drosophila erecta, Drosophila ananassae, Drosophila pseudoobscura, Drosophila willistoni, Drosophila mojavensis, Drosophila virilis, and Drosophila grimshawi. Muller element F (the dot chromosome) was ignored in this analysis because of its small size and because its heterochromatic composition makes it difficult to assemble. Additionally, D. yakuba and D. erecta share a pericentric inversion of chromosome 2 (Muller elements B and C), and a large block of genes from Muller element A are now found XR in D. pseudoobscura (the arm of the X chromosome homologous to Muller element D). We took these large changes into account when counting individual gene duplications between Muller elements.
Identification of Nonlineage-Specific “Retroposed” Duplications in the D. melanogaster Genome
We identified FRB gene families containing two D. melanogaster genes. These were screened for families in which one gene contains multiple exons and the other contains a single exon. The multi-exon gene was inferred to be the ancestral copy, and the single-exon gene the derived copy that arose via retroduplication. Although this method ignores many other signatures of retroposed duplications (Kaessmann et al. 2009), we are limited in our ability to detect those signatures because many of the duplications are very old, and we do not have appropriate outgroups for comparisons.
Identification of Lineage-Specific Duplicated Genes with Both Copies Retained
Lineage-specific gene duplications were identified in two ways. The first method (phylogenetic method) used the phylogenies constructed for each FRB family, with gene duplication events identified by NOTUNG. If a duplication event occurred along one of the lineages of interest (fig. 1), it was retained in the analysis. All duplications that occurred ancestral to the lineages of interest were ignored. Genes on scaffolds that had not been assigned to a Muller element were excluded. Ancestral and derived copies were inferred for inter-Muller element duplications using the chromosomal locations of the orthologs from the other species in the same family. The ancestral copy is the paralog located on the same Muller element as the orthologs from the other species. If neither copy is on the same Muller element as the orthologous genes, the ancestral and derived copies could not be inferred. Finally, duplicated genes were retained only if a homolog is present in the FRB family from both subgenera. A subset of these data was extracted consisting of lineage-specific duplications in which the family had only a single duplication event along a particular lineage (phylogenetic method—two copies).
The second approach (counting method) took advantage of the finding that a maximum likelihood method for identifying duplicated genes using the number of members in each FRB family for each species performs remarkably similar to a phylogenetic method (Hahn et al. 2007). In this method, duplications were identified if two genes from a species—or multiple species derived from a lineage of interest—are in an FRB family, and no more than one gene from each of the other species is in that FRB family (supplementary fig. S1, Supplementary Material online). More details on the counting method can be found in the supplementary methods (Supplementary Material online).
Identifying Lineage-Specific Inter-Chromosome-Arm Duplicated Genes in Which the Ancestral Copy was Lost
A variation of the counting method was used to identify genes that were relocated between Muller elements along the lineages leading to D. melanogaster, D. pseudoobscura, and D. willistoni (fig. 1). Drosophila mojavensis, D. virilis, and D. grimshawi were used as outgroups in all the analyses. Additionally, D. pseudoobscura and D. willistoni were used as outgroups for D. melanogaster, but only D. melanogaster was used as an outgroup for D. pseudoobscura and D. willistoni. A gene relocation was said to have occurred if all genes in an FRB family from the species of interest are located on the same Muller element, whereas all homologs in the outgroup species are found on a different Muller element (supplementary fig. S1 Supplementary Material online). The most likely explanation for this pattern is an interarm duplication event that occurred after the divergence of that species from all five outgroups, followed by the loss of the ancestral copy. Alternatively, the gene could have relocated via a translocation event along the lineage of interest. However, the mechanism of relocation is irrelevant to our analysis. The ancestral location of the gene prior to duplication was inferred based on the location of the homologs in the outgroup species.
Statistical Tests for Excess Numbers of Interarm Duplications and Relocations in Certain Classes
The expected number of genes duplicated between each Muller element in each species was determined based on the number of genes on the Muller element that contains the ancestral copy, the length of the Muller element that contains the derived copy (in sequenced nucleotides), and whether that Muller element is hemizygous in males (these values were estimated for each species individually). We estimate the expected frequency of genes duplicated between any two Muller elements as
 |
where i is the index of the Muller element containing the ancestral copy, j is the index of the Muller element containing the derived copy, Ni is the number of genes on Muller element i, Lj is the length of Muller element j, and Xi and Xj are equal to one if the Muller element is autosomal and 0.75 if it is X linked (cf., Betrán et al. 2002). These frequencies were used to estimate the expected number of interchromosome arm–duplication events in three different ways. First, we estimated the expected number of autosome-to-autosome (A → A) duplications, ancestral X-to-autosome (X → A) duplications, and autosome-to-ancestral X (A → X) duplications. Additionally, the expected numbers of autosome-to-neo-X (A → neo-X), neo-X-to-autosome (neo-X → A), and neo-X-to-ancestral X (neo-X → X) duplications were calculated for D. pseudoobscura and D. willistoni. G-tests for goodness of fit were performed to test whether the observed counts of duplications in each class deviated significantly from the calculated expectations. Second, we estimated the expected number of genes duplicated from the autosomes (both to other autosomes and to the X chromosome, denoted “A→”) and the expected number of genes duplicated from the X chromosome to any other chromosome (denoted “X→”), and we determined if the observed data fit our expectations using G-tests for goodness of fit. For species with neo-X chromosomes, we also included the observed and expected counts of genes duplicated from the neo-X (neo-X→) in our tests for goodness of fit. Third, we estimated the expected number of genes duplicated on to autosomes (→A), on to the ancestral X (→X), and on to the neo-X (→neo-X). These expectations were compared with the observed data using G-tests for goodness of fit.
Inferring the Mechanism of Duplication
Lineage-specific duplicated genes were assigned to one of three classes based on the number of introns in the ancestral and derived copies. Duplications in which both copies have multiple exons were classified as DNA duplications. Those in which the derived copy is a single exon gene and the ancestral copy has at least one intron were classified as retroposed. And duplicated genes in which the ancestral copy has a single exon were classified as ambiguous. We also determined the mechanism of duplication using gene structure in the outgroup species (see supplementary methods, Supplementary Material online). Both methods for determining the mechanism of duplication yielded similar results, and only the results from the first method are presented. Finally, relocated genes were classified as single-exon and multi-exon genes because we were unable to determine whether the ancestral copy was single- or multi-exon.
Tissue-Expression and Sex-Biased Expression Data
Expression data for D. melanogaster genes were taken from FlyAtlas (http://www.flyatlas.org), which has expression data from multiple body parts (Chintapalli et al. 2007). We considered the signal of each gene sampled in brain, thoracicoabdominal ganglia, salivary gland, crop, midgut, Malpighian tubule, hindgut, ovary, testis, accessory gland, larval salivary gland, larval midgut, larval Malpighian tubule, and larval fat body. Tissue specificity of expression for each gene was measured as
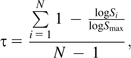 |
where N is the number of tissues, Si is the signal intensity in tissue i, and Smax is the maximum signal intensity of that gene in all tissues (Yanai et al. 2005; Larracuente et al. 2008); larger τ indicates more tissue specificity. We also performed the same calculations excluding only sex-specific tissues (ovary, testis, and accessory gland), excluding only larval tissues, and excluding both sex-specific and larval tissues. Additionally, we considered genes to be testis expressed if they had a signal >100 when measured in testis. We assigned D. melanogaster and D. pseudoobscura genes to one of three classes of sex-biased expression (male biased, female biased, or not sex biased) based on previously published data from whole adult flies (Sturgill et al. 2007; Zhang et al. 2007). No genome-wide expression data are available for D. willistoni, and tissue-specific expression data are not available for D. pseudoobscura.
Results
Drosophila melanogaster Retroposed Genes
It has previously been reported that retrotransposed genes in the D. melanogaster genome tend to arise from X-linked ancestral copies and duplicate to the autosomes (Betrán et al. 2002; Dai et al. 2006). We also observe an excess of X → A retrotransposed duplicates in the D. melanogaster genome (table 1), validating our methods. The observed counts of A → A, A → X, and X → A retroposed duplications do not fit those expected based on the sizes of the chromosomes (Gadj = 37.8, P < 6.1 × 10−9). Additionally, there is an excess of X→ retroposed duplications when compared with A→ retropositions (Gadj = 37.9, P < 7.4 × 10−10). There is not a significant difference between the observed and expected counts of →A and →X retroposed duplications in the D. melanogaster genome (Gadj = 2.79, P = 0.095).
Table 1.
Retroposed Genes between Chromosome Arms in the D. melanogaster Genome
| A → A | A → X | X → A | |
| Observed | 15 | 3 | 22 |
| Expected | 28.03 | 6.59 | 5.38 |
| Anc-testis | 8 | 3 | 11 |
| Dup-testis | 12 | 2 | 21 |
NOTE.—The direction of retroposition is given (A, autosome; X, X chromosome). Anc-testis, ancestral copy is testis expressed; Dup-testis, derived copy is testis expressed.
Two main explanations have been given for the excess retroposition from the D. melanogaster X chromosome: escape from spermatogenic X-inactivation (Betrán et al. 2002) and sexually antagonistic selection against male-favorable genes on the X chromosome (Wu and Xu 2003). If genes retropose from the X to the autosomes to escape X-inactivation in spermatogenesis, we would expect the derived copies on the autosomes to be expressed in the testis. Indeed, 21 of 22 derived copies of X → A retroposed genes are testis expressed (table 1). However, we also observe that testis expression is a common feature of the derived copies of all retroposed duplications, regardless of whether they originate from the X chromosome or the autosomes (table 1). Additionally, single exon genes across the D. melanogaster genome are more likely to be testis expressed than expressed in other body parts (P < 2.2 × 10−16, Fisher’s exact test [F.E.T.]), whereas multiple exon genes do not show such a dramatic excess of testis-expressed genes (supplementary fig. S2 Supplementary Material online). This suggests that new retroposed genes and small genes are preferentially expressed in the testis regardless of whether they arose from an X-linked copy.
Lineage-Specific Duplication from the Ancestral X Chromosome
The previous analysis included all retroposed duplications in the D. melanogaster genome regardless of when the duplication events occurred. We also identified gene duplication events that occurred along lineages that arose after the most recent common ancestor (MRCA) of the genus (fig. 1; supplementary tables S1 and S2 Supplementary Material online). Approximately, 100–300 duplicated genes were identified along each lineage, of which 9–38% were duplicated between Muller elements—different counts were obtained for each lineage and when using different methods to identify duplicated genes (supplementary table S3, Supplementary Material online). Both the phylogenetic and counting methods of identifying lineage-specific duplications yield similar results; therefore, our findings are not an artifact of the method used to identify duplicated genes. The results presented from hereon are from the phylogenetic method only (because this method yields the largest sample sizes of duplicated genes), unless otherwise stated. One possible concern regarding our data is the lack of independence of some of the lineages examined (fig. 1). Indeed, genomes with shared lineages (e.g., D. yakuba and D. erecta or D. mojavensis and D. virilis) often show the same patterns of duplication between the X chromosome and the autosomes (see below). However, we also recover similar patterns from completely independent lineages, suggesting that our results are robust to the minimal lineage overlap in our sample.
We first examined lineages without neo-X chromosomes. There are more X → A duplications than expected based on the sizes of the chromosomes in all lineages (fig. 2; supplementary table S4, Supplementary Material online). To assess whether these excesses are significant, we performed G-tests for the goodness of fit of the observed and expected counts of A → A, A → X, and X → A gene duplications. The D. mojavensis and D. virilis lineages do not have a sufficient number of events to assess significance. There is a significant excess of X → A duplications in the D. melanogaster genome, regardless of the method used to identify the duplicated genes. A significant excess of X → A duplications is also observed using data from most of the methods used for identifying duplicated genes in the D. yakuba, D. erecta, D. ananassae, and D. grimshawi genomes (supplementary table S4, Supplementary Material online).
FIG. 2.—
Lineage-specific interarm duplications. Duplicated genes were identified using the phylogenetic method. Gray bars are the observed counts and black bars are the expected counts (based on chromosome sizes and hemizygosity). X, ancestral X chromosome; A, autosome; neo-X, neo-X chromosome. Arrows indicated direction of duplication event.
We also determined whether a significant excess of genes was duplicated from the X chromosome along the lineages without neo-X chromosomes by comparing the observed and expected counts of A→ and X→ gene duplications (supplementary table S5, Supplementary Material online). There is a significant excess of X→ duplications along the lineages leading to D. melanogaster, D. yakuba, D. erecta, D. ananassae, and D. grimshawi using most methods of identifying duplicated genes (supplementary table S5, Supplementary Material online). There is not a significant difference between the observed and expected counts of →A and →X duplications (supplementary table S6, Supplementary Material online).
We examined the expression profiles of the ancestral and derived copies of lineage-specific duplicated genes in the D. melanogaster genome (Chintapalli et al. 2007). The duplicated genes were identified using the phylogenetic method, limited to a single duplication event per lineage. The derived copies tend to have more tissue-specific expression than the ancestral copies (as determined by a Wilcoxon test using paired samples), regardless of whether we consider all tissues (P < 0.0005), only tissues found in adults (P < 0.0005), only tissues found in both sexes (P < 0.001), or only adult tissues found in both sexes (P < 0.0005). The majority of the derived copies (19/33) are expressed at higher levels in testis than in any other body part examined, whereas only nine of the ancestral copies are expressed most highly in testis (P < 0.05, F.E.T.). Additionally, a significant excess of derived copies of lineage-specific X → A duplicated genes in the D. melanogaster genome is testis expressed relative to A → A duplicated genes (P < 0.05, F.E.T.); the same pattern is not observed for the ancestral copies (P = 0.26, F.E.T.) (table 2). There are also data on sex-biased expression in whole flies from D. melanogaster (Sturgill et al. 2007). There is no evidence that the derived copies of X → A duplicated genes in D. melanogaster are more likely to have male-biased expression than the derived copies of A → A duplications (supplementary table S7, Supplementary Material online).
Table 2.
Mechanism of Duplication and Testis Expression of D. melanogaster Lineage-Specific Inter-Chromosome-Arm Duplicated Genes
| DNA Duplication |
Ambiguous |
Retroposed |
||||||||||
| Ancestral |
Derived |
Ancestral |
Derived |
Ancestral |
Derived |
|||||||
| N | Y | N | Y | N | Y | N | Y | N | Y | N | Y | |
| A → A | 4 | 2 | 5 | 1 | 1 | 3 | 2 | 2 | 1 | 2 | 0 | 3 |
| A → X | 1 | 1 | 2 | 0 | 3 | 0 | 3 | 0 | 0 | 2 | 1 | 1 |
| X → A | 1 | 3 | 0 | 4 | 0 | 1 | 0 | 1 | 5 | 6 | 1 | 10 |
NOTE.—Duplicated genes were identified using the phylogenetic method, limited to two copies per lineage. N, not testis expressed; Y, testis expressed.
The molecular mechanism by which a gene was duplicated can be inferred based on the number of exons in the two copies (see Materials and Methods). Duplicated genes were identified using the phylogenetic method, but we limit our analysis to those families with a single duplication event along the lineage. In most lineages without neo-X chromosomes, retroposition accounts for the majority of X → A duplications, whereas A → A duplications are driven by a combination of the two mechanisms (fig. 3; supplementary table S8, Supplementary Material online). To assess whether these deviations are significant, a G-test for goodness of fit was performed using the observed counts of duplications and those expected based on chromosome sizes. We examine all lineages without neo-X chromosomes, but again the D. virilis and D. mojavensis lineages were not tested because of small numbers of duplicated genes. When looking at retroposed duplications only, there is a significant excess of X→ duplicated genes in all of the lineages examined. However, for DNA duplications, there is not a significant excess of X→ duplications in any lineage. This result indicates that the excess duplication from the X chromosome in these lineages is due to retroposition. Consistent with our earlier analysis of all retroposed duplications in the D. melanogaster genome, nearly all of the lineage-specific retroposed genes in D. melanogaster have derived copies that are testis expressed, regardless of whether they were A→ or X→ duplications (table 2). Interestingly, there is also a significant excess of testis-expressed derived copies of X→ DNA duplications when compared with A→ DNA duplications (P < 0.05, F.E.T.). Therefore, it appears that different dynamics influence the testis expression of derived copies of retroposed and DNA duplications that originate from X-linked ancestral copies.
FIG. 3.—
Mechanisms of lineage-specific gene duplications. Observed counts of the number of DNA duplications (black), retroposed duplications (white), and ambiguous duplications (gray) are shown for interarm duplications between X chromosomes and autosomes. Duplicated genes were identified using the phylogenetic method, limited to two copies per lineage.
Gene Relocation from the D. melanogaster X Chromosome
If a gene is duplicated from one Muller element to another, and the ancestral copy is subsequently lost, it will appear as if that gene was relocated along the lineage upon which the duplication and loss occurred. The observed numbers of A → A, A → X, and X → A relocated genes in the D. melanogaster genome do not fit the expected counts based on the sizes of the chromosome (Gadj = 23.2, P < 9.2 × 10−6) because of an excess of X → A relocations (fig. 4). Additionally, there is an excess of X→ relocated genes relative to A→ relocations (Gadj = 21.7, P < 3.2 × 10−6), and there is a deficiency of →X relocated genes (Gadj = 4.54, P < 0.05). A higher frequency of single-exon genes, relative to multi-exon genes, are X→ relocations when compared with single- and multiexon A→ relocated genes (P < 0.005, F.E.T.) (supplementary table S9, Supplementary Material online). There is an excess of single-exon X→ relocated genes when compared with the expected counts of X→ and A→ relocated genes (Gadj = 27.4, P < 1.7 × 10−7), but there is not a significant difference between the observed and expected counts of multi-exon X→ and A→ relocated genes (Gadj = 2.38, P = 0.12). This suggests that retroposition may drive the excess relocation off the D. melanogaster X chromosome.
FIG. 4.—
Lineage-specific gene relocation between chromosome arms. The observed (gray) and expected (black) counts of relocated genes are shown for all possible directions of relocation. A, autosome; X, ancestral X chromosome; neo-X, neo-X chromosome.
Our analysis of retroposed duplicates indicates that the derived copies tend to be testis expressed. We interrogated the relocated genes to see if the X → A relocated genes are more likely to be testis expressed or have male-biased expression than A → A relocated genes. We found that X→ relocated genes are no more likely to be testis expressed than A→ relocated genes (P = 0.51, F.E.T.) (table 3; supplementary table S9, Supplementary Material online). Additionally, relocated genes have significantly broader expression than the derived copies of duplicated genes (P < 0.0005, Wilcoxon test), whereas there is not a significant difference in tissue specificity between relocated genes and ancestral copies of duplicated genes (P = 0.904, Wilcoxon test). This makes intuitive sense as relocated genes must be able to perform all of the functions carried out by the original single-copy gene. We also find that there are more male-biased X→ relocated genes than female-biased X→ relocated genes, while there is an equal number of male- and female-biased A→ relocated genes (table 3); however, this difference is not significant (P = 0.144, F.E.T.).
Table 3.
Testis- and Sex-Biased Expression of Lineage-Specific Relocated Genes
| Testis Expr |
Sex Bias |
||||
| N | Y | F | M | NA | |
| D. melanogaster | |||||
| A → A | 20 | 17 | 6 | 6 | 25 |
| A → X | 5 | 0 | 0 | 0 | 5 |
| X → A | 15 | 9 | 1 | 6 | 17 |
| D. pseudoobscura | |||||
| A → A | 0 | 4 | 6 | ||
| A → neo-X | 0 | 3 | 2 | ||
| A → X | 4 | 1 | 6 | ||
| neo-X → A | 1 | 7 | 10 | ||
| neo-X → X | 0 | 0 | 4 | ||
| X → A | 2 | 3 | 6 | ||
| X → neo-X | 0 | 0 | 0 | ||
NOTE.—Testis expr, whether or not the gene is testis expressed; sex-bias, whether the gene has female-biased expression (F), male-biased expression (M), or no significant sex-biased expression (NA). A, autosome; X, ancestral X chromosome; neo-X, neo-X chromosome; arrows indicate direction of relocation.
Gene Duplication and Relocation from Neo-X Chromosomes
Drosophila pseudoobscura and D. willistoni each have neo-X chromosome arms that arose via the independent fusion of Muller element D with the ancestral X chromosome along the lineages leading to those species (fig. 1). We refer to Muller element D in these species as the neo-X chromosome. There are more neo-X→ duplicated genes in the D. pseudoobscura and D. willistoni genomes than expected based on the sizes of those chromosomes (fig. 2; supplementary tables S10 and S11 Supplementary Material online). To determine whether these excesses are significant, we performed G-tests for goodness of fit comparing the observed and expected counts of A → A, A → neo-X, A → X, neo-X → A, and X → A duplications (we ignore the neo-X → X and X → neo-X duplications because of small observed and expected counts). There is a poor fit between the observed and expected counts in D. pseudoobscura (Gadj = 45.9, P < 1.3 x 10-11) and D. willistoni (Gadj = 15.2, P < 0.0005) (supplementary table S10, Supplementary Material online). Additionally, the observed counts of A→, X→, and neo-X→ duplicated genes do not fit the expected counts in D. pseudoobscura (Gadj = 32.3, P < 9.7 x 10-8) or D. willistoni (Gadj = 9.59, P < 0.01) (supplementary table S11, Supplementary Material online). To determine whether the poor fit is because of the excess X→ or neo-X→ duplications, we compared the observed and expected counts of X→ and neo-X→ duplications individually with the A→ duplications. If we only look at A→ and X→ gene duplications, there is a significant excess of X→ duplicated genes in both D. pseudoobscura (Gadj = 9.31, P < 0.005) and D. willistoni (Gadj = 7.60, P < 0.01). There is also a significant excess of neo-X→ gene duplications in D. pseudoobscura (Gadj = 29.4, P < 6.1 x 10-8) and D. willistoni (Gadj = 4.10, P < 0.05) when compared with the observed and expected number of A→ duplications. Finally, the observed numbers of →A, →X, and →neo-X duplicated genes fit our expectations using most methods of identifying duplicated genes (supplementary table 12, Supplementary Material online). In summary, there is a significant excess of X→ and neo-X→ duplicated genes in D. pseudoobscura and D. willistoni but no deficiency of genes duplicated on to those chromosomes.
D. pseudoobscura belongs to the obscura group, which has three subgroups: obscura, pseudoobscura, and affinis. All species in the pseudoobscura and affinis subgroups share the same X-autosome fusion, whereas none of the obscura subgroup species have the fusion (Patterson and Stone 1952; Steinemann et al. 1984). Therefore, the X-autosome fusion occurred after the divergence of the obscura subgroup from the pseudoobscura and affinis subgroups but prior to the split between the pseudoobscura and affinis subgroups. Phylogenies built from the sequences of Adh and Gpdh give conflicting branching orders of the three subgroups (Russo et al. 1995; Wells 1996), indicating that the lineage upon which the X-autosome fusion occurred is quite short. These two genes have approximately 0.2 < dS < 0.4 between the subgroups. Therefore, duplicated genes that arose after the X-autosome fusion should have dS < 0.4. The median synonymous divergence between neo-X → A duplications is significantly less than that of X → A duplications (P < 0.01, Mann–Whitney test) (fig. 5). There is an excess of neo-X → A duplicated genes with dS < 0.3 relative to X → A duplications in the D. pseudoobscura genome (P < 0.05, F.E.T.) (fig. 5). This seems to indicate that the burst of duplication from the D. pseudoobscura neo-X followed the X-autosome fusion. The same analysis cannot be performed for genes duplicated from the D. willistoni neo-X because this X-autosome fusion is shared by all species in the willistoni group, precluding an accurate dating of the event (Ehrman and Powell 1982).
FIG. 5.—
A burst of duplication followed the creation of the neo-X chromosome in Drosophila pseudoobscura. The number of paralogs with divergence between copies is graphed for three classes of duplicated genes in the D. pseudoobscura genome: autosome-to-autosome (large dashes, hollow diamonds), ancestral X-to-autosome (solid line, solid diamonds), and neo-X-to-autosome (small dashes, hollow squares). Duplicated genes were identified using the phylogenetic method.
To determine what factors drive the excess duplication from the D. pseudoobscura neo-X chromosome, we used previously published analyses of genes with sex-biased gene expression measured in whole flies (Sturgill et al. 2007). There is no evidence that the derived copies of X → A or neo-X → A duplications have a disproportionate frequency of genes with male-biased expression (supplementary table S7, Supplementary Material online). However, →A duplicated genes are more likely to have derived copies with sex-biased expression than →X or →neo-X duplications (P < 0.0001, F.E.T.). Some factor appears to prevent the accumulation of new sex-biased genes on the D. pseudoobscura X chromosome.
The mechanisms of duplication also reveal information regarding the forces driving X→ and neo-X→ gene duplication. In the species without neo-X chromosomes, we observed that retroposition is primarily responsible for the excess X→ duplication. In D. pseudoobscura and D. willistoni, there are approximately equal numbers of X→ DNA duplications and retroposed duplications (fig. 3; supplementary table S13, Supplementary Material online). However, the majority of neo-X → A duplications in D. pseudoobscura arose via a DNA-based mechanism (fig. 3). In D. willistoni, there are more neo-X→ retroposed genes than neo-X→ DNA duplications (fig. 3). To assess the significance of these differences, we tested whether the observed counts of A→, X→, and neo-X→ retroposed and DNA duplications fit those expected based on the sizes of the chromosomes. The observed counts of A→, X→, and neo-X→ retroposed duplications significantly differ from the expected counts in both D. pseudoobscura (Gadj = 16.6, P < 0.0005) and D. willistoni (Gadj = 16.0, P < 0.0005) because of an excess of X→ and neo-X→ retroposed genes. In D. pseudoobscura only, the observed counts of A→, X→, and neo-X→ DNA duplications significantly differ from the expected counts (Gadj = 18.1, P < 0.0005) because of an excess of neo-X→ DNA duplications. The large amount of DNA duplications along the D. pseudoobscura lineage may be the result of a repeat sequence unique to the D. pseudoobscura genome (Richards et al. 2005), which appears to be involved in generating DNA duplications (Meisel 2009b).
We also identified genes that had been relocated between chromosome arms in D. pseudoobscura and D. willistoni, and we compared the observed counts with those expected based on the sizes of the chromosomes (fig. 4). There are more neo-X→ relocated genes than expected in the D. pseudoobscura and D. willistoni genomes and more X→ relocated genes in the D. pseudoobscura genome (fig. 4). However, there are fewer X→ genes than expected in D. willistoni (fig. 4). To determine if any of these differences are significant, we again performed G-tests for goodness of fit between the observed and expected counts of A → A, A → neo-X, A → X, neo-X → A, and X → A (we exclude X → neo-X and neo-X → X relocated genes because of low counts). The observed counts significantly differ from the expected counts in both D. pseudoobscura (Gadj = 29.5, P < 5.6 × 10−8) and D. willistoni (Gadj = 10.5, P < 0.005). Additionally, the observed counts of A→, X→, and neo-X→ relocated genes differ from the expected counts in D. pseudoobscura (Gadj = 22.8, P < 1.2 × 10−5) and D. willistoni (Gadj = 11.3, P < 0.005) because of an excess of neo-X→ relocated genes in both species. If we compare the observed and expected counts of X→ and A→ relocated genes, there is a significant excess of X→ relocated genes in D. pseudoobscura (Gadj = 3.99, P < 0.05) and a deficiency of X→ relocated genes D. willistoni (Gadj = 4.24, P < 0.05). It is unclear why there would be excess relocation off the ancestral X in D. melanogaster and D. pseudoobscura but not in D. willistoni.
We can also use the calls of sex-biased gene expression in D. pseudoobscura (Sturgill et al. 2007) to examine the forces responsible for the excess neo-X→ relocation. Neo-X→ relocated genes are no more or less likely to have sex-biased expression than X→ or A→ relocated genes (table 3). However, whether a relocated gene has male-biased expression is not independent of whether it is the result of →A or →X relocation (P < 0.05, F.E.T.) because of a deficiency of →X relocated genes with male-biased expression (table 3). Additionally, the mechanism of relocation may also reveal insights into the factors driving the relocation of genes from the neo-X to the autosomes. The majority of genes relocated between chromosome arms in D. pseudoobscura and D. willistoni have multiple exons (supplementary table S14, Supplementary Material online), and there is no evidence that single-exon genes are more likely to be neo-X→ relocations in the D. pseudoobscura genome. This suggests that DNA intermediates, and not retroposition, are responsible for most of the relocation events. However, there is a higher frequency of single-exon neo-X→ relocated genes in D. willistoni when compared with the number of single- and multi-exon A→ relocations (P < 0.05, F.E.T.). Therefore, the excess neo-X→ relocation in D. willistoni may be driven by excess neo-X→ retroposition relative to A→ retroposition. These results are consistent with the inferred mechanisms of duplication between chromosome arms, where retroposed and DNA duplications are responsible for the excess neo-X→ duplication in D. pseudoobscura, and only retroposition is responsible for the excess neo-X→ duplication in D. willistoni neo-X (fig. 3).
The X-autosome fusions in D. pseudoobscura and D. willistoni are most likely the result of independent events because a large number of lineages that arose after the MRCA of those two species do not have the same neo-X chromosome. There would be strong evidence that the excess neo-X→ duplication and relocation in these two species is driven by a common mechanism or evolutionary force if homologous genes were duplicated or relocated from the neo-X chromosome in both species. We considered genes from D. pseudoobscura and D. willistoni homologous if they are in the same FRB family. Of the 50 genes which were either neo-X → A duplications or relocations along the D. pseudoobscura lineage (combining all methods of identifying duplicated genes), nine have a homolog that is a neo-X → A duplicated or relocated gene in the D. willistoni genome. Additionally, one gene that was neo-X → A duplicated gene in D. pseudoobscura has a neo-X → X relocated homolog in D. willistoni. Genes can also be lost from a neo-X chromosome without ever being duplicated. The change in cellular dynamics accompanying X-linkage—that is, dosage compensation (Straub and Becker 2007) and spermatogenic X-inactivation (Hense et al. 2007)—may not be favorable for certain genes. Additionally, X-linked genes are under different selection pressures than autosomal genes (Vicoso and Charlesworth 2006), which may allow for the neutral or selective loss of neo-X-linked genes. If those genes are not duplicated to an autosome, they will be lost from the genome if they are lost from the neo-X chromosome. Seven neo-X → A duplicated or relocated genes in the D. pseudoobscura genome do not have homologs in D. willistoni, indicating that they may have been lost from the D. willistoni neo-X. In comparison, none of the 32 X → A duplicated or relocated genes in D. pseudoobscura have homologs that are X → A duplications or relocations in D. willistoni. And only two X → A duplicated or relocated genes along the D. pseudoobscura lineage were lost from the D. willistoni ancestral X chromosome arm. In summary, there is a significant excess of genes that independently moved from the D. pseudoobscura neo-X and the D. willistoni neo-X relative to those that moved from the ancestral X chromosomes in these lineages (P < 0.005, F.E.T.).
Our inference of homologous genes moving from independent neo-X chromosomes in D. pseudoobscura and D. willistoni may be the result of events that occurred prior to the divergence of the two species. In this case, we would be falsely inferring the duplications and relocations as lineage specific. However, eight of common the neo-X→ duplicated and relocated genes have a derived copy on a different chromosome arm in the two species, whereas only three of the common neo-X→ duplicated and relocated genes have derived copies on the same chromosome arm in the two species. This indicates that most or all the common duplication and relocation events were independent. Additionally, the genes missing from the D. willistoni neo-X chromosome may be the by-product of 25% lower sequencing coverage of the X chromosome. However, if this were the case, we expect that many of the X→ duplicated and relocated genes in D. pseudoobscura would be missing from the D. willistoni ancestral X, but they are not. Therefore, it is unlikely that the observed patterns are the result of sequencing coverage.
Discussion
It has been previously observed that the D. melanogaster X chromosome is a disproportionate source of retroposed genes (Betrán et al. 2002; Dai et al. 2006), and two selective mechanisms have been presented to explain this pattern. The autosomal derived copies may be retained because they are expressed during spermatogenic X-inactivation (Betrán et al. 2002). Alternatively, male-favorable genes may preferentially accumulate on the autosomes because of sexually antagonistic selection (Wu and Xu 2003). We examined patterns of lineage-specific gene duplication between X chromosomes and autosomes throughout the Drosophila genus to gain a better understanding of forces responsible for the excess duplication from the X chromosome. Many of the sampled lineages have an excess of genes duplicated from the ancestral X chromosome (fig. 2), but the magnitude of that excess varies substantially between lineages. There has also been excess duplication from the D. pseudoobscura and D. willistoni neo-X chromosomes (fig. 2). An excess of genes was relocated off the ancestral X chromosome in D. melanogaster and D. pseudoobscura and the neo-X chromosome in D. pseudoobscura and D. willistoni but not the ancestral X in D. willistoni (fig. 4). The excess duplication from the X chromosome is due mostly to retrotransposition, although there is also an excess of DNA duplications from the D. pseudoobscura neo-X (fig. 3). The excess duplication from the X chromosomes along many of the lineages examined gives us the power to identify the forces responsible for this pattern.
X chromosomes are inactivated, premeiotically, in D. melanogaster spermatogenesis (Hense et al. 2007). X-linked genes with potentially beneficial functions if expressed during the period of X-inactivation must be duplicated or relocated to autosomes for the organism to realize the benefit. The derived copies of retroposed duplications from the X chromosome in the D. melanogaster genome have been shown to be testis expressed (Betrán et al. 2002). However, we find that testis expression is a general property of the derived copies of all retroposed genes in D. melanogaster, not just those that are derived from the X chromosome (tables 1 and 2). Given the propensity for all retroposed genes to be testis expressed, how can we explain the marked excess of X → A retroposed genes? The possibility that the X → A bias is the result of mutational pressure has been rejected previously (Betrán et al. 2002), but that study did not consider the role that hypertranscription of the X chromosome in the male germ line (Gupta et al. 2006) may play in X → A retroposition. Consistent with the mutational hypothesis, DNA-based duplicated genes do not show as striking a bias for X → A events as retroposed duplications in D. melanogaster and most other lineages (fig. 3). However, there is an excess of DNA duplications from the D. pseudoobscura neo-X chromosome, suggesting that hypertranscription is not a likely explanation for the X → A bias in all species. Additionally, retroposed genes in D. melanogaster do not have an excess of testis-expressed ancestral copies (table 1), which would be predicted if hypertranscription during spermatogenesis were responsible for the excess X→ duplication. Finally, an independent collection of recently duplicated genes in the D. pseudoobscura genome contains both apparently functional and pseudogenized duplications (Meisel 2009a). If we look only at inter-chromosome-arm duplications, there are six X→ and neo-X→ functional duplicated genes, two functional A→ duplicated genes, seven X→ or neo-X→ pseudogenes, and 30 A→ pseudogenes (P < 0.005, F.E.T.). Because the pseudogenes do not show any bias for duplication from the X chromosome, the excess duplication from the D. pseudoobscura X chromosome is unlikely to be driven by mutational pressure.
We hypothesize that the bias in favor of X → A duplication in D. melanogaster may be driven by selection favoring escape from X-inactivation, despite the fact that the derived copies of A → A retroposed genes are also testis expressed. In mammals, there is an excess of X → A retroposed genes (Emerson et al. 2004), and the expression profile of many of these genes also suggests that the autosomal derived copies were preferentially retained because they escape spermatogenic X-inactivation (Bradley et al. 2004; Dass et al. 2007; Potrzebowski et al. 2008). Additionally, young retroposed genes in humans tend to be testis expressed (Vinckenbosch et al. 2006), indicating that testis expression is a common feature of all new retroposed genes. Furthermore, de novo genes in Drosophila are also testis expressed and, surprisingly, often X linked (Levine et al. 2006; Begun et al. 2007). If new genes (especially small genes and those that have been retroposed) and single exon genes tend to be testis expressed, this could explain the excess of X → A retroposed genes in Drosophila. The derived autosomal copy would have a high likelihood of testis expression and would be preferentially retained if the X-linked ancestral copy would have conferred a fitness benefit if expressed during spermatogenic X-inactivation. Also, the derived copies of X → A DNA duplications and ambiguous duplications tend to be testis expressed (table 2), suggesting that escape from X-inactivation may drive all of the excess duplication from the D. melanogaster X chromosome. Autosomal genes could also be selectively retained when retroposed to another autosome if they would confer a selective benefit if testis expressed. However, because the autosomes are not inactivated during meiosis, autosomal genes do not require retroposition to gain expression during the period of X-inactivation (they can gain testis expression by obtaining a testis-specific promoter). Therefore, because testis expression is so common for the derived copies of retroposed genes, it offers a way for X-linked genes that would have beneficial functions in spermatogenesis to gain such a function. In a sense, the testis acts as a “proving ground” for new genes (Vinckenbosch et al. 2006), and X → A retroposed genes are more likely to be selectively retained because they perform a unique function unavailable to the ancestral copy.
The forces driving the relocation of genes off the X chromosome (i.e., duplication followed by loss of the ancestral copy) appear to differ from those that drive the duplication of genes from the X chromosome to the autosomes (i.e., duplication with retention of the ancestral copy). There is a significant excess of X → A gene relocations in D. melanogaster and D. pseudoobscura, but not in D. willistoni (fig. 4), consistent with a paper that appeared while our manuscript was under review (Vibranovski et al. 2009). Additionally, both the D. pseudoobscura and D. willistoni neo-X chromosome are a disproportionate source of relocated genes (fig. 4). Whereas X→ relocations in D. melanogaster and neo-X→ relocations in D. willistoni tend to be intronless, suggesting they may have been retroposed, the neo-X→ relocations in D. pseudoobscura have multiple exons. This suggests that retroposition drives gene relocation from the X chromosome in most lineages, but DNA duplications are responsible for relocation off the X in D. pseudoobscura, consistent with other results (Vibranovski et al. 2009).
The X→ relocated genes in D. melanogaster do not show any bias for testis expression (table 3), and they have broader expression profiles than the derived copies of duplicated genes. This suggests that gene relocation is not driven by spermatogenic X-inactivation. That does not rule out the possibility that these genes were relocated to the autosomes because of selection against X-linked genes that are favorable to males but harmful to females (Wu and Xu 2003; Vicoso and Charlesworth 2006). However, there is not a significant excess of X → A male-biased relocated genes, relative to female-biased genes, when compared with A → A relocated genes (table 3). Interestingly, few of the →X relocated genes in D. pseudoobscura have male-biased expression (table 3), providing some evidence that there is selection against male-biased genes on the X chromosome. Additionally, when we look at duplicated genes in which both copies are retained, derived copies of →neo-X duplicated genes in D. pseudoobscura are less likely to have sex-biased expression (both male and female biased) than →A duplications (supplementary table S7, Supplementary Material online). This is interesting because models of sexually antagonistic selection predict more genes responsible for sexually dimorphic traits on the X chromosome (Rice 1984). It is possible that sex-biased gene expression is not an adequate proxy for whether a gene performs a sexually antagonistic function.
The independently derived neo-X chromosomes of D. pseudoobscura and D. willistoni provide an opportunity to examine the forces responsible for the excess duplication from Drosophila X chromosomes. Homologous genes were independently duplicated from, relocated off, and lost from these neo-X chromosomes. Mutational pressures cannot explain this phenomenon because the mechanisms driving the duplication and relocation of these genes differ—D. willistoni neo-X → A duplications and relocations appear to be retroposed, whereas in D. pseudoobscura, there is an excess of neo-X→ duplication for both DNA-based and retroposed duplications (fig. 3). Furthermore, homologous genes were duplicated from the neo-X in one species and then lost from the neo-X in the other species (without ever being duplicated). Gene duplication and gene loss are not expected to affect the same genes by mutational pressure alone. However, it is unclear what selection pressures are acting on these genes to favor their duplication and/or loss from the neo-X.
There is no evidence that genes with male-biased expression are preferentially relocated from the D. pseudoobscura neo-X chromosome (table 3). This seems to contradict a previous analysis of the same expression data (Sturgill et al. 2007), which found that genes with male-biased expression are more likely to be relocated or lost from the D. pseudoobscura neo-X chromosome than male-biased genes on an autosome. Sturgill et al. (2007) began by identifying genes with ancestrally sex-biased expression using the extant outgroup expression profiles to infer the ancestral expression profile prior to the X-autosome fusion. They then determined if these genes are more likely to be relocated or lost from the D. pseudoobscura neo-X than autosomal genes with ancestrally sex-biased expression. In contrast, we began by identifying genes that had been relocated from the neo-X chromosome and then determined whether those genes currently have sex-biased expression. The methodological differences between our study and that of Sturgill et al. (2007) could explain our apparently conflicting results.
Does mutational pressure, X-inactivation, or sexual antagonism drive the traffic of genes from Drosophila X chromosomes? The answer most likely depends on the lineage in question, the retention of the ancestral copy, the molecular mechanism by which the initial duplication occurs, and the age of the X chromosome. We find no evidence that mutational pressure drives the excess duplication from X chromosomes. The X → A retroposed duplications in D. melanogaster provide strong evidence in favor of selection for escape from spermatogenic X-inactivation favoring the retention of the autosomal derived copies. However, X → A relocated genes do not show a bias for testis expression. Furthermore, DNA duplications, not retroposition, drive the excess neo-X → autosome duplications along the D. pseudoobscura lineage. Either genes driven off the D. pseudoobscura neo-X chromosome are favored for some other reason than escape from X-inactivation or the derived copies can gain testis expression without being retroposed—similar to the X → autosome DNA duplications in D. melanogaster. Unfortunately, not much is known about X-inactivation in D. pseudoobscura (Lifschytz and Lindsley 1972), and no testis-expression data are available for this species. Finally, it is also possible that the X → A and neo-X → A relocated genes are selectively retained because they perform male-favorable functions (Vicoso and Charlesworth 2006). If these genes are under selection for functions that are beneficial to males but detrimental to females, selection in males could favor their relocation to the autosomes. Although there is no evidence that X → A relocated genes have male-biased expression, sex-biased gene expression is a very coarse measure of sex differences. Further experimentation is needed to examine the functions of genes relocated from Drosophila X chromosomes to determine if they are under sexually antagonistic selection. We therefore conclude that duplication via retroposition from Drosophila X chromosomes with retention of both copies is often driven by selection for testis expression in the derived copy because the X-linked ancestral copy is silenced in spermatogenesis. Relocated genes are more pleiotropic in their expression, and the excess relocation off the X chromosome is driven by a process other than selection for testis expression.
Supplementary Material
Supplementary methods, figures S1 and S2, and tables S1–S14 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by National Science Foundation Doctoral Dissertation Improvement [DEB-0608186 to R.P.M.]; National Institutes of Health [R01-GM076643 to S. Nuzhdin and M.W.H., F32-GM087611 to R.P.M.]; and National Science Foundation [DBI-0543586 to M.W.H.].
Supplementary Material
Acknowledgments
We would like to thank Stephen W. Schaeffer, Hiroshi Akashi, and Leonie Moyle for helpful discussion. Arjun Bhutkar and Stephen W. Schaeffer provided data that allowed us to map the scaffolds to chromosomes arms. Chung-I Wu and three anonymous reviewers provided useful comments that greatly improved the manuscript. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
References
- Bai Y, Casola C, Feschotte C, Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics. 2007;176:1131–1137. doi: 10.1534/genetics.106.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Baltus A, Skaletsky H, Royce-Tolland M, Dewar K, Page DC. An X-to-autosome retrogene is required for spermatogenesis in mice. Nat Genet. 2004;36:872–876. doi: 10.1038/ng1390. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Dai H, Yoshimatsu TF, Long M. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene. 2006;385:96–102. doi: 10.1016/j.gene.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, MacDonald CC. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci USA. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Durand D, Halldorsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol. 2006;13:320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrman L, Powell JR. The Drosophila willistoni species group. In: Ashburner M, Carlson HL, Thompson JN Jr, editors. The genetics and biology of Drosophila. Vol. 3b. London: Academic Press; 1982. pp. 193–225. [Google Scholar]
- Elsik C, Mackey A, Reese J, Milshina N, Roos D, Weinstock G. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko O, Malley J, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han S-G. Gene family evolution across 12 Drosophilagenomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophilaspermatogenesis. PLoS Genet. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee M-H, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Evolutionary dynamics of recently duplicated genes: selective constraints on diverging paralogs in the Drosophila pseudoobscuragenome. J Mol Evol. 2009a;69:81–93. doi: 10.1007/s00239-009-9254-1. [DOI] [PubMed] [Google Scholar]
- Meisel RP. Repeat mediated gene duplication in the Drosophila pseudoobscura genome. Gene. 2009b;438:1–7. doi: 10.1016/j.gene.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Bearings of the ‘Drosophila’ work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JT, Stone WS. Evolution in the genus Drosophila. New York: The Macmillan Company; 1952. [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jégou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann M, Pinsker W, Sperlich D. Chromosome homologies within the Drosophila obscura group probed by in situ hybridization. Chromosoma. 1984;91:46–53. [Google Scholar]
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS. Nucleotide variation at the Gpdh locus in the genus Drosophila. Genetics. 1996;143:375–384. doi: 10.1093/genetics/143.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Xu EY. Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.