Abstract
In plants, an oligogene family encodes NADP-malic enzymes (NADP-me), which are responsible for various functions and exhibit different kinetics and expression patterns. In particular, a chloroplast isoform of NADP-me plays a key role in one of the three biochemical subtypes of C4 photosynthesis, an adaptation to warm environments that evolved several times independently during angiosperm diversification. By combining genomic and phylogenetic approaches, this study aimed at identifying the molecular mechanisms linked to the recurrent evolutions of C4-specific NADP-me in grasses (Poaceae). Genes encoding NADP-me (nadpme) were retrieved from genomes of model grasses and isolated from a large sample of C3 and C4 grasses. Genomic and phylogenetic analyses showed that 1) the grass nadpme gene family is composed of four main lineages, one of which is expressed in plastids (nadpme-IV), 2) C4-specific NADP-me evolved at least five times independently from nadpme-IV, and 3) some codons driven by positive selection underwent parallel changes during the multiple C4 origins. The C4 NADP-me being expressed in chloroplasts probably constrained its recurrent evolutions from the only plastid nadpme lineage and this common starting point limited the number of evolutionary paths toward a C4 optimized enzyme, resulting in genetic convergence. In light of the history of nadpme genes, an evolutionary scenario of the C4 phenotype using NADP-me is discussed.
Keywords: gene duplication, molecular convergence, evolutionary constraint, genetic adaptation, multiple origins
Introduction
C4 photosynthesis is an improvement over the classical C3 carbon acquisition, which evolved more than 50 times independently in at least 18 flowering plant families (Sage 2004; Muhaidat et al. 2007). In the C4 pathway, atmospheric CO2 is fixed in the mesophyll cells by the phosphoenolpyruvate carboxylase (PEPC). The resulting four-carbon acids are then transformed and transported into the bundle–sheath layer cells, where their decarboxylation releases CO2 for the Calvin–Benson cycle. This creates a CO2 pump that, by concentrating CO2 around Rubisco, decreases photorespiration rates and is thus beneficial, especially under high air temperature and low CO2 concentrations (Ehleringer et al. 1997; Sage 2004). Despite being overall convergent, the C4 photosynthetic trait greatly varies among plant taxa, both anatomically and biochemically (Sinha and Kellogg 1996; Dengler and Nelson 1999; Muhaidat et al. 2007). Three different C4 biochemical subtypes are traditionally defined according to the decarboxylating enzyme they use (Gutierrez et al. 1974; Prendergast et al. 1987): the NADP-malic enzyme (NADP-me), NAD-malic enzyme (NAD-me) or phosphoenolpyruvate carboxykinase (PCK). The NADP-me subtype is the most widespread (Sage et al. 1999), being present both among dicots and monocots. In the grass family (Poaceae), which contains 60% of all C4 species, this subtype is present in all C4 lineages defined in Christin et al. (2008a) except in subfamily Chloridoideae (lineages 3 and 4).
C4 photosynthesis is an evolutionary puzzle, having emerged independently a high number of times despite its apparent complexity. In leaves of maize, a C4 grass, 18% of the genes are differentially expressed in M and BS cells, suggesting that C4 evolution involved important adaptation of gene regulatory elements (Sawers et al. 2007; Majeran and van Wijk 2009). In addition, several enzymes of the C4 pathway, such as PEPC, have been shown to have different biochemical properties compared with the non-C4 ancestral enzymes (e.g., Svensson et al. 2003; Gowik et al. 2006). The C4-specific kinetic optimization resulted in important parallel genetic changes between the different C4 origins as recently demonstrated for PEPC- and PCK-encoding genes (Christin et al. 2007; Besnard et al. 2009; Christin, Petitpierre, et al. 2009). Therefore, despite variation in the C4 pathway of extant plant species, a high number of convergent genetic changes recurrently led to the same evolutionary innovation. The high number of C4 evolutions in some lineages suggests that C3 to C4 transition has a relatively high probability in these plant groups. This could be due to the presence in their genome of genes that can rapidly acquire a C4 function through a low number of key genetic changes in their regulatory and coding regions (Christin et al. 2007). More generally, the large populations and short generation times that characterize plant groups containing C4 species likely favored the constitution of a reservoir of duplicated genes, which could have contributed to rapid genomic diversification and finally C4 evolution (Monson 2003). The high number of distinct gene duplicates encoding some C4-related enzymes (Paterson et al. 2009), such as PEPC in grasses and sedges (Besnard et al. 2009), supports this view. Unfortunately, our understanding of C4 evolution at the genetic level is hampered by the small number of studies that addressed molecular evolution of C4 enzymes in multiple species. In addition, the first genome of a C4 plant having come out only very recently (Paterson et al. 2009), the number of genes encoding C4-related enzymes and their genomic localization remained poorly known. In particular, genes encoding NADP-me have been the focus of relatively few investigations in grasses, despite their high economical importance, being a key element of the C4 pathway of major crops, such as maize, sorghum, sugarcane, and several millets. NADP-me enzymes are not restricted to C4 plants, but exist in both eukaryotes and prokaryotes (Drincovich et al. 2001). In plants, genes encoding NADP-me form a small multigene family, whose different gene lineages encode various isoforms involved in nonphotosynthetic functions as well as in CAM or C4 pathways (Cushman 1992; Edwards and Andreo 1992; Honda et al. 2000; Drincovich et al. 2001; Lai, Tausta et al. 2002; Lai, Wang et al. 2002; Gerrard Wheeler et al. 2005, 2008; Müller et al. 2008). Some NADP-me isoforms are expressed in the cytosol, whereas others, among which stand the C4 ones, act in the chloroplasts (Edwards and Andreo 1992; Drincovich et al. 2001). Nonphotosynthetic NADP-me plastid isoforms seem to be constitutively expressed and have been suggested to be involved in plastid biogenesis, fatty acid synthesis, defence pathways, and other nonphotosynthetic housekeeping functions (Maurino et al. 2001; Lai, Wang et al. 2002; Tausta et al. 2002; Fu et al. 2009). On the other hand, plastid isoforms of NADP-me acting in the C4 pathway are highly expressed and upregulated by light in bundle–sheath cells of C4 plants that use the NADP-me pathway (Maurino et al. 1996; Drincovich et al. 1998; Tausta et al. 2002). Biochemical and structural differences are also observed between non-C4 and C4 NADP-me enzymes (Drincovich et al. 1998; Tausta et al. 2002; Detarsio et al. 2003, 2007, 2008; Estavillo et al. 2007), suggesting that the evolution of a C4-specific NADP-me isoform may have implied key adaptive modifications, as observed for other changes of NADP-me function (Gerrard Wheeler et al. 2008). However, genetic processes linked to the emergence of C4-specific NADP-me are still not resolved. This enzyme has been studied in very few species, and the limited number of sequences currently available disables comparative studies, which are necessary to capture the diversity of the C4 pathway linked to its multiple origins (Christin, Salamin, et al. 2009). In particular, the number of evolutionary transitions towards C4-specific NADP-me enzymes are still unknown despite species phylogenies pointing to complex transitions between the different C4 biochemical subtypes in grasses (Giussani et al. 2001; Vicentini et al. 2008). Similarly, the evolutionary relationships between non-C4 and C4-specific nadpme genes are poorly resolved because genomes of C3 taxa sister to C4 species have never been screened. Recently, sequencing of both C3 and C4 genomes have been completed in the grass family (i.e., rice and sorghum) offering new perspectives for a genomic study of C4 genes (Yu et al. 2002; Paterson et al. 2009; Wang et al. 2009) and more specifically for a better understanding of the C4 NADP-me molecular evolution. Functional and genomic information available for such model species should be now coupled to a phylogenetic approach of the nadpme multigene family based on a dense species sampling of grasses.
The present study addresses the genetic mechanisms linked to the evolution of C4-specific NADP-me enzymes in grasses. The distribution and characteristics of genes encoding NADP-me (nadpme) in genomes of model grasses is analyzed and used to design a comparative phylogenetic analysis of the nadpme evolutionary history from a wide sample of both C3 and C4 grasses. This combination of genomic and phylogenetic approaches aims to 1) assess the diversity of nadpme genes in grasses, 2) identify the independent C4-nadpme origins, and 3) test for the occurrence of positive selection and genetic convergence linked to the acquisition of the C4-NADP-me function.
Materials and Methods
Genomics of the nadpme Multigene Family
NADP-me encoding genes (nadpme) annotated in GenBank were blasted against complete genomes of rice and sorghum as well as the draft sequence of Brachypodium distachyon genome (www.brachybase.org), and nadpme genes were retrieved. The delimitation of exons available for these genomes was refined by comparison with available transcript sequences. Exon homology was established through alignment using ClustalW (Thompson et al. 1994). The genetic structure as well as their genomic location was then reported for each nadpme gene. The presence of plastid transit peptides on nadpme sequences and the localization of their cleavage site were predicted using the ChloroP software (Emanuelsson et al. 1999).
Amplification of nadpme Genes
Sequences of grass nadpme available in GenBank were retrieved and added to the data set from grass genomes. The coding sequences were aligned and oligonucleotide primers were defined in conserved regions as distant as possible. A forward primer (nadpme-491-for; AYGAGAGGCTBTTCTACAAG) was defined in the fourth exon and a reverse primer (nadpme-1606-rev; GGGAARATGTAGGCRTTGTT) in the 17th exon (fig. 1). This primer pair was used to polymerase chain reaction (PCR) amplify nadpme genes from either genomic DNA (gDNA) or complementary DNA (cDNA) isolated from green leaves for a sample of grasses chosen to represent both several independent C4 origins and a diversity of biochemical subtypes as defined from the literature (supplementary table 1 [supplementary material online]; Sage et al. 1999; Christin, Salamin, et al. 2009). Both gDNA and cDNA were obtained from previous studies (Christin et al. 2007, 2008a). PCR amplification, purification, cloning, and sequencing were carried out as described for PCK-encoding genes (Christin, Petitpierre, et al. 2009), but the annealing temperature was lowered to 52 °C. The extension time of PCR amplifications from cDNA was lowered to 2 min. Later, a modified forward primer (nadpme-494-for; AGAGGCTBTTCTACAAGCTT) was used to preferentially amplify the nadpme-IV gene lineage, which was shown to contain genes encoding C4-related enzymes (see Results).
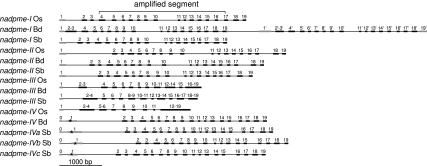
FIG. 1.—
Genomic organization of nadpme genes in model grasses. For each gene present in the genomes of rice (Os), Brachypodium distachyon (Bd) and sorghum (Sb), exons are represented by thick bars and introns by thin bars. Exons homologous among gene lineages are in black and have the same number in all sequences. Exons in grey are not homologous in all gene lineages. Asterisks represent the predicted cleavage site of the plastid transit peptides. The localization of the gene segment amplified through PCR is indicated on rice nadpme-I.
The nadpme gene encoding the C4 isoform is highly transcribed in green leaves of C4 species from the NADP-me biochemical subtype (Maurino et al. 1996; Drincovich et al. 1998; Tausta et al. 2002). To identify this gene in a subset of C4 species, PCR was carried out on green leaf cDNAs using primers nadpme-491-for/nadpme-1606-rev. The size of the amplified region was exactly the same whatever the nadpme gene lineage. PCR products were purified and directly sequenced with the primers used for the PCR amplification. The sequence dominating in the chromatogram was reported as the most transcribed gene, that is, in C4 species using the NADP-me subtype, the C4-specific isoform.
Sequences Analyses
Introns of nadpme isolated from gDNA were identified through comparisons with the cDNAs and following the GT–AG rule. Coding sequences of genes and those obtained from cDNA were translated into amino acids and aligned using ClustalW (Thompson et al. 1994). For Brachypodium nadpme-I, which is composed of two repeats of the standard coding sequence (fig. 1), each repeat was treated as a separate sequence in phylogenetic analyses. Once translated back into nucleotides, the alignment was manually refined. Bayesian inference, as implemented in MrBayes 3.2 (Ronquist and Huelsenbeck 2003), was used to construct a phylogenetic tree based on coding sequences of all grass nadpme genes and a sample of other monocot and dicot sequences retrieved from GenBank (supplementary table 1, Supplementary Material online). The best-fit model was the HKY substitution model with a gamma shape parameter and a proportion of invariant sites (HKY + G + I) as determined through hierarchical likelihood ratio tests (hLRT). All model parameters were optimized independently for first, second, and third positions of codons. Two analyses, each of four chains, were run for 10,000,000 generations. Trees were sampled every 1,000 generations after a burn-in period of 3,000,000.
Coding sequences can be phylogenetically misleading due to adaptive evolution (Christin et al. 2007). To prevent such a bias, a phylogenetic tree was also inferred from combined introns and third positions of codons. This analysis was performed on genomic sequences belonging to the nadpme-IV gene lineage only because introns alignment of very divergent sequences was problematic. In addition, this gene lineage contains several C4 isoforms (see Results) and is therefore prone to phylogenetic biases due to adaptive changes linked to functional switches (Christin et al. 2007). Sequences isolated from gDNA, still containing introns and exons, were aligned with ClustalW (Thompson et al. 1994) with gap opening and gap extensions penalties set to 15.0 and 6.66, respectively, for both pairwise and multiple alignments. Exons boundaries were refined manually, and all exons were removed from this data set. Introns alignment was visually checked but not manually edited to avoid subjectivity. Best-fit substitution models were determined through hLRT for the introns and third positions separately. For both data sets, the best-fit model was the general time reversible substitution model with a gamma shape parameter (GTR + G). A phylogenetic tree was obtained through Bayesian inference with analysis parameters as described above. All model parameters were optimized separately for introns and third positions.
Positive Selection Analyses
To test for the occurrence of positive selection during the evolution of C4-specific NADP-me, three codon models were optimized using the software codeml, implemented in the PAML package (Yang 2007). A description of the three models, M1a, A, and A′, is available elsewhere (Yang et al. 2000; Yang and Nielsen 2002; Zhang et al. 2005). Only nadpme-IV genes isolated from gDNA were considered. The topology inferred from introns and third positions of nadpme-IV gene lineage was used because it is more likely to represent the evolutionary history of nadpme genes (Christin et al. 2007) and better reflects the evolutionary history of grasses deduced from plastid markers (see Results). For branch-site models (models A and A′), branches on which positive selection might have occurred (foreground branches) must be defined a priori. Branches basal to each group of nadpme sequences belonging to C4-NADP-me species and which were shown to be the most highly transcribed in green leaves were used as foreground branches. This included branches leading to Digitaria, Echinochloa, Paspalum, the Stenotaphrum–Pennisetum–Spinifex cluster and nadpme-IVc of Andropogoneae (see Results). It was not possible to determine whether nadpme sequences belonging to C4 species using the NADP-me subtype were involved in the C4 pathway when cDNA was not available. The presence of unidentified C4 genes could bias the positive selection analyses. Therefore, seven sequences (from the C4 NADP-me genera Aristida, Arundinella, Mesosetum, Stipagrostis, Streptostachys, and Tatianyx) were removed from the data set and manually pruned from the topology. Similarly, it is not known whether Andropogoneae lineages nadpme-IVa and nadpme-IVb are linked to C4 evolution (see Results). Thus, the 16 sequences of these groups were also removed. Positive selection tests were done on the 32 remaining sequences.
Results
Genomics of nadpme Multigene Family
Four genes were retrieved from B. distachyon and rice genomes (fig. 1). The lineages I, II, and IV of rice nadpme are located on chromosome 1, whereas nadpme-III is on chromosome 5. Sorghum genome contains six genes and not only five as previously reported (Wang et al. 2009). Its nadpme lineages I, II, IVb, and IVc are on chromosome 3 and its lineages III and IVa lie on chromosome 9. Two of its nadpme-IV genes (IVb and IVc) are organized in tandem and separated by approximately 15 kbp. Lineages III and IVb-IVc are located on duplicated chromosomal regions in both rice and sorghum (Paterson et al. 2004, 2009).
The structure of nadpme genes is generally well conserved with 18 exons homologous among all sequences (exons 2–19; fig. 1), except exon 1, which is not homologous among lineages. In addition, nadpme-IV genes have a supplementary exon (numbered 0; fig. 1). Genes from lineage III have a reduced and variable number of introns leading to the fusion of several exons but without significant alteration of coding sequences (fig. 1). Gene nadpme-I of Brachypodium is composed of a repeat of the 19 exons, which probably appeared through tandem gene duplication followed by merging of the two genes, similarly to what happened in sorghum carbonic anhydrase-encoding genes (Wang et al. 2009).
A plastid transit peptide was significantly predicted in the four nadpme-IV genes but not in other genes. According to this prediction, the cleavage site lies in the exon 1 of these genes (fig. 1).
Phylogenetic Patterns
Sixty-four sequences were isolated from gDNA (supplementary table 1, Supplementary Material online). The size of the isolated fragments ranged from 1,850 to 3,060 bp, generally including 13 introns, but with a range between 6 and 13. The exons provided an average of 1,095 bp of coding sequences. Twenty-two additional sequences isolated from cDNA and 65 nadpme genes taken from GenBank and genomes were added for a total of 151 sequences (supplementary table 1, Supplementary Material online).
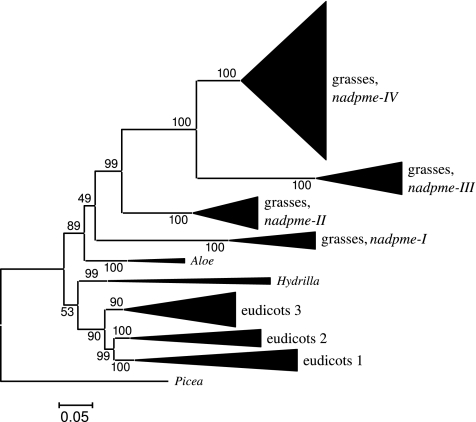
According to the phylogenetic tree inferred from coding sequences, three main gene lineages are present in eudicots, named 1, 2, and 3 (fig. 2; supplementary fig. 1, Supplementary Material online). Eudicot lineage 1 corresponds to group II as previously circumscribed (Gerrard Wheeler et al. 2005), whereas eudicot lineages 2 and 3 were part of groups I and IV in Gerrard Wheeler et al. (2005). Lineage 1 of eudicots contains all described and predicted eudicot genes encoding plastidic isoforms of NADP-me (Lipka et al. 1994; Gerrard Wheeler et al. 2005; Müller et al. 2008). In grasses, the existence of four main gene lineages (nadpme-I to IV) is supported by phylogenetic analyses (fig. 2). Each of these lineages was isolated from representatives of the main grass subfamilies (supplementary figs. 1 and 2, Supplementary Material online) but nadpme-IV was never isolated from Chloridoideae (i.e., Dactyloctenium, Lepturus, and Sporobolus). All grass genes clustered together as sister groups of eudicot genes (fig. 2). Species relationships deduced from each grass lineages are congruent with those deduced from plastid markers (Christin et al. 2008a). However, in gene lineage nadpme-IV, sequences of NADP-me C4 Paniceae belonging to three putatively independent C4 lineages (7-Stenotaphrum clade, 9-Echinochloa, and 11-Digitaria; Christin et al. 2008a) clustered together. In the tribe Andropogoneae, up to three distinct nadpme-IV genes were isolated from the same species (i.e., Sorghum bicolor, Hyparrhenia rufa, and Bothriochloa saccharoides), indicating the presence of three distinct nadpme-IV lineages in this tribe. These were named nadpme-IVa, b, and c (figs. 1 and 3, supplementary fig. 2, Supplementary Material online). Lineage nadpme-IVa corresponds to sorghum gene Sb09g017550, lineage nadpme-IVb contains sorghum gene Sb03g003220, whereas C4 gene of sorghum Sb03g003230 belongs to nadpme-IVc. Two sequences (isolated from Coix and Arthraxon) have an unclear position, being neither in lineage IVa nor in lineage IVb. Because at least two of the Andropogoneae duplicates are in tandem (nadpme-IVb and IVc of sorghum), it is possible that in Coix and Arthraxon, tandem repeats were subject to gene conversion, which blurred the phylogenetic signal.
FIG. 2.—
Phylogenetic tree of the nadpme multigene family. The phylogenetic tree was inferred from all available coding sequences using Bayesian analyses. The main gene lineages are compressed and designated by their name. Bayesian posterior probabilities are indicated next to each branch. The full tree is available in supplementary figs. 1 and 2 (Supplementary Material online).
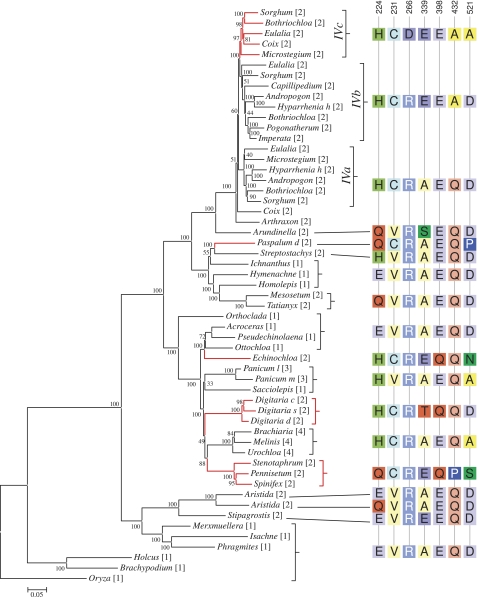
FIG. 3.—
Phylogenetic tree of nadpme-IV deduced from introns and third positions. Bayesian posterior probabilities are given next to the branches. Branches of putative C4-related groups are in red and Andropogoneae duplicates are specifically named. Numbers in square brackets after species names indicate photosynthetic types and subtypes. [1]: C3, [2]: C4 NADP-me, [3]: C4 NAD-me, and [4]: C4 PCK. Amino acids that predominate in each gene cluster are indicated on the right for each position under C4-linked positive selection. For visual clarity, C4-specific amino acids are brightened.
A phylogenetic tree was also inferred to include the sequences obtained through direct sequencing of PCR products obtained on cDNA from NADP-me C4 species. It showed that the most highly transcribed genes all belonged to the nadpme-IV gene lineage (supplementary fig. 2, Supplementary Material online). In the four Andropogoneae whose cDNA was screened, the nadpme-IVc lineage was always the highest amplified sequence. However, nadpme-IVb gene lineage was also detectable in Pogonatherum paniceum and H. rufa, suggesting that this gene is also expressed significantly in green leaves of some Andropogoneae.
The phylogenetic tree of nadpme-IV inferred from introns, and third positions only (fig. 3) was globally congruent with that inferred from all coding sequences (supplementary fig. 2, Supplementary Material online). However, genes from NADP-me species of grass lineages 7, 9, and 11 did not cluster together, congruently with plastid DNA phylogeny (Christin et al. 2008a). This phylogeny inferred from neutral markers confirms these three grass groups as independent C4-NADP-me lineages and is likely more reliable than the phylogenetic tree based on the whole coding sequence.
Positive Selection Tests
The model implementing positive selection on branches basal to each C4 nadpme group was significantly better than the model with constant rates across the phylogeny (models A vs M1a: chi squared = 61.8, degrees of freedom [df] = 2, P value < 0.0001) and the model with relaxed selection in C4 branches (models A vs A′: chi squared = 28.6, df = 1, P value < 0.0001). Seven sites had a posterior probability of being under positive selection greater than 0.95, at positions 224, 231, 266, 339, 398, 432, and 521 (numbered based on Zea mays sequence, AY271262). Most of the sites under positive selection are conserved in non-C4 nadpme of grasses (supplementary table 2, Supplementary Material online) but mutated one to several times independently in C4 nadpme genes, often to an identical residue (fig. 3).
Discussion
Diversification of the nadpme Multigene Family
Four main nadpme gene lineages were identified in distant grass subfamilies (e.g., Pooideae, Ehrhartoideae, and Panicoideae). According to the phylogenetic inferences (fig. 2), recurrent duplications involved in the diversification of grass nadpme genes have occurred after the split between eudicots and monocots, contradicting phylogenetic patterns deduced from amino acid sequences (Gerrard Wheeler et al. 2005) but confirming previous analyses on nucleotide sequences (Estavillo et al. 2007). Lineages III and IV of grasses are located on duplicated chromosome segments in both rice and sorghum (Paterson et al. 2004, 2009). Their duplication is thus probably linked to the suggested whole-genome duplication that occurred before or early during grass diversification (Paterson et al. 2004). All nadpme duplications were followed by changes of exon 1, which could have promoted functional diversification. For instance, nadpme-IV has acquired a plastid localization after gene duplication, apparently via the acquisition of an exon 1 containing a plastid transit peptide. According to phylogenetic patterns, NADP-me localized in plastids clearly evolved independently in grasses (nadpme-IV; fig. 2) and in eudicots (eudicots 1; fig. 2), as already suggested by the lack of similarity in their transit peptides (Börsch and Westhoff 1990). The newly evolved plastid localization of nadpme-IV likely allowed a diversification of NADP-me functions, including, among others, a role in the photosynthetic pathway of some C4 plants (Tausta et al. 2002).
The gene lineage nadpme-IV was further duplicated before divergence of the tribe Andropogoneae. A first duplication probably gave nadpme-IVa and the ancestral copy of napdme-IVb and nadpme-IVc. A second event consisted in tandem duplication of one of these copies giving rise to napdme-IVb and nadpme-IVc, which are in tandem in the sorghum genome (Paterson et al. 2009). These duplications of genes with plastid expression could have further favored a diversification of NADP-me functions in plastids (see below).
Identification of C4 nadpme
For the five C4 NADP-me grass lineages whose cDNAs were screened, the predominant transcripts all belonged to nadpme-IV and nadpme-IVc for Andropogoneae. These genes are thus likely to be involved in C4 photosynthesis of these species, the C4 isoform of NADP-me being strongly transcribed in green leaves (Maurino et al. 1996; Drincovich et al. 1998; Tausta et al. 2002). This is perfectly congruent with the previous classification of the nadpme-IVc gene of maize (AY271262) and sorghum (Sb03g003230) as encoding the C4 isoform (Tausta et al. 2002; Paterson et al. 2009; Wang et al. 2009).
In addition, five other NADP-me C4 grass species representing four additional C4 origins were included in this study (lineages 1-Stipagrostis, 2-Aristida, 15-Streptostachys, and 17-Mesosetum-Tatianyx; Christin et al. 2008a). The unavailability of green leaf cDNAs for these species prevented the identification of the C4 nadpme. The presence of putative C4-adaptive amino acids in genes of some of these species (fig. 3; supplementary table 2, Supplementary Material online) could suggest that some of the nadpme-IV sequences that were sampled in this study are involved in the C4 pathway. Nevertheless, further investigations, such as screening of cDNAs from these species, are necessary to confirm the C4 specificity of these genes.
Genetic Convergence
The recurrent recruitments of nadpme-IV for the C4 pathway, out of the four gene lineages present in the grass family (fig. 2), emphasizes the predispositions of this gene lineage to become C4 specific. This lineage encodes the only plastidic isoform in rice (Chi et al. 2004), maize (Tausta et al. 2002), and wheat (Fu et al. 2009). This is also the only gene lineage with a plastid transit peptide in Brachypodium and sorghum (fig. 1). Because these taxa belong to different subfamilies which have diverged early during grass diversification (Christin et al. 2008a; Vicentini et al. 2008), it is very likely that all nadpme-IV of grasses are expressed specifically in plastids and that this lineage was already active in plastids of C3 ancestral grasses. This probably strongly facilitated the acquisition of a C4-specific gene, the chloroplast localization of the C4 isoform being necessary for the CO2 pump of C4 photosynthesis to be efficient. On the other hand, nadpme-IV was never isolated from the Chloridoid species sampled. If confirmed, the possible absence of this gene lineage from Chloridoid genomes could have prevented the evolution of the C4 NADP-ME subtype in this C4 grass subfamily, largely explaining the absence of this biochemical pathway from this speciose C4 lineage. The evolutionary transition to a C4-optimized NADP-me must later have implied adaptation of the regulatory sequences to confer a light-induced expression specifically in leaf bundle–sheath layer cells. Key changes in the amino acid sequences probably optimized the kinetic properties of the encoded enzyme for the C4 function, as shown by positive selection tests.
The multiple recruitment of nadpme-IV for the C4 function also means that all grass C4-specific nadpme derived from genes with highly similar amino acid sequences and kinetic properties. This common starting point potentially strongly limited the possible paths to C4-specific kinetics (Weinreich et al. 2006), explaining that the same positions were recurrently mutated in different grass C4 lineages (fig. 3). On the other hand, C4 nadpme from the eudicot genus Flaveria evolved from a different non-C4 gene lineage also expressed in plastids (Lipka et al. 1994; lineage 1 of eudicots in fig. 2). These different starting points implied that the protein changes required to acquire C4-specific characteristics were different in Flaveria and grasses (supplementary table 2, Supplementary Material online).
All positions detected as under positive selection in grass C4 nadpme except codon 266 mutated several times independently in different C4 groups, often to an identical residue (fig. 3), pointing to convergent evolution at the genetic level, as shown for other C4 enzymes (Christin et al. 2007, 2008b; Christin, Petitpierre, et al. 2009). The codon at position 231 presents an especially striking pattern. This position is occupied by a Valine in all non-C4 nadpme monocot and eudicot sequences (supplementary table 2, Supplementary Material online), indicating strong purifying selection. However, it mutated five times independently to a Cysteine, an amino acid with very different biochemical properties. Most parallel changes demonstrated for other genes were due to single-nucleotide mutations (Christin et al. 2007, 2008b; Christin, Petitpierre, et al. 2009). On the other hand, the transition from a Valine (codon GTN) to a Cysteine (codon TGY) requires at least two nucleotide mutations. The C4-adaptive value of a Cysteine at this position must have been very important to recurrently lead to the fixation of the mutants, which were probably rare due to the double-nucleotide mutation required. This highlights the putatively crucial function of this residue for the C4-specific characteristics of NADPme enzymes.
The exact effects of the amino acid changes observed on the seven codons under positive selection are difficult to precisely predict. However, they are likely responsible for the biochemical differences observed between C4 and non-C4 NADP-me, such as substrate affinity, allosteric regulation (e.g., malate inhibition), and oligomeric state stability of the enzyme (i.e., dimer or tetramer). For instance, residue 231 is located in a highly conserved motif likely involved in NADP binding (Drincovich et al. 2001), and the transition from a Valine to a Cysteine observed on this site could alter this function. By reconstructing chimerical enzymes from maize nadpme-Va and nadpme-IVc, residues between 248 and the C-terminal part were also shown to be involved in malate inhibition (Detarsio et al. 2007). Changes on residues 266, 339, 432, and 521 could thus be involved in the optimization of the C4 enzyme allosteric regulation. These hypotheses on the functional significance of the observed amino acid transitions should be tested through site-directed mutagenesis.
Evolution of the NADP-me Subtype in Core C4 Paniceae
The core C4 Paniceae lineage (lineage 7 in Christin et al. 2008a) is intriguing because it is composed of three strongly supported monophyletic subgroups, each using a different C4 subtype (Giussani et al. 2001; Christin, Salamin, et al. 2009). These three clades apparently acquired their C4 PEPC from a common ancestor (Christin et al. 2007). Thus, the presence of the three subtypes results probably from switches between the subtypes, but their direction cannot be determined based solely on species trees (Giussani et al. 2001).
Analysis of PCK-encoding genes unequivocally demonstrated that the group composed of Brachiaria, Urochloa, and Melinis acquired the PCK subtype after they diverged from the NAD-me and NADP-me clades (Christin, Petitpierre, et al. 2009). Interestingly, the present study showed that species from the NAD-me (Panicum laetum and Panicum miliaceum) and PCK (Brachiaria, Melinis, and Urochloa) C4 subtypes exhibit two to three C4-adaptive amino acids on nadpme-IV genes (fig. 3). This could suggest that a C4 NADP-me activity exists in these species. However, their NADP-me expression levels do not differ from those of C3 plants (Gutierrez et al. 1974; Prendergast et al. 1987). The most likely explanation is that the NADP-me subtype is the ancestral state of this core Paniceae C4 lineage. NAD-me and PCK cycles would then have added to the NADP-me pathway (see Muhaidat et al. 2007; Christin, Petitpierre, et al. 2009) and progressively became dominant in some lineages. The C4 nadpme genes would have kept evolving under positive selection only in the group still using the NADP-me subtype, explaining the larger amount of C4-adaptive changes in the Stenotaphrum clade (fig. 3). This evidence of numerous switches between C4 biochemical subtypes questions their different adaptive values (for a discussion on this issue, see Christin, Petitpierre, et al. 2009). Further comparative physiological studies are needed to address this issue, and the phylogenetic framework developed here and in the study of PCK-encoding genes (Christin, Petitpierre, et al. 2009) should help designing the species sampling.
Diversification of Plastid nadpme in Andropogoneae
Out of the six detected C4-adaptive amino acids present in nadpme-IVc, four are shared with nadpme-IVb and two with nadpme-IVa (fig. 3). This could indicate either that the three gene lineages are or have been involved in the C4 function, which would explain the amplification of several gene lineages from cDNA in two Andropogoneae species (i.e., P. paniceum and H. rufa) or that the C4-adaptive residues appeared before the gene duplication and the subsequent neofunctionalization (Aharoni et al. 2005). The nadpme-IVa gene of maize is constitutively expressed (Tausta et al. 2002; Detarsio et al. 2008) and displays non-C4 kinetic properties (Saigo et al. 2004; Detarsio et al. 2007), suggesting that it is not currently involved in C4 photosynthesis. However, a previous link to the C4 pathway (e.g., in the ancestral copy, before gene duplication) cannot be excluded. The presence of several duplicates after the evolution of C4 photosynthesis could have allowed fine tuning of the NADP-me C4 and non-C4 functions through recurrent neofunctionalization or subfunctionalization, as suggested for genes encoding malate dehydrogenase in Andropogoneae (Rondeau et al. 2005).
All the species of the Andropogoneae–Arundinella group (lineage 12 in Christin et al. 2008a) are reported to mainly use the NADP-me subtype (Sage et al. 1999), although some of them complete their carbon acquisition with a PCK shuttle (e.g., Wingler et al. 1999; Calsa and Figueira 2007). Interestingly, nadpme-IV of Arundinella displays amino acid changes on two codons that underwent adaptive changes during C4 evolution, but these changes are not shared with those observed in Andropogoneae genes (fig. 3). This suggests that either core Andropogoneae evolved their C4-specific nadpme gene after they diverged from Arundinella or (at least) that these two grass lineages optimized their C4 nadpme independently. These two taxa seem to have acquired some of their C4 characteristics, such as their C4-specific PEPC (Christin et al. 2007), from their common ancestor. Others, such as their C4-tuned NADP-me, were acquired independently at a later stage of their evolutionary history. The atypical Kranz anatomy of Arundinella (Dengler and Dengler 1990; Dengler et al. 1997) could suggest that some anatomical characters were also acquired independently in Arundinella and other Andropogoneae. This demonstrates that the different traits which together create the CO2 pump characterizing C4 plants did not evolve simultaneously but were gradually acquired during a slow transition toward an optimized and fully efficient C4 pathway.
Conclusions
Using phylogenetic analyses and genomic information, this study showed that the main grass subfamilies share four nadpme gene lineages. Duplications of these genes occurred before grass diversification and were followed by shifts of the first exon, which at least once converted to a plastidic isoform through the acquisition of a transit peptide. These events were likely followed by genetic diversification (sometimes with subsequent duplications like in tribe Andropogoneae) and partially helped the evolution of a C4-specific NADP-me. The gene lineage already encoding a plastidic enzyme was hence recurrently recruited for the C4 pathway through successive amino acid adaptive changes in its coding region. Our study therefore confirms the constitution of a reservoir of gene duplicates as an important predisposition for C4 genetic evolution (Monson 2003; Wang et al. 2009). Regarding other C4-related genes in grasses, there is a minimum of six distinct PEPC encoding gene lineages (Christin et al. 2007). On the other hand, genes encoding PCK form one or two lineages, but in five of the C4-specific PCK origins, its evolution was directly preceded by a gene duplication (Christin, Petitpierre, et al. 2009). In most cases, the evolution of C4-specific enzymes can be linked to the presence of gene duplicates, which in grasses can be especially numerous due to ancient whole-genome duplication (Paterson et al. 2004, 2009) as well as recent and frequent polyploidizations and gene-specific duplications. The genomic richness of grasses is thus likely a key to understanding the recurrence of C4 evolution in this diversified family. The future release of several C4 grasses genomes (Buell 2009) will provide an exceptional opportunity to understand the genomic characteristics linked to the rise of C4 photosynthesis, one of the most successful innovations in flowering plant history.
Supplementary Material
Supplementary tables 1 and 2 and figures 1 and 2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by Swiss National Science Foundation [grant 3100AO-105886].
Supplementary Material
Acknowledgments
We thank Nicole Galland and Rowan Sage for their useful comments on an earlier version of this manuscript.
References
- Aharoni A, et al. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Besnard G, et al. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol Biol Evol. 2009;26:1909–1919. doi: 10.1093/molbev/msp103. [DOI] [PubMed] [Google Scholar]
- Börsch D, Westhoff P. Primary structure of NADP-dependent malic enzyme in the dicotyledonous C4 plant Flaveria trinervia. FEBS Lett. 1990;273:111–115. doi: 10.1016/0014-5793(90)81063-t. [DOI] [PubMed] [Google Scholar]
- Buell CR. Poaceae genomes: going from unattainable to becoming a model clade for comparative plant genomics. Plant Physiol. 2009;149:111–116. doi: 10.1104/pp.108.128926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsa T, Figueira A. Serial analysis of gene expression in sugarcane (Saccharum spp.) leaves revealed alternative C4 metabolism and putative antisense transcripts. Plant Mol Biol. 2007;63:745–762. doi: 10.1007/s11103-006-9121-z. [DOI] [PubMed] [Google Scholar]
- Chi W, Yang J, Wu N, Zhang F. Four rice genes encoding NADP malic enzyme exhibit distinct expression profiles. Biosci Biotechnol Biochem. 2004;68:1865–1874. doi: 10.1271/bbb.68.1865. [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr Biol. 2007;17:1241–1247. doi: 10.1016/j.cub.2007.06.036. [DOI] [PubMed] [Google Scholar]
- Christin PA, et al. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr Biol. 2008a;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Christin PA, et al. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Mol Biol Evol. 2008b;25:2361–2368. doi: 10.1093/molbev/msn178. [DOI] [PubMed] [Google Scholar]
- Christin PA, Petitpierre B, Salamin N, Büchi L, Besnard G. Evolution of C4 phosphoenolpyruvate carboxykinase in grasses, from genotype to phenotype. Mol Biol Evol. 2009;26:357–365. doi: 10.1093/molbev/msn255. [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Vicentini A, Kellogg EA, Besnard G. Integrating phylogeny into studies of C4 variation in the grasses. Plant Physiol. 2009;149:82–87. doi: 10.1104/pp.108.128553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC. Characterization and expression of NADP-malic enzyme cDNA induced by salt stress from the facultative crassulacean acid metabolism plant, Mesembryanthemum crystallinum. Eur J Biochem. 1992;208:259–266. doi: 10.1111/j.1432-1033.1992.tb17181.x. [DOI] [PubMed] [Google Scholar]
- Dengler RE, Dengler NG. Leaf vascular architecture in the atypical C4 NADP-malic enzyme grass Arundinella hirta. Can J Bot. 1990;68:1208–1221. [Google Scholar]
- Dengler NG, Nelson T. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, editors. C4 plant biology. San Diego (CA): Academic Press; 1999. pp. 133–172. [Google Scholar]
- Dengler NG, Woodvine MA, Donnelly PM, Dengler RE. Formation of vascular pattern in developing leaves of the C4 grass Arundinella hirta. Int J Plant Sci. 1997;158:1–12. [Google Scholar]
- Detarsio E, Gerrard Wheeler MC, Bermudez VAC, Andreo CS, Drincovich MF. Maize C4 NADP-malic enzyme—expression in Escherichia coli and characterization of site-directed mutants at the putative nucleotide-binding sites. J Biol Chem. 2003;278:13757–13764. doi: 10.1074/jbc.M212530200. [DOI] [PubMed] [Google Scholar]
- Detarsio E, Alvarez CE, Saigo M, Andreo CS, Drincovich MF. Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme. J Biol Chem. 2007;282:6053–6060. doi: 10.1074/jbc.M609436200. [DOI] [PubMed] [Google Scholar]
- Detarsio E, et al. Maize cytosolic NADP-malic enzyme (ZmCytNADP-ME): a phylogenetically distant isoform specifically expressed in embryo and emerging roots. Plant Mol Biol. 2008;68:355–367. doi: 10.1007/s11103-008-9375-8. [DOI] [PubMed] [Google Scholar]
- Drincovich MF, et al. Evolution of C4 photosynthesis in Flaveria species—isoforms of NADP-malic enyzme. Plant Physiol. 1998;117:733–744. doi: 10.1104/pp.117.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drincovich MF, Casati P, Andreo CS. NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 2001;490:1–6. doi: 10.1016/s0014-5793(00)02331-0. [DOI] [PubMed] [Google Scholar]
- Edwards GE, Andreo CS. NADP-malic enzyme from plants. Phytochemistry. 1992;31:1845–1857. doi: 10.1016/0031-9422(92)80322-6. [DOI] [PubMed] [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR. C4 photosynthesis, atmospheric CO2, and climate. Oecologia. 1997;112:285–299. doi: 10.1007/s004420050311. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Rao SK, Reiskind JB, Bowes G. Characterization of the NADP malic enyzme gene family in the facultative, single-cell C4 monocot Hydrilla verticillata. Photosynth Res. 2007;94:43–57. doi: 10.1007/s11120-007-9212-y. [DOI] [PubMed] [Google Scholar]
- Fu ZY, Zhang ZB, Hu XJ, Shao HB, Ping X. Cloning, identification, expression analysis and phylogenetic relevance of two NADP-dependent malic enzyme genes from hexaploid wheat. C R Biol. 2009;332:591–602. doi: 10.1016/j.crvi.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Gerrard Wheeler MC, et al. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 2005;139:39–51. doi: 10.1104/pp.105.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard Wheeler MC, et al. Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties. Plant Mol Biol. 2008;67:231–242. doi: 10.1007/s11103-008-9313-9. [DOI] [PubMed] [Google Scholar]
- Giussani LM, Cota-Sánchez JH, Zuloaga FO, Kellogg EA. A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. Am J Bot. 2001;88:1993–2012. [PubMed] [Google Scholar]
- Gowik U, Engelmann S, Bläsing OE, Raghavendra AS, Westhoff P. Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characteristics of the C4 isozyme and its orthologues in C3 and C3/C4 Alternantheras. Planta. 2006;223:359–368. doi: 10.1007/s00425-005-0085-z. [DOI] [PubMed] [Google Scholar]
- Gutierrez M, Gracen VE, Edwards GE. Biochemical and cytological relationships in C4 plants. Planta. 1974;119:279–300. doi: 10.1007/BF00388331. [DOI] [PubMed] [Google Scholar]
- Honda H, Akagi H, Shimada H. An isozyme of the NADP-malic enzyme of a CAM plant, Aloe arborescens, with variation on conservative amino acid residues. Gene. 2000;243:85–92. doi: 10.1016/s0378-1119(99)00556-9. [DOI] [PubMed] [Google Scholar]
- Lai LB, Tausta SL, Nelson TM. Differential regulation of transcripts encoding cytosolic NADP-malic enzyme in C3 and C4Flaveria species. Plant Physiol. 2002;128:140–149. [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Wang L, Nelson TM. Distinct but conserved functions for two chloroplastic NADP-malic enzyme isoforms in C3 and C4Flaveria species. Plant Physiol. 2002;128:125–139. [PMC free article] [PubMed] [Google Scholar]
- Lipka B, Steinmüller K, Rosche E, Börsch D, Westhoff P. The C3 plant Flaveria pringlei contains a plastidic NADP-malic enzyme which is orthologous to the C4 isoform of the C4 plants F. trinervia. Plant Mol Biol. 1994;26:1775–1783. doi: 10.1007/BF00019491. [DOI] [PubMed] [Google Scholar]
- Majeran W, van Wijk KJ. Cell-type-specific differentiation of chloroplasts in C4 plants. Trends Plant Sci. 2009;14:100–109. doi: 10.1016/j.tplants.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Maurino VG, Drincovich MF, Andreo CS. NADP-malic enzyme isoforms in maize leaves. Biochem Mol Biol Int. 1996;38:239–250. [PubMed] [Google Scholar]
- Maurino VG, Saigo M, Andreo CS, Drincovich MF. Non-photosynthetic ‘malic enzyme’ from maize: a constitutively expressed enzyme that responds to plant defence inducers. Plant Mol Biol. 2001;45:409–420. doi: 10.1023/a:1010665910095. [DOI] [PubMed] [Google Scholar]
- Monson RK. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int J Plant Sci. 2003;164:S43–S54. [Google Scholar]
- Muhaidat R, Sage RF, Dengler NG. Diversity of Kranz anatomy and biochemistry in C4 eudicots. Am J Bot. 2007;94:362–381. doi: 10.3732/ajb.94.3.362. [DOI] [PubMed] [Google Scholar]
- Müller GL, Drincovich MF, Andreo CS, Lara MV. Nicotiana tabacum NADP-malic enzyme: cloning, characterization and analysis of biological role. Plant Cell Physiol. 2008;49:469–480. doi: 10.1093/pcp/pcn022. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Prendergast HDV, Hattersley PW, Stone NE. New structural/biochemical associations if leaf blades of C4 grasses (Poaceae) Aust J Plant Physiol. 1987;14:403–420. [Google Scholar]
- Rondeau P, Rouch C, Besnard G. NADP-malate dehydrogenase gene evolution in Andropogoneae (Poaceae): gene duplication followed by sub-functionalization. Ann Bot. 2005;96:1307–1314. doi: 10.1093/aob/mci282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sage RF, Li M, Monson RK. The taxonomic distribution of C4 photosynthesis. In: Sage RF, Monson RK, editors. C4 Plant Biology. San Diego (CA): Academic Press; 1999. pp. 551–584. [Google Scholar]
- Sage RF. The evolution of C4 photosynthesis. New Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- Saigo M, et al. Maize recombinant non-C4 NADP-malic enzyme: A novel dimeric malic enzyme with high specificity. Plant Mol Biol. 2004;55:97–107. doi: 10.1007/s11103-004-0472-z. [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Liu P, Anufrikova K, Hwang JTG, Brutnell TP. A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics. 2007;8:12. doi: 10.1186/1471-2164-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NR, Kellogg EA. Parallelism and diversity in multiple origins of C4 photosynthesis in the grass family. Am J Bot. 1996;83:1458–1470. [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. Evolution of C4phosphoenolpyruvate carboxylase. Arch Biochem Biophys. 2003;414:180–188. doi: 10.1016/s0003-9861(03)00165-6. [DOI] [PubMed] [Google Scholar]
- Tausta SL, Coyle HM, Rothermel B, Stiefel V, Nelson T. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol Biol. 2002;50:635–652. doi: 10.1023/a:1019998905615. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. The age of the grasses and clusters of origins of C4 photosynthesis. Glob Chang Biol. 2008;14:2963–2977. [Google Scholar]
- Wang XY, et al. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 2009;10:R68. doi: 10.1186/gb-2009-10-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen ZH, Leegood RC. Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol. 1999;120:539–545. doi: 10.1104/pp.120.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.