Abstract
Glutaredoxins (GRXs) are ubiquitous glutathione-dependent oxidoreductase enzymes implicated in redox homeostasis, particularly oxidative stress response. Three major classes of GRX genes exist, the CPYC, CGFS classes are present in all pro- and eukaryote species, whereas the CC-type class GRXs are specific to land plants. In the basal land plant Physcomitrella patens, only two CC-type GRXs are present, compared with 21 in Arabidopsis. In contrast, sizes of the CPYC and CGFS classes remained rather similar throughout plant evolution, raising the interesting question as to when the CC-type GRXs first originated and how and why they expanded during land plant evolution. Recent evidence suggests that CC-type GRXs may have been recruited during evolution into diverse plant-specific functions of flower development (ROXY1, ROXY2) and pathogenesis response (ROXY19/GRX480). In the present study, GRX genes from the genomes of a range of green algae and evolutionarily diverse land plant species were identified; Ostreococcus, Micromonas, Volvox, Selaginella, Vitis, Sorghum, and Brachypodium. Previously identified sequences from Chlamydomonas, Physcomitrella, Oryza, Arabidopsis, and Populus were integrated to generate a more comprehensive understanding of the forces behind the evolution of various GRX classes. The analysis indicates that the CC-type GRXs probably arose by diversification from the CPYC class, at a time coinciding with colonization of land by plants. This strong differential expansion of the CC-type class occurred exclusively in the angiosperms, mainly through paleopolyploidy duplication events shortly after the monocot–eudicot split, and more recently through multiple tandem duplications that occurred independently in five investigated angiosperm lineages. The presented data suggest that following duplications, subfunctionalization, and subsequent neofunctionalization likely facilitated the sequestration of land plant-specific CC-type GRXs into novel functions such as development and pathogenesis response.
Keywords: GRX, CC-type GRX class, ROXY, gene duplication, land plant evolution, tandem arrays of genes
Introduction
Glutaredoxins (GRX; EC 1.20.4.1) are glutathione-dependent oxidoreductase proteins and are members of the thioredoxin (TRX) fold superfamily (Hoog et al. 1983). Like TRXs (EC 1.8.4.10), GRXs exhibit cysteine (Cys) thiol disulfide exchange catalytic properties (Mannervik and Axelsson 1980). The GRX pathway requires a supply of reduced glutathione (GSH) to maintain cellular protein redox stability in the presence of reactive oxygen species (ROS). ROS are generated in the cellular respiration and photosynthesis processes and increase during biotic and abiotic stresses (Doke 1985; Prasad et al. 1994; Tsugane et al. 1999; Bolwell et al. 2002), but their release is also controlled by hormones such as gibberellin and abscisic acid (Schopfer et al. 2001). Under oxidative and nitrosative conditions, free thiols are oxidized and disulfide bond formation occurs between Cys residues. Additionally, disulfide bridges are also formed between Cys residues and GSH to produce mixed disulfides (S-glutathionylation). These mechanisms both bear the potential to alter protein conformation and activity and also act as a protective mechanism to prevent irreversible oxidation under severe oxidative stress in prokaryotes and eukaryotes (reviewed in Dalle-Donne et al. [2009]).
Plant GRXs are a diverse group of proteins, commonly divided into three different classes based on homology and composition of the active site motifs. In Arabidopsis thaliana and rice, the CPYC class GRXs possess a C(P/G/S)(Y/F)(C/S) active site, their putative proteins are 110–180 amino acids in length and share ∼41% identity, and the genes generally possess three or four introns. These GRXs operate predominantly by the dithiol mechanism involving both active site thiols to perform disulfide bond reduction on target proteins at the expense of GSH as an electron donor (Holmgren 1979). CPYC proteins are localized throughout the cell, particularly in the chloroplast, endoplasmic reticulum, and cytoplasm (Rouhier et al. 2004; Heazlewood et al. 2007; Meyer et al. 2008; Zybailov et al. 2008). The proposed roles of CPYC class GRXs are in protein redox balance as evidence shows that AtGrxC1 (At5g63030) is able to reduce disulfide bonds of a redox sensitive green fluorescent protein reporter (Meyer et al. 2007). They are also able to reduce ribonucleotide reductase in Escherichia coli (Aslund et al. 1994), peroxiredoxin II in poplar (Rouhier et al. 2002), TRX in poplar (Gelhaye et al. 2003), and dehydroascorbate in Chlamydomonas (Zaffagnini et al. 2008). The CGFS class exhibits an absolutely conserved CGFS motif and these GRXs operate via a monothiol reaction (Bushweller et al. 1992), which involves the reduction of S-glutathionylated Cys residues using ferredoxin TRX reductase as an electron donor. The intron exon structures are highly variant, the CGFS genes in Arabidopsis having 0–6 introns, comprised of 170–300 amino acids and sharing 28–37% protein identity. Most plant CGFS class GRXs resemble yeast GRX5 in biochemical properties and are found predominantly in the chloroplast and mitochondria (Herald et al. 2003; Cheng et al. 2006; Zybailov et al. 2008). These proteins have been shown to be important for protein redox homeostasis and stress response in plants (Cheng et al. 2006) as well as iron sulfur cluster assembly in yeast (Rodriguez-Manzaneque et al. 2002).
The CC-type GRXs possess distinctive CC(M/L)(C/S) conserved active site motifs in Arabidopsis isoforms. The motif is extended to C(C/G/F/Y/P)(M/L)(C/S/I/A) in rice, the N-terminal Cys being strictly conserved (Xing et al. 2006). Arabidopsis CC-type putative proteins share ∼49% identity, and all described CC-type GRX genes are intronless. The T-DNA insertion mutants of ROXY1 and ROXY2 exhibited the first plant GRX phenotypes, which, surprisingly, revealed crucial roles for these genes in governing petal initiation and morphogenesis as well as anther development (Xing et al. 2005; Xing and Zachgo 2008). Furthermore, the roxy1 mutant provides a most suitable system to determine the crucial functional domains of CC-type GRXs. Directed mutagenesis of the ROXY1 coding sequence (CDS) coupled with complementation studies of roxy1 led to the identification of an expendable nonconserved N-terminal extension of ROXY1, whereas its four C-terminal residues (ALWL) were absolutely essential for in planta function of ROXY1. All but 5 of the 21 Arabidopsis CC-type GRXs exhibit this conserved C-terminus, the lack of which causes a failure to complement roxy1 (Li et al. 2009). Further, these four residues were demonstrated to be crucial for physical interaction of ROXY1 with its target proteins (Li et al. 2009).
With numerous CC-type GRX CDSs now shown to be capable of complementing roxy1 when expressed under the control of ROXY1 regulatory elements (Li et al. 2009), it is becoming clear that the temporal and spatial mRNA expression patterns could be important for determination of ROXY gene function. Even the orthologues OsROXY1 and OsROXY2 from the distantly related monocot, rice, are capable of mediating petal morphogenesis in Arabidopsis, although the lodicules, that is, second whorl floral organs of rice, differ in shape and function from the eudicot petals (Wang et al. 2009). The expression patterns of eudicot and monocot ROXY1/2 were found to be tightly regulated temporally and spatially (Xing et al. 2005; Xing and Zachgo 2008; Wang et al. 2009), the expression being generally transient and restricted to young stages of floral meristem and organ primordia formation. In addition to floral function, another CC-type gene, ROXY19 (GRX480), has been shown to be upregulated by salicylic acid (SA) in Arabidopsis. Ectopic overexpression of ROXY19 causes a reduction of the jasmonic acid (JA)–dependent PLANT DEFENSIN 1.2A (PDF1.2) mRNA level, representing an ROXY19 regulated ROS-linked interface between SA and JA signaling pathways involved in biotic stress response (Ndamukong et al. 2007). However, ROXY19, when expressed in a floral context, can also rescue roxy1 and thus modify target proteins participating in flower development (Li et al. 2009). It has been demonstrated that both the floral ROXY1 and the stress-related ROXY19 physically interact with TGACG motif binding (TGA [Lam et al. 1989]) transcription factors, implicating TGAs as posttranslational targets of ROXY protein activity. Indeed, recent studies on intracellular localization coupled with roxy1 complementation have proven that nuclear interaction of ROXY1 with TGAs is crucial for its function during petal development (Li et al. 2009). Thus, a conserved ROS-linked molecular mechanism may exist for CC-type GRXs, wherein they act in the nucleus to modify Cys residues of their interactors such as TGAs, thereby driving changes in downstream gene expression in diverse functions such as development and pathogen response.
Previous work (Xing et al. 2006) has highlighted that the numbers of CPYC and CGFS class GRXs have remained relatively constant (four to six members) among evolutionarily distant plants such as the bryophyte moss (Physcomitrella patens) and Arabidopsis. In comparison, CC-type GRXs appear to have increased dramatically in numbers during the evolution of higher land plants. To highlight this fact, in previous expressed sequence tag (EST) searches, CC-type genes were noted to be absent in unicellular green algae (Lemaire 2004), with the most basal CC-type genes identified to date being in Physcomitrella (which has two such genes) followed by the gymnosperm Pinus taeda (which has at least 5; Xing et al. 2006). In higher plants, however, these genes are much more numerous, with A. thaliana having 21 (Lemaire 2004) and rice 16 (Rouhier et al. 2004). The aim of this study was to determine how, when, and possibly why the CC-type class specifically underwent an increase in numbers and if/how this process correlates to the evolution of more complex plant functions. To achieve this, advantage was taken of the recent avalanche of whole-genome data by defining new GRX genes from further evolutionarily informative members of the Viridiplantae: Ostreococcus lucimarinus and Micromonas pusilla (both marine prasinophyte picophytoplanktons), Volvox carteri (a colony forming alga from the class Chlorophyceae), Selaginella moellendorffii (a lycophyte from the class Isoetopsida), Brachypodium distachyon (a wild grass related to barley and wheat), Sorghum bicolor (cultivated sorghum), Vitis vinifera (grapevine), and Arabidopsis lyrata (an outcrossing cousin of A. thaliana). A set of 195 GRX sequences was compared that comprises 91 newly identified GRXs from the above genomes and 104 previously reported ones from Chlamydomonas reinhardtii (a chlorophyte alga), P. patens (a moss), Oryza sativa (rice), A. thaliana, and Populus trichocarpa (poplar). The results have led to new insights into the evolution of the GRX family in the green lineage, wherein it appears that the CC-type class exclusively underwent independent, parallel expansions in the mono- and eudicot lineages, with members likely sequestered into flower development and stress response processes.
Materials and Methods
Genome-Wide Search for GRX Genes in Ostreococcus, Micromonas, Volvox, Selaginella, Brachypodium, Sorghum, Vitis, and A. lyrata
Reported GRX peptide sequences (Lemaire 2004; Xing et al. 2006) were used to conduct TBlastN searches with the following rationale. Chlamydomonas reinhardtii GRXs were used as queries to search the O. lucimarinus v2.0, M. pusilla v2.0, and V. carteri v1.0 genomes (http://genome.jgi-psf.org/). Sequences from P. patens were used to search the S. moellendorffii v1.0 genome database (http://www.phytozome.net/). As the individual Selaginella plant sequenced was not of a repetitively inbred line, two alleles were identified for each locus. Oryza sativa (rice) GRX sequences were used to conduct Blast searches in the following databases: Brachybase for B. distachyon draft sequence (http://www.brachybase.org/; accessed February 2009) and Phytozome for S. bicolor v1.0 (http://www.phytozome.net/). Arabidopsis thaliana GRX sequences were used as queries to search for GRX genes in A. lyrata v1.0 at the US Department of Energy, Joint Genome Institute (http://genome.jgi-psf.org/Araly1/Araly1.home.html) and those in V. vinifera (grapevine) in the Phytozome database. TBlastN hits with E values <10−5 were collected and redundancies removed. NCBI's conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to screen out artifactual hits such as TRXs. ESTs corresponding to GRX genes were identified in the tracks of the above genome browse resources, and these ESTs were employed as the primary method for gene structure determination. In the absence of EST data, BlastX alignments (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi) were used to identify conserved exon sequences between orthologs, which were assembled into contig gene models. In cases of low conservation of exon sequences and lack of transcript information, FGENESH hidden Markov model–based gene structure prediction (http://linux1.softberry.com/berry.phtml) was utilized. Values for % amino acid identity were determined in a pairwise fashion with Jalview. All sequences cited in this article are available in the Supplementary Material online. However, as a number of the genomes investigated in this work are in draft or early versions, there is a possibility that further GRX genes may lie in unassembled or unsequenced regions.
Phylogenetic Reconstructions
Novel GRX protein sequences acquired as above from Micromonas, Ostreococcus, Volvox, Selaginella, Vitis, Brachypodium, and Sorghum were aligned with reported Chlamydomonas, Physcomitrella, Oryza, A. thaliana, and poplar (P. trichocarpa) GRX sequences in ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Phylogenetic reconstructions were inferred using the Neighbor-Joining method (Saitou and Nei 1987). The evolutionary distances, expressed as number of amino acid substitutions per site, were computed using the Dayhoff matrix–based method (Schwarz and Dayhoff 1979), and positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons. Bootstrap values are given beside the branch points, when >50 (1,000 iterations; Felsenstein 1985). Phylogenetic analyses were undertaken in MEGA4.0 (Tamura et al. 2007). Graphical representations depicting tandemly arrayed clusters of CC-type GRX genes (TAGs) were produced based on data from the following sources: A. thaliana (http://www.Arabidopsis.org/cgi-bin/gbrowse/Arabidopsis/), A. lyrata (http://genome.jgi-psf.org/Araly1/Araly1.home.html), Oryza (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), Sorghum and Populus (http://www.phytozome.net), and Brachypodium (http://www.modelcrop.org/cgi-bin/gbrowse/brachy4x/). The cluster phylogenies were produced as above, with branches collapsed for nodes with bootstrap values of <50. GENECONV (Sawyer 1999) was used to scan CDSs for evidence of gene conversion events especially in the TAGs. No gene conversion events could be detected using the default settings (http://www.lifesci.sussex.ac.uk/CSE/test/gcv.php). It should be noted that GENECOV has detection limitations when genes share >70% identity or when conversion events are masked by later mutation.
Determination of Ks, Ka, and Time Since Divergence
For determination of the time of gene duplication, Ks (number of synonymous substitutions per site) was used as a proxy measure of time since divergence assuming a clock-like rate. Protein coding nucleotide sequences were aligned using ClustalW2. The analysis was performed based on a rate of the number of synonymous substitutions per site per year of between 4.9 × 10−9 (Ramakrishna et al. 2002) and 6.5 × 10−9 (Gaut et al. 1996; Yu et al. 2005) for Oryza, Sorghum, and Brachypodium. The rate used for Arabidopsis was 1.5 × 10−8 (Blanc and Wolfe 2004) and that for Populus was 9.1 × 10−9 (Lynch and Conery 2000). Ks values >2.0 were eliminated due to possible saturation of synonymous substitutions. The mode of selection was determined by using the Ka/Ks statistic, that is, the ratio of nonsynonymous (Ka) to synonymous (Ks) substitutions. The program K-estimator 6.1 (Comeron 1999) was used to calculate all values for Ks and Ka for the GRX sequence comparisons using the Kimura 2-p correction method for multiple possible pathways (Kimura 1980).
Results
Identification of GRX Genes in Ostreococcus, Micromonas, Volvox, Selaginella, Brachypodium, Sorghum, Vitis, and A. lyrata
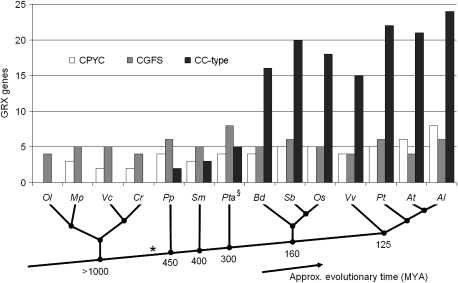
Novel GRX genes were identified through Blast searches on the evolutionarily informative genomes: Ostreococcus, Micromonas, Volvox, Selaginella, Vitis, Brachypodium, Sorghum, and A. lyrata. Gene structure was determined primarily with the aid of EST resources. Where absent, pairwise BlastX alignments to known GRX orthologues were used to identify conserved exons. In cases of ambiguous exon conservation, an ab initio gene prediction algorithm was employed to determine gene structure. A summary of newly identified and known GRX genes in algae and plant genomes is seen in figure 1, where CC-type GRXs appear to have originated before the divergence of Bryophyta from vascular plants. The complete results of the sequence searches along with relevant information used in the determination of gene structures are found in supplementary table S1 (Supplementary Material online).
FIG. 1.—
The sizes of the three classes of GRXs in green algae and plants. Although CPYC and CGFS GRXs are present in all organisms including green algae, CC-type GRXs have only been observed in land plants. The first CC-type gene is likely to have arisen 1,000–450 Ma (*), and in angiosperms, their numbers have increased dramatically in comparison to CPYC and CGFS GRXs. (§) Denotes the use of EST library sequencing, and as such, more GRX genes might exist in the Pinus genome. Abbreviations: Ol, Ostreococcus lucimarinus; Mp, Micromonas pusilla; Vc, Volvox carteri; Cr, Chlamydomonas reinhardtii; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii, Pta, Pinus taeda; Bd, Brachypodium distachyon; Sb, Sorghum bicolor, Os, Oryza sativa; Vv, Vitis vinifera; Pt, Populus trichocarpa; At, A. thaliana; and Al, A. lyrata. The emergence of the bryophytes has been estimated at 450 Ma (Lang et al. 2008; Rensing et al. 2008). The divergence of lycophytes is estimated at 400 Ma, with the first distinctive lycophyte fossil dated at ∼345 Ma (Banks 2008). The gymnosperms are estimated to have diverged from angiosperms some 300 Ma (Theissen and Becker 2004). The divergence of monocots and eudicots has been estimated to be 140–150 Ma (Chaw et al. 2004). The divergence of the Vitales (Vitis) from the rosids (Arabidopsis/Populus) occurred approximately 125 Ma (Wikstrom et al. 2001).
In the full genome assemblies of green algae, CC-type GRX genes were absent, consistent with previous EST searches in Chlamydomonas that had yielded two CPYC class and four CGFS class GRXs (Lemaire 2004). Searches revealed two CPYC and five CGFS genes in the V. carteri genome, whereas M. pusilla possesses three CPYC and five CGFS isoforms. Ostreococcus lucimarinus, possibly possessing the smallest genome of any free-living eukaryote (Palenik et al. 2007), has four CGFS GRXs, and no conventional CPYC GRX could be identified. However, the dithiol function performed by CPYC GRX could have been adopted by a distinct GRX-like gene (NCBI gene accession: OSTLU_27341) that possesses a CPHC active site motif and homology to a family of described redoxin genes in higher plants (Navrot et al. 2006).
Inclusion of Selaginella (spike moss) in the analysis is informative in an evolutionary context as Selaginella is a member of the most basal group of the vascular plants, the lycophytes. Two alleles, one representing each haplotype (see Materials and Methods), were identified for each Selaginella GRX gene. Three CPYC genes were apparent, possessing three introns each and encoding putative proteins from 103 to 131 amino acids in length. Of the five CGFS genes identified, SmGRX7 and SmGRX8 had no corresponding ESTs and the position of their start codons could not be inferred confidently. SmGRX11, an intronless CGFS gene, was found to have two considerably different alleles, one containing two tandem gene copies (SmGRX11b and SmGRX11c) separated by a 17-kb retrotransposon, whereas the alternate allele (SmGRX11a) lacked such a duplicate. Three intronless CC-type genes were noted and were predicted to encode proteins ranging in size from 124 to 132 amino acids in length. The longer isoforms, SmGRX1 and SmGRX2, display higher homology with the CC-type GRXs PpGRX1 and PpGRX2 from Physcomitrella (∼60% protein identity) including conservation of the C-terminal ALWL motif, whereas SmGRX3 has more limited protein identity (∼38%) and lacks this motif (fig. 2a).
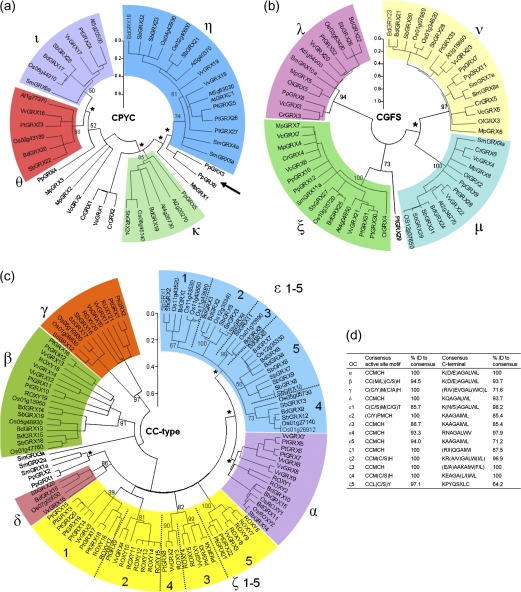
FIG. 2.—
Active site and C-terminal motifs in 78 newly analyzed CC-type GRXs from Selaginella, Brachypodium, Sorghum, Vitis, and A. lyrata. Sequences were aligned with Physcomitrella CC-types PpGRX1 and PpGRX2 in ClustalW, and only the active sites and C termini are shown in (a), (b and c) shows the frequency of each particular amino acid in the putative active site (b) and C-terminal protein-binding motif (c) among all 117 CC-type GRX genes from Physcomitrella, Selaginella, A. thaliana, Populus, Vitis, Brachypodium, Oryza, and Sorghum.
Brachypodium distachyon and S. bicolor along with O. sativa were selected to assess the variation in gene family composition in the economically significant grasses (Poaceae). These species are estimated to have diverged some 50–60 Ma (Paterson et al. 2003). In the draft Brachypodium sequence, 4 CPYC (120–178 amino acids, 3 introns each), 5 CGFS (145–478 amino acids, 1–5 introns), and 16 CC-type genes could be identified (103–145 amino acids in length, intronless). In Sorghum, 5 genes encoding CPYC class GRXs (115–170 amino acids, 3–4 introns), 6 CGFS (147–499 amino acids, 0–5 introns), and 20 CC-types (101–166 amino acids, intronless) were noted. Additionally, a potential pseudogene was identified (SbGRX32), it exhibits limited homology to CPYC genes but lacks a CPYC active site and does not have a corresponding EST. These figures were comparable to numbers found in rice (5 CPYC, 5 CGFS, 18 CC-type; Rouhier et al. 2006), demonstrating that grasses have similarly large complements of CC-type genes akin to eudicots.
Vitis vinifera was selected to aid in the comparative investigation of GRXs within core eudicots because the order Vitales (containing Vitis) is situated at a basal position within the rosids, compared with the Brassicaceae (containing Arabidopsis) belonging to the rosidsII and the Salicaceae (containing Populus) belonging to the rosidsI. The Vitis genome is thus helpful in elucidating events in the rosid lineage after Vitales divergence some ∼125 Ma (Wikstrom et al. 2001). The Vitis genome contained 4 CPYC (114–136 amino acids, 3–4 introns), 4 CGFS (170–403 amino acids, 1–4 introns), and 15 CC-type (83–150 amino acids, intronless) genes, the smallest complement of GRX genes among these angiosperms. Arabidopsis lyrata is of special interest because it is an outcrossing relative of A. thaliana, and comparisons of these genomes are likely to uncover selective pressures and genomic rearrangements that have occurred in the relatively short period since their divergence. The A. lyrata genome was found to contain 8 CPYC (114–181 amino acids, 3–4 introns), 6 CGFS (133–489 amino acids, 0–5 introns), and 24 CC-type (99–150 amino acids, intronless) genes, all displaying at least 80% identity on the amino acid level to A. thaliana orthologues. This interspecies variation demonstrates that the number of GRXs in a genome is dynamic, likely attributable to chromosomal rearrangements.
Novel CC-type GRX sequences identified in this work were aligned with Physcomitrella CC-types and trimmed to illustrate sequence diversity in the active site and C-terminal motifs as in figure 2. As expected, prominent active site motifs included CC(M/L)(C/S), although novel active site motifs were identified including YPMC of BdGRX2 and BdGRX3. The ALWL functional motif at the C-terminus was conserved in 60 of the 78 analyzed CC-type GRXs presented here, emphasizing the importance of this motif in an evolutionary context.
CPYC and CGFS GRX Classes Reveal Two Vastly Different Evolutionary Histories
In an effort to trace the growth in complexity of GRX families through evolution, protein sequences from Ostreococcus, Micromonas, Chlamydomonas, Volvox, Physcomitrella, Selaginella, A. thaliana, Populus, Vitis, Oryza, Sorghum, and Brachypodium were divided into respective classes and aligned with ClustalW2. Neighbor-Joining trees were used to identify ancestral genes through orthologous clusters (OCs). Phylogenic investigation of CPYC GRXs (fig. 3a) shows that there are four OCs (labeled η, θ, ι, and κ) in common with all angiosperms. In contrast, the genes belonging to the unicellular green algae are linked to the root of the tree, with the exception of MpGRX1, which is positioned between OC-κ and OC-η; however, the position is tenuous given the low bootstrap value (36) of the node in question. In all, there is sufficient evidence from this phylogeny, especially from Chlamydomonas and Volvox, to conclude that there was only one CPYC GRX in ancestral green algae.
FIG. 3.—
Phylogenetic Neighbor-Joining trees of Viridiplantae GRXs of CPYC (a), CGFS (b), and CC-type (c) GRX protein sequences from Micromonas (Mp), Ostreococcus (Ol), Chlamydomonas (Cr), Volvox (Vc), Physcomitrella (Pp), Selaginella (Sm), Oryza (Os), Sorghum (Sb), Brachypodium (Bd), Populus (Pt), Vitis (Vv), and A. thaliana (At). (d) Illustrates the signature active site and C-terminus of each CC-type OC and the degree of agreement expressed as average % identity to consensus. The trees are drawn to scale in units of number of amino acid substitutions per site using the Dayhoff matrix–based method, positions containing alignment gaps were eliminated in pairwise sequence comparisons. OCs are shaded, and bootstrap values are given, key bootstrap values which were found to be <50 are denoted by (*). The arrow indicates the position of PpGRX1 (CC-type) when integrated into the phylogeny. Arbitrarily, only one allele for each Selaginella GRX gene was selected. CPYC and CGFS trees each clearly demonstrate four OCs at the divergence of monocots and eudicots. In the CC-type class, four OCs in common with monocots and eudicots are shown (OC-α, β, γ, and δ), as well as monocot-specific (OC-ε) and eudicot-specific groups (OC-ζ). The phylogenetic methods employed could not confidently establish whether OC-ζ and OC-ε groups shared a common ancestral gene. Similar analyses using Neighbor-Joining and minimum evolution methods with alternative substitution scoring methods (no. of differences, p distance, Poisson correction, Equal input, JTT matrix) yielded similar results.
Physcomitrella has a clear representative in OC-κ, whereas an ancestral form of PpGRX4 appears to have founded OC-ι and θ. An ancestral form of the apparent paralogues PpGRX3 and PpGRX6 could have founded OC-η, but the phylogeny could not be determined conclusively. Two similar Selaginella genes (SmGRX4 and SmGRX5) are clearly part of OC-η, whereas a further Selaginella CPYC gene (SmGRX6) appears to be part of OC-ι. Despite the existence of a Physcomitrella CPYC gene in OC-κ, there appears to be no Selaginella isoform, indicating a gene loss event. Interestingly, OC-κ genes are also absent in Vitis and Populus lineages but present in Arabidopsis. OC-κ is typified by its CS(Y/F)(S/C) active site motif, notably all isoforms lack the proline at the second active site position in favor of a serine. OC-θ seems to have arisen in the time between the origin of the ferns and the divergence of the monocot–eudicot lineage. OC-η is the largest group in this class and consists of GRXs with the active site motifs CPYC, CGYC, and CPFC. In monocot plants, there appear to be four CPYC class OCs, whereas the last common ancestors of Oryza and Sorghum share five CPYC OCs.
In stark contrast, CGFS GRXs appear to have established their four OCs extremely early in the evolution of the green lineage, with isoforms from Micromonas, Chlamydomonas, Volvox, and Ostreococcus grouping clearly into each of the four OCs (λ, μ, ν, and ξ) despite large evolutionary distances involved (fig. 3b). Among angiosperms, OC-ν was observed to be the largest of the CGFS class with nine members compared with OC-μ (seven members), OC-ξ (seven members), and OC-λ (six members).
Evolutionary History of the CC-Class
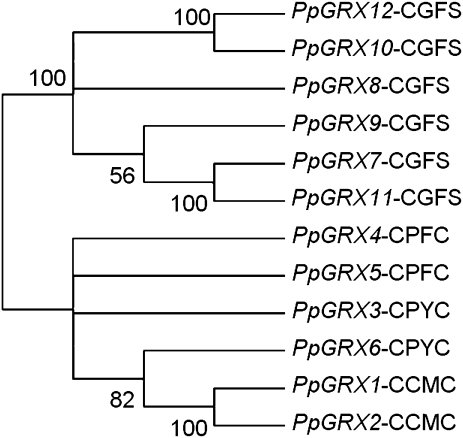
The innovation of the initial CC-type GRX in basal plants is a particularly interesting event, especially in light of functional data showing roles for some CC-type GRXs in development and pathogen response in Arabidopsis. In terms of sequence homology, CC-type genes are more related to CPYC genes than CGFS genes as phylogeny shows a strong grouping of PpGRX1 and PpGRX2 with the CPYC gene PpGRX6 (fig. 4). Interestingly, whereas related CPYC genes PpGRX3, PpGRX6, and CrGRX2 possess two introns, CC-type PpGRX1 and PpGRX2 are intronless, suggesting the involvement of an mRNA-mediated mode of gene duplication for the initial CC-type isoform.
FIG. 4.—
A phylogenic Neighbor-Joining tree of Physcomitrella GRX sequences. The tree demonstrates the evolutionary relationship of CC-type genes PpGRX1 and PpGRX2 with PpGRX6, a CPYC gene. Neighbor-Joining comparisons were conducted using predicted amino acid sequences, nodes with bootstrap scores <50 had branches collapsed.
In a phylogenic investigation of 117 CC-type GRXs, genes from Physcomitrella and Selaginella were, as expected, located close to the root of the Neighbor-Joining tree (fig. 3c). Only four OCs (designated α, β, γ, and δ) are clearly demonstrated to be shared between monocots and eudicots, indicating that early angiosperms, before the monocot–eudicot split, possessed only four CC-type GRX genes. The presence of an OC without an Arabidopsis or Populus ortholog (OC-δ) implies a gene loss event in the eudicots after the divergence of the Vitales.
Five additional OCs are specific to monocots (labeled ϵ 1–5) and to eudicots (ζ 1–5), respectively, and phylogeny indicates that these two groups arose within a narrow evolutionary time window (Ka < 0.05). Bootstrap values of <50 imply a significant degree of uncertainty in the positioning of those groups with respect to OC-α. Similar analyses using both Neighbor-Joining (Saitou and Nei 1987) and minimum evolution methods (Rzhetsky and Nei 1992) with alternative substitution scoring methods p distance, Poisson correction (Zuckerkandl and Pauling 1965), Equal input (Tajima and Nei 1984), and JTT matrix (Jones et al. 1992) yielded comparable tree topology and bootstrap values at these key positions. Whether these monocot- and eudicot-specific groups shared a single ancestral gene or whether they came about in independent events cannot be determined with confidence due to the narrow evolutionary time and uncertainty indicated by the bootstrap analysis. The proliferation of OC-ζ into five sister OCs caused CC-type gene number in early core eudicots to increase from 4–5 to 8–9 by the time of Vitales divergence. In parallel, it seems OC-ε also experienced duplication in early monocots to produce five sister OCs, although the number of OCs in common with grasses (Poaceae) is estimated at 11–12 due to further duplications in OC-α and OC-β.
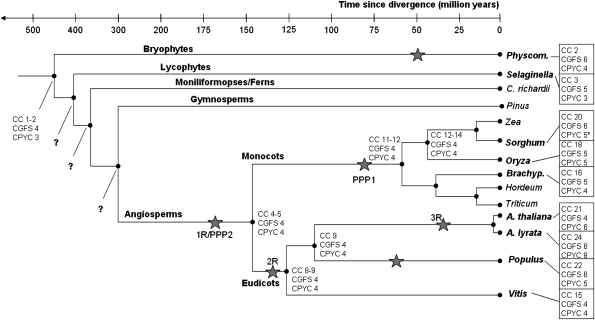
The predicted GRX gene family expansion from Physcomitrella to angiosperms using the OC method is summarized in figure 5, showing that early angiosperms, the common ancestors of monocots and eudicots, possessed just four to five CC-type genes some 140 Ma. In the Poaceae, that number had increased to 11 by 65 Ma, with present-day grass and cereal species possessing 16–20 CC-type genes. Similarly, the last common ancestor of the rosids likely shared eight to nine CC-type genes, the number growing to 15–24 in the eudicot species investigated here.
FIG. 5.—
The expansion of CC-type GRX class compared with the CPYC and CGFS classes in land plants. The predicted number of genes in the last common ancestor was estimated by orthologous clustering and is given at each node. Genome duplications denoted by stars are mentioned in the following articles (Simillion et al. 2002; Maere et al. 2005; Zhang et al. 2005; Rensing et al. 2008). (*) Denotes the omission of the Sorghum CPYC-like gene (SbGRX32), which might be a pseudogene. Abbreviations: CC, CC-type GRX; Physcom., Physcomitrella; C. richardii, Ceratopteris richardii; and Brachyp., Brachypodium. The origin of the ferns is estimated to have occurred ∼365 Ma (Pryer et al. 2004). The Zea mays/Sorghum/Oryza lineage diverged from the Brachypodium/Hordeum/Triticum lineage approximately 50–60 Ma (Paterson et al. 2003), the Zea/Sorghum lineage diverged from the Oryza lineage ∼42 Ma (Paterson et al. 2003), and the Zea lineage diverged from Sorghum 10–14 Ma (Gaut et al. 1996). The Brachypodium lineage diverged from the Hordeum/Triticum lineage 35–40 Ma (Bossolini et al. 2007), and the Hordeum lineage diverged from the Triticum lineage ∼12 Ma (Wolfe et al. 1989). The divergence of Populus from Arabidopsis occurred 100–120 Ma (Tuskan et al. 2006). Divergence of A. thaliana and A. lyrata is estimated at 3.0–5.8 Ma (Clauss and Koch 2006).
Selective Pressures at Work in the GRX Family
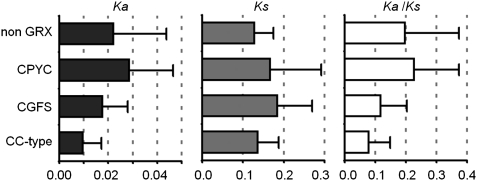
Alignment of A. lyrata and A. thaliana GRX genes led to the identification of orthologous gene pairs, where all but one A. lyrata GRX (AlROXY18) had a corresponding orthologue. Values for Ka, Ks, and Ka/Ks were determined for each orthologue pair and averages were found for each class, for comparative purposes, a survey of 22 previously reported A. thaliana–A. lyrata gene pairs (Wright et al. 2002) was employed to calculate the average and standard deviation values for non-GRX genes. This non-GRX set yielded Ka values of 0.022 ± 0.0217, Ks values of 0.127 ± 0.047, and Ka/Ks values of 0.198 ± 0.176. The analyses show that Ka, the rate of accumulation of nonsynonymous substitutions is lower for the CC-type class (0.010 ± 0.008) as compared with CPYC class (0.029 ± 0.018), CGFS class (0.018 ± 0.010), and the non-GRX set, summarized in figure 6. The difference between the Ka of CC-type genes compared with CPYC genes was determined to be statistically significant (P = 0.020), as was the comparison of CC-type genes with the non-GRX sample (P = 0.017).
FIG. 6.—
Estimates of sequence divergence between A. lyrata and A. thaliana orthologous GRX sequences (nCPYC = 8, nCGFS = 6 and nCC-type = 24). Reported values from a survey of 22 non-GRX gene pairs are described in Wright et al. (2002).
The Ka/Ks values also varied, the CPYC class recording 0.226 ± 0.147, similar to the non-GRX set, compared with 0.118 ± 0.086 for the CGFS class and 0.078 ± 0.069 for CC-type class. The differences were found to be significant in the comparison of the CPYC and CC-type genes (P = 0.025) and of the non-GRX set with CC-type genes (P = 0.006). Thus, CC-type GRXs were likely undergoing significantly greater purifying selection compared with CPYC and representative non-GRX genes, suggestive of crucial functions for CC-type class genes.
Recent Lineage-Specific Tandem Duplications of CC-Type GRX Genes
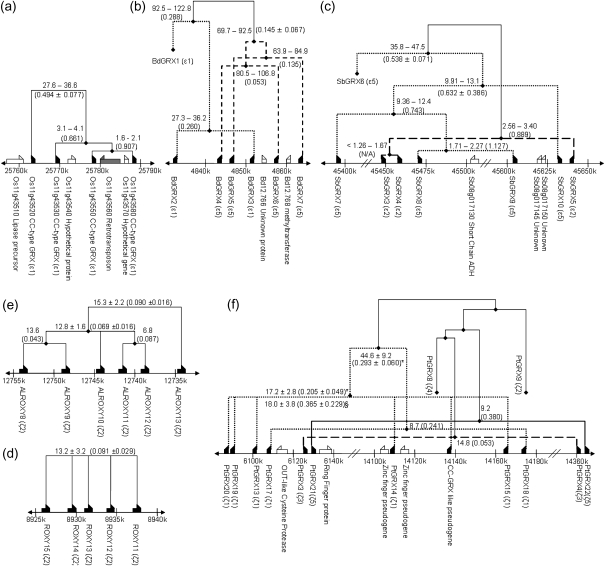
Upon further investigation of CC-type genes in angiosperms, it is apparent that many are found arranged in tandem array of genes (TAGs), identified in Oryza, Brachypodium, Sorghum, Arabidopsis, and Populus but not Vitis. Figure 7 shows the phylogenic relationships between the tandemly arrayed genes at the amino acid level, the estimated time of duplication, and the form of selection experienced (using Ka/Ks). However, gene conversion events could lead to a distortion of the apparent rate of substitution of tandem duplicates to some extent, as observed for the NBS–LRR gene family (Mondragon-Palomino and Gaut 2005). GENECONV was employed for the detection of such events, which yielded a negative result in these TAG regions. This does not, however, rule out the possibility of gene conversion events for duplicates that share high sequence homology or where a gene conversion event is hidden by later substitutions. In Oryza, a 26-kb TAG on chromosome 11 contains four CC-type genes Os11g43520, Os11g43530, Os11g43550, and Os11g43580 in uniform transcriptional orientation that appear to have originated from a single gene (OC-ε1) present at the time of branching from Sorghum. Using Ks information, duplications were approximated to have occurred ∼30 Ma, followed by duplication events 3–4 Ma and, later, 1.6–2.1 Ma. Although the last two duplications are relatively recent, other genes are found interspersed at this locus, including a putative retrotransposon. GRX genes in the TAG appear to be under relaxed selection, with the most recent duplicate pair recording a value for Ka/Ks of 0.907. For older duplicates, values for Ka/Ks vary between 0.494 and 0.661. In Brachypodium, a TAG containing six CC-type genes, a methyltransferase-encoding gene, and a gene of unknown function are observed in a 120-kb section of scaffold 12. The dates estimated for the duplications appear large as accumulation of silent mutations was more pronounced in this lineage. The data suggest duplications at 30, 70, 90 (two instances), and 110 Ma; however, this may be spurious due to the fact that Brachypodium and Oryza/Sorghum lineages are approximated to have diverged only 50–60 Ma (Paterson et al. 2003) and that phylogenetic data (fig. 3c) show that this tandem duplication behavior to have occurred independently in the three lineages. The Ka/Ks values for the Brachypodium cluster were low, ranging between 0.053 and 0.288, indicating purifying selection. In Sorghum, a cluster of seven CC-type genes was identified in a 250-kb section of chromosome 8, with one gene (SbGRX5) in an inverted transcriptional orientation. The seven genes belong to two phylogenetically distinct ancestral genes. Recent duplication of SbGRX3 and SbGRX4 (estimated at <1.7 Ma) resulted in the two CDSs being identical, and other duplications are approximated to have occurred 2.0, 10.9, and 11.5 Ma. Despite these apparently recent duplications, unrelated genes are noted to be interspersed within this TAG. The Ka/Ks values were in the range of 1.127 for recent duplicates compared with 0.538 for older pairs.
FIG. 7.—
Phylogenetic relationship between CC-type GRX genes presents as TAGs. The Neighbor-Joining comparisons were conducted using predicted amino acid sequences, nodes with bootstrap scores <50 had branches collapsed. TAGs were identified on Oryza chromosome 11 (a), Brachypodium superscaffold 12 (b), Sorghum chromosome 8 (c), A. thaliana chromosome 4 (d), A. lyrata superscaffold 7 (e), and Populus chromosome 14 (f). No GRX gene–containing TAG clusters were identified in the Vitis genome. The two dashed line styles indicate the phylogenetically distinct genes at the loci. The estimated time of duplication is given at the branch point (Ma), and in parentheses, the Ka/Ks values are given. Although none were detected, gene conversion events could cause an underestimation of the actual time of tandem duplication. (*) Denotes the exclusion of the pseudogene from the analysis, and § denotes the inclusion of the pseudogene.
In A. thaliana, the GRX cluster consists of five tandemly arrayed genes from OC-ζ2 in the same transcription orientation on a 12-kb section of chromosome 4, which is free of any intervening non-GRX genes. There is limited variation among the peptide sequences, sharing an average of 93.2% identity at the amino acid level. Duplications are estimated to have occurred 6.6, 11.4, 15.4, and 13.8 Ma, consistent with data from A. lyrata. The Ka/Ks statistic for the duplicate genes in the TAG cluster was in the range of 0.06–0.15 in the two species. The A. lyrata TAG contains six gene copies; a corresponding A. thaliana gene has been lost since divergence of the two species. In Populus, a region containing 11 CC-type genes and a GRX-like pseudogene are present on chromosome 14 at three loci separated by some 8.3 Mb. These 11 genes originated from three distinct ancestral genes at the time of divergence from Arabidopsis. The clade containing five CC-type genes and pseudogene is estimated to have come about 15–20 Ma. The presence of paralogous pairs at this locus, PtGRX17 and PtGRX18 (CCMS), PtGRX3 and PtGRX4 (CCMC), and PtGRX21 and PtGRX22 (CCLC), implies a tandem segmental duplication dating 8–15 Ma. Together with phylogenetic data, the absence of GRX TAGs in the Vitis genome further demonstrates that tandem duplications observed in Populus and Arabidopsis were independent to tandem duplication events in the monocots.
Discussion
The Expansion of the GRX Family in the Green Lineage
Expansion of gene families through gene duplications has been found to be widespread in plant evolution; in fact, most genes in Arabidopsis are members of large gene families (Cannon et al. 2004). Previous work has shown that the GRX gene family underwent an expansion in numbers in the evolutionary time period between the occurrence of bryophytes and angiosperms. However, this expansion was limited only to CC-type GRXs (fig. 1) that have recently been demonstrated to exert crucial function during floral organogenesis and stress response (Wang et al. 2009). In order to gain a deeper understanding of the mechanism and timeline of this expansion, 195 GRX genes were collated, including 91 newly identified sequences for phylogenetic analysis from Ostreococcus, Micromonas, Volvox, Chlamydomonas, Physcomitrella, Selaginella, Vitis, A. thaliana, Sorghum, Brachypodium, and Populus.
The analysis of O. Micromonas, Volvox, and Chlamydomonas GRX genes (fig. 3) indicates that the CGFS class consisted of four genes in early algae, the ancestors of the Prasinophyceae and the Chlorophyceae. In contrast, algal CPYC class genes do not segregate into clear OCs, which is suggestive of the existence of only one isoform in ancestral green algae. The presence of four CPYC genes in Physcomitrella demonstrates that a modest increase in CPYC isoforms occurred in the transition of plants to land. This observation, together with significant sequence homology between the basal CC-type genes PpGRX1/PpGRX2 and the CPYC class PpGRX6 (fig. 4), shows that CPYC gene duplications were likely key events in the genesis of the initial CC-type GRX.
The two reported paralogous CC-type GRXs in Physcomitrella both possess the C-terminal ALWL motif, which had been shown to be important for CC-type function in the case of ROXY1, due to its role in mediating interaction with TGA factors (Li et al. 2009). In light of this experimental data, the nuclear localized CC-type GRX–TGA interaction could be an ancient characteristic of CC-type class GRXs. Of the three CC-type genes in the Selaginella genome, two have retained the ALWL motif and one isoform SmGRX3 appears to lack the C-terminal ALWL motif (fig. 2), which is likely to have implications for TGA-binding properties. This C-terminal motif is conserved in most angiosperm CC-type genes with the exception of members of OC-ζ5 and several genes from Brachypodium, Sorghum, and Vitis, which do not cluster together and are unlikely to be capable of complementing the roxy1 mutant.
Phylogenetic analysis indicates that the last common ancestor of monocots and eudicots possessed four to five CC-type genes. Following the monocot–eudicot split, data show that OC-ε of the monocots and OC-ζ of the eudicots underwent rapid gene duplications to produce the five existing sub-OCs (fig. 3). Chromosomal positions of OC-ε and ζ members point to ancient tandem duplications as being partly responsible for OC-ε and ζ dividing into five subgroups as members of ζ1, ζ3, and ζ5 are dispersed along a syntenic segment in Arabidopsis, Populus, and Vitis (data not shown). Similarly, ε2 and ε5 orthologues are seen dispersed along another syntenic segment in the monocots, Oryza, Sorghum, and Brachypodium. Members of OC-ε and ζ were also the subjects of tandem duplications, which were detected in five of the six angiosperm genomes in this study. TAG clusters make up an estimated 17% of genes in Arabidopsis (Zhang and Gaut 2003) and are important because they are suggested to supply the genetic redundancy that can lead to the evolution of novel gene functions (Ohno 1970). Analysis of the functional bias in TAG gene ontology showed that genes involved in response to both biotic and abiotic stress and those which had “other enzymatic” functions were overrepresented in Oryza and Arabidopsis TAG clusters (Rizzon et al. 2006). Examples include proteins in the mitogen-activated kinase pathways (abiotic stress) and disease-resistance proteins of the TIR–NBS–LRR classes (biotic stress; Rizzon et al. 2006). Both phylogeny and molecular clock analyses point to the establishment of TAGs to be unrelated, separate events in the five plant lineages investigated. No recent CC-type TAG cluster was identified in the Vitis genome, indicating that TAG cluster formation in the rosids likely started less than 125 Ma, consistent with our molecular clock analyses.
The Driving Forces behind CC-Type Gene Duplications and Functional Recruitment
Since the monocot–eudicot split some 140 Ma, the rate of CC-type gene duplication is approximately 15-fold higher than for CPYC and CGFS GRXs and nearly 3-fold higher than the Arabidopsis genome-wide average (Lynch and Conery 2000). Reported consequences of gene duplication include nonfunctionalization of one copy, neofunctionalization of one copy, or deterioration of both copies to a point where they are subfunctionalized and share a single function (Lynch and Conery 2000). Limited subfunctionalization has been suggested as an important step in eventual neofunctionalization (He and Zhang 2005; Rastogi and Liberles 2005). This particular mode of adaptive evolution has likely occurred repeatedly in the CC-type class of GRXs, where initially redundant duplicates (in expression pattern and peptide properties) undergo a period of subfunctionalization caused by genetic drift. This phase of genetic drift, measured by Lynch and Conery (2000), is consistent with nearly neutral selection detected in recent CC-type gene duplicates in Oryza and Sorghum (fig. 7). Following this period, duplicate copies are guided by natural selection on cis-acting elements or other gene expression defining features to adopt distinct domains of gene expression thereby establishing individual functions.
This duplication mechanism is consistent with functional data from Li et al. (2009), where it has been shown that several CC-type GRX CDSs are virtually interchangeable with ROXY1. The secondary structures, active site Cys, and GSH-binding residues appear to be preserved in most CC-type isoforms and many share the C-terminal ALWL motif crucial for TGA interaction (supplementary fig. S1, Supplementary Material online). ROXY19, which has been shown to function in pathogen defense by affecting defensin gene repression (Ndamukong et al. 2007), shares all these features with ROXY1. Thus, if ROXY19 is expressed properly in a floral context, it complements the roxy1 mutant and can exert the same target protein modifications as ROXY1 in the nucleus, that is, the interaction and likely modification of TGA transcription factors (Li et al. 2009). Interestingly, this cross complementation property is not limited only to ROXY1 and ROXY19, with five further CC-type class genes capable of complementing roxy1. The similarity of the tightly regulated, dynamic floral expression patterns exhibited by the Arabidopsis ROXY1, ROXY2 (Xing et al. 2005; Xing and Zachgo 2008), as well as the rice OsROXY1 and OsROXY2 (Wang et al. 2009) CC-type GRXs also support this model. Additionally, overexpression of ROXY1 as well as OsROXY1 in Arabidopsis interferes with cellular redox balance causing an increase in H2O2 levels and an increased susceptibility to the necrotrophic pathogen Botrytis cinerea in leaves (Wang et al. 2009) further emphasizing that their biochemical activity depends on their expression regulation. Importantly, this mechanism explains in part how apparently interchangeable CC-type CDSs can be beneficially recruited, over sufficiently long evolutionary times, to different ROS-linked biological functions such as development (ROXY1 and ROXY2) and pathogen response (ROXY19).
Conclusion
This analysis has shown comprehensively that CC-type GRXs are absent in the genomes of unicellular green algae, and the most basal CC-type gene encountered so far belongs to the moss Physcomitrella. The described findings show that the CC-type class likely arose from the CPYC class in a period when the CPYC class itself was undergoing moderate increase in numbers and diversity. From an evolutionary standpoint, the sequestration of redox homeostasis proteins (GRXs) to signaling functions in development (OC-α, ROXY1, ROXY2) and pathogen stress response (OC-β, ROXY19) is fascinating. The CC-type class GRXs underwent a period of rapid expansion after the monocot–eudicot split (fig. 5). Some duplicates from this period continued to undergo tandem duplications less than 125 Ma in a number of separate angiosperm lineages until recently. Significantly slow evolutionary changes to CC-type proteins in Arabidopsis (fig. 6), together with previously demonstrated cross complementation of several isoforms, indicate that the main benefit in having a large complement of similar CC-type GRXs could lie in the increased capacity to control temporal and spatial gene expression.
Supplementary Material
Supplementary table S1 and figure S1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by an Australian Postgraduate Award Scholarship at Swinburne University of Technology to M.Z.
Supplementary Material
Acknowledgments
The authors wish to thank Peter Gollan (Faculty of Life and Social Sciences, Swinburne University of Technology, Hawthorn, Victoria 3122, Australia) for informative discussions and comments on the manuscript.
References
- Aslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA. Selaginella and 400 million years of separation. Annu Rev Plant Biol. 2008;60:223–238. doi: 10.1146/annurev.arplant.59.032607.092851. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, et al. The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot. 2002;53:1367–1376. [PubMed] [Google Scholar]
- Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. doi: 10.1111/j.1365-313X.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- Bushweller JH, Aslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem. 2006;281:26280–26288. doi: 10.1074/jbc.M601354200. [DOI] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA. Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci. 2006;11:449–459. doi: 10.1016/j.tplants.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Comeron JM. K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Doke N. NADPH-dependent O2-generation in membrane fractions isolated from wounded potato tubers inoculated with Phytophtora infestans. Physiol Plant Pathol. 1985;27:311–322. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP. Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett. 2003;555:443–448. doi: 10.1016/s0014-5793(03)01301-2. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis subcellular database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herald VL, Heazlewood JL, Day DA, Millar AH. Proteomic identification of divalent metal cation binding proteins in plant mitochondria. FEBS Lett. 2003;537:96–100. doi: 10.1016/s0014-5793(03)00101-7. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. Characterization of the enzymatic mechanism of Escherichia coli glutaredoxin. J Biol Chem. 1979;254:3672–3678. [PubMed] [Google Scholar]
- Hoog JO, Jornvall H, Holmgren A, Carlquist M, Persson M. The primary structure of Escherichia coli glutaredoxin. Distant homology with thioredoxins in a superfamily of small proteins with a redox-active cystine disulfide/cysteine dithiol. Eur J Biochem. 1983;136:223–232. doi: 10.1111/j.1432-1033.1983.tb07730.x. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lam E, Benfey PN, Gilmartin PM, Fang RX, Chua NH. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA. 1989;86:7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Zimmer AD, Rensing SA, Reski R. Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 2008;13:542–549. doi: 10.1016/j.tplants.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Lemaire SD. The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth Res. 2004;79:305–318. doi: 10.1023/B:PRES.0000017174.60951.74. [DOI] [PubMed] [Google Scholar]
- Li S, et al. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell. 2009;21:429–441. doi: 10.1105/tpc.108.064477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik B, Axelsson K. Role of cytoplasmic thioltransferase in cellular regulation by thiol-disulphide interchange. Biochem J. 1980;15:125–130. doi: 10.1042/bj1900125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- Meyer Y, et al. Glutaredoxins and thioredoxins in plants. Biochim Biophys Acta. 2008;1783:589–600. doi: 10.1016/j.bbamcr.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Gaut BS. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol. 2005;22:2444–2456. doi: 10.1093/molbev/msi241. [DOI] [PubMed] [Google Scholar]
- Navrot N, Gelhaye E, Jacquot JP, Rouhier N. Identification of a new family of plant proteins loosely related to glutaredoxins with four CxxC motives. Photosynth Res. 2006;89:71–79. doi: 10.1007/s11120-006-9083-7. [DOI] [PubMed] [Google Scholar]
- Ndamukong I, et al. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by genome duplication. Heidelberg (Germany): Springer-Verlag; 1970. [Google Scholar]
- Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA. Structure and evolution of cereal genomes. Curr Opin Genet Dev. 2003;13:644–650. doi: 10.1016/j.gde.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell. 1994;6:65–74. doi: 10.1105/tpc.6.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot. 2004;91:1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Ramakrishna W, et al. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics. 2002;162:1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Jacquot JP. Genome-wide analysis of plant glutaredoxin systems. J Exp Bot. 2006;57:1685–1696. doi: 10.1093/jxb/erl001. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J Biol Chem. 2002;277:13609–13614. doi: 10.1074/jbc.M111489200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci. 2004;61:1266–1277. doi: 10.1007/s00018-004-3410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. GENECONV: a computer package for the statistical detection of gene conversion. 1999. Distributed by the author. St Louis (MO): Department of Mathematics, Washington University. [Google Scholar]
- Schopfer P, Plachy C, Frahry G. Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol. 2001;125:1591–1602. doi: 10.1104/pp.125.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Dayhoff M. Matrices for detecting distant relationships. In: Dayhoff M, editor. Atlas of protein sequences. Washington (DC): National Biomedical Research Foundation; 1979. pp. 353–358. [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide-sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A. Gymnosperm orthologues of class B floral homeotic genes and their impact on understanding flower origin. Crit Rev Plant Sci. 2004;23:129–148. [Google Scholar]
- Tsugane K, et al. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell. 1999;11:1195–1206. doi: 10.1105/tpc.11.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xing S, Birkenbihl RP, Zachgo S. Conserved functions of Arabidopsis and rice CC-type glutaredoxins in flower development and pathogen response. Mol Plant. 2009;2:323–335. doi: 10.1093/mp/ssn078. [DOI] [PubMed] [Google Scholar]
- Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc Biol Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Rates of synonymous substitution in plant nuclear genes. J Mol Evol. 1989;29:208–211. [Google Scholar]
- Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol Biol Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- Xing S, Lauri A, Zachgo S. Redox regulation and flower development: a novel function for glutaredoxins. Plant Biol (Stuttg) 2006;8:547–555. doi: 10.1055/s-2006-924278. [DOI] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development. 2005;132:1555–1565. doi: 10.1242/dev.01725. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gaut BS. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 2003;13:2533–2540. doi: 10.1101/gr.1318503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu GH, Guo XY, Fan LJ. Two ancient rounds of polyploidy in rice genome. J Zhejiang Univ Sci B. 2005;6:87–90. doi: 10.1631/jzus.2005.B0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson H, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–116. [Google Scholar]
- Zybailov B, et al. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.