Abstract
Olfactory receptor (OR) is a large family of G protein–coupled receptors that can detect odorant in order to generate the sense of smell. They constitute one of the largest multiple gene families in animals including primates. To better understand the variation in odor perception and evolution of OR genes among primates, we computationally identified OR gene repertoires in orangutans, marmosets, and mouse lemurs and investigated the birth-and-death process of OR genes in the primate lineage. The results showed that 1) all the primate species studied have no more than 400 intact OR genes, fewer than rodents and canine; 2) Despite the similar number of OR genes in the genome, the makeup of the OR gene repertoires between different primate species is quite different as they had undergone dramatic birth-and-death evolution with extensive gene losses in the lineages leading to current species; 3) Apes and Old World monkey (OWM) have similar fraction of pseudogenes, whereas New World monkey (NWM) have fewer pseudogenes. To measure the selective pressure that had affected the OR gene repertoires in primates, we compared the ratio of nonsynonymous with synonymous substitution rates by using 70 one-to-one orthologous quintets among five primate species. We found that OR genes showed relaxed selective constraints in apes (humans, chimpanzees, and orangutans) than in OWMs (macaques) and NWMs (marmosets). We concluded that OR gene repertoires in primates have evolved in such a way to adapt to their respective living environments. Differential selective constraints might play important role in the primate OR gene evolution in each primate species.
Keywords: olfactory receptor, birth-and-death evolution, accelerated evolution, primates
Introduction
Olfaction enables an animal to find food, mate, and detect danger. In vertebrates, the ability of odor perception is mediated by olfactory receptors (ORs) that are embedded in olfactory epithelia in nasal cavities, and ORs are the primary receptors in the main olfactory epithelium system (Buck and Axel 1991; Mombaerts 2004b). OR genes are members of G protein–coupled receptors (GPCRs) and constitute one of the largest multigene families in vertebrates (Buck and Axel 1991; Mombaerts 2004a).
OR genes were first identified in rats (Rattus norvegicus) in 1991 (Buck and Axel 1991), which opened the door for subsequently molecular analyses. At the time of this study, OR gene repertoires had been described in many vertebrate species, such as human (Homo sapiens), mouse (Mus musculus), dog (Canis familiaris), chicken (Gallus gallus), some teleost fishes, etc. (Niimura and Nei 2003, 2005a, 2005b, 2007). Comparative analysis of these repertories can help understand the mechanism and species-specific evolution of olfaction systems (Nei et al. 2008), as it is known that the importance and sensitivity of smell varies significantly among different vertebrate species, even among species that are phylogenetically close to each other (Niimura and Nei 2007; Go and Niimura 2008; Nei et al. 2008). Recent studies also revealed that the number of OR genes and the fraction of pseudogenes are quite different between macrosmatic (having keen olfactory sense) and microsmatic (having poor olfactory sense) species (Niimura and Nei 2007). For example, humans have far fewer number of functional OR genes than rodents and dogs, and the fraction of pseudogenes in humans is much larger than that in rodents and in dogs (Quignon et al. 2003; Niimura and Nei 2005a). It is generally believed that this has contributed to the degeneration of the odor perception in humans.
Based on the interpretation of neuroanatomical features, it was believed that the olfactory portion of the primate brain has declined significantly in comparison to other mammals; therefore, primates are viewed as microsmatic animals (King and Fobes 1974; Baron et al. 1983). A number of OR genes were also cloned, and detailed molecular analyses had been carried out. Rouquier et al. (1998, 2000) found an increase in the pseudogenization process from New World monkeys (NWMs) to Old World monkeys (OWMs) and apes, with humans having the highest fraction of pseudogenes (∼70%). From analysis of a small number of OR genes, Gilad, Bustamante, et al. (2003; Gilad, Man, et al. 2003) suggested that the fractions of pseudogenes differed remarkably among humans and nonhuman primates, and it is likely that there was a relaxation of evolutionary constraint on OR genes in humans. In another paper, they randomly sequenced ∼100 OR genes from each of 19 primate species and found that the fraction of OR pseudogenes in apes, OWMs, and one NWM (the howler monkey) is higher than those in NWMs and prosimians, which coincided with the acquisition of full trichromatic color vision in primates (Gilad et al. 2004). It was suggested that “advanced” primates have reduced sense of olfaction and rely more on vision than olfaction for survival. However, the OR gene sequences they obtained were not the full length of coding region, and some potential nonsense or frameshift mutations located near the ends of the open reading frame could not have been identified. With the availability of whole-genome sequences, complete OR gene repertoires were identified in human, chimpanzee, and macaques. Contrary to previous reports by Gilad et al. (2005), surprisingly, it was found that these three primates had almost the same fraction of OR pseudogenes; therefore, the difference in olfaction cannot be fully explained by the pseudogenization process (Niimura and Nei 2003; Gimelbrant et al. 2004; Go and Niimura 2008). However, these three primate species all have trichromatic vision, which still cannot address the “vision priority hypothesis.”
To date, the evolutionary trajectory of OR genes in primate lineages is far from clear. In this paper, we identified the OR gene repertoires from the complete genome sequences of three additional primates: orangutans (ape), marmosets (NWM), and mouse lemurs (prosimian). These OR gene repertoires provide an opportunity to detect the underlying genetic basis of the evolution and diversity of odor perception in the entire primate lineage. We then performed comparative analyses and illustrated the pattern of gains and losses of OR genes. In parallel, we also estimated the selective pressures on OR genes in each lineage. As the results, we found that the fractions of pseudogenes might be not the same as reported by Gilad et al. (2004) both in trichromatic and dichromatic primates, and the OR repertoires in primates surveyed have been shaped by substantial birth-and-death processes, and some OR genes in apes have undergone significant accelerated evolution.
Methods
Genome Sequences
The draft genome sequences of orangutan (Pongo pygmaeus abelii; ponAbe2, released in July 2007; 6× coverage) and marmoset (Callithrix jacchus; calJac1, released in June 2007; 6× coverage) were downloaded from the University of California–San Cruz Genome Bioinformatics Web site (http://genome.ucsc.edu). The low-coverage genome sequence of mouse lemur (Microcebus murinus, 2× coverage) was downloaded from Ensembl Genome Brower (http://www.ensembl.org/).
OR Gene Identification
We used the same data mining method as previously reported in Niimura and Nei (2007) and Go and Niimura (2008). Briefly, it involves three steps. First, we use previous published OR genes in vertebrates as query sequences and conducted a TBlastN (Altschul et al. 1990) search against the genome sequences with a cutoff E value of 1 × 10−20 to detect the OR gene repertoires. Second, the nonredundant sequences were extended the regions of Blast hit sequences to 5′ and 3′ directions along the genome sequence, and the potential coding regions were extracted from these sequences. Third, we reconducted a TBlastN against the genome sequences using potential coding sequences to detect the OR pseudogenes that contain interrupting stop codons or frameshifts. If the coding regions without either start codons or stop codons, or both of them in the amino acid sequences, they were assumed as partial OR genes.
Phylogenetic Analysis
For multiple alignments of translated amino acid OR sequences, the program FFT-NS-I nested in Mafft version 5 (Katoh et al. 2005) was used with default parameters. The phylogenetic OR gene tree was constructed using MEGA3 software (Kumar et al. 2004) and the Neighbor-Joining (Saitou and Nei 1987) method with the Poisson correction distances and was carried out by 1,000 bootstrap replications.
Estimation of OR Gene Birth-and-Death Evolution
The evolutionary processes such as gene duplication and gene losses can introduce incongruence between gene phylogeny and species phylogeny. To estimate the number of OR genes in ancestral species and gains and losses of genes during primate evolution, we used the modified reconciled tree method as described by Nam and Nei (2005). Briefly, by comparing with the bootstrap condensed gene tree and the species tree under the parsimony principle, the number of ancestral genes can be estimated, with the information of the past occurrence of gene expansion and contraction. We used a 70% condensed tree of OR genes for analyses.
Identification of OR Gene Orthologous
We generated the one-to-one OR gene orthologous in primates using best reciprocal BlastP method. Alignments of primate OR orthologous were performed using ClustalX (Thompson et al. 1997) with default parameters. To further identify orthologous relationships, we constructed a phylogenetic tree using 1,771 OR intact genes in primates and identified phylogenetic clades (with at least 90% bootstrap value support) containing genes from five species. If the OR genes in primates have one-to-one relationship and also involved in the same phylogenetic clade, they were regarded as orthologous. At last, a total of 70 primate OR gene orthologous were obtained.
Selective Pressure on Primate Lineages
The ratio between nonsynonymous substitutions and the synonymous substitutions (ω) can measure the selective pressure acting on protein coding genes. The ω < 1, ω = 1, and ω > 1 represent negative selection, neutral selection, and positive selection, respectively. To test the selective pressure acting on OR genes in primates, we performed codon-based maximum likelihood method using CODEML nested in PAML package (Yang 2007). The free-ratio model (Yang and Nielsen 1998) assumes independent ω ratio for all branches in the phylogeny. Compared with one-ratio model (Goldman and Yang 1994) by likelihood ratio test (LRT), we can test the heterogeneity of ω ratio among primate species. Next, each species was, respectively, designated as foreground branch. By comparing two-ratio model (Yang 1998) with one-ratio model, we derived the genes under accelerated or relaxed evolution. To identify putative cases of positive selection in each branch, branch site model was applied. Benjamini et al. (2001) correction and a false discovery rate of 5% were applied to adjust the significance level.
Results
OR Gene Repertoires in Primates
Even though the nearly complete OR gene repertoires had been previously described in humans, chimpanzees, and macaques (Niimura and Nei 2003; Go and Niimura 2008), their evolutionary trajectory in the primate lineages has not been fully investigated and is still poorly understood. In order to obtain a complete picture of OR evolution, we sought to identify the OR repertoire in three other primate species: orangutans (P. pygmaeus abelii), marmosets (C. jacchus), and mouse lemurs (M. murinus). To avoid result discrepancy, we used the same data mining method and standard as previously described in Niimura and Nei (2007) and Go and Niimura (2008). Because some of these genome sequences were still incomplete or of low coverage at the time of this work, it is likely that the number of OR genes may be underestimated. Therefore, we also carefully cataloged partial OR genes from the draft genome sequences, which may be incomplete due to sequencing or assembly error. This type of sequences is treated separately from the obvious functional OR genes or pseudogenes (see below). Table 1 shows the number of OR genes identified from these three primate species as well as previously documented number of OR genes from human, chimpanzee, and macaque.
Table 1.
Number of OR Genes and Pseudogenes in Primates
| Apes |
OWM | NWM | Prosimian | |||
| Species | Humana | Chimpanzee b | Orangutan | Macaque b | Marmoset | Mouse lemur |
| Intact genes | 387 | 380 | 312 | 309 | 383 | 371 |
| Partial genes | 0 | 19 | 9 | 17 | 12 | 226 |
| Pseudogenes | 415 | 414 | 366 | 280 | 258 | 177 |
| Fraction of pseudogenes (%) | 51.7 | 50.9∼53.3 | 53.3∼54.6 | 46.2∼49 | 39.5∼41.3 | Unknown |
OR gene number is from Niimura and Nei (2007).
OR gene number is from Go and Niimura (2008).
In contrast to nonprimate mammals, which reportedly have very different number of OR genes among them (Niimura and Nei 2005a, 2005b, 2007), the total number of OR genes generally do not show dramatic variation among primates (table 1). Except for macaques and orangutans, which have the fewest number of intact OR genes (309 and 312, respectively), the rest of the primates have similar number of OR genes. The NWMs (marmosets) has 383 intact OR genes, almost the same as in humans (387) and chimpanzees (380). We also identified 371 intact OR genes from the mouse lemurs. However, the mouse lemur genome sequence has not been fully completed, which consists many short contigs and is estimated to cover only ∼80% of the whole genome. It is possible that we had missed some potential intact OR genes. To remedy for this, OR partial genes with length greater than 250 codons were regarded as potential intact genes. In the mouse lemur genome, there were 80 of such potential intact OR genes. Therefore, we estimated that mouse lemurs had the highest number of intact OR genes among primates.
Previous studies have reported that the fraction of pseudogenes in OR gene repertoire varies substantially among primates species, which was interpreted as the cause of variation in olfaction abilities (Rouquier et al. 2000; Gilad, Man, et al. 2003). For example, Rouquier et al. (2000) found that the fraction of pseudogenes in human is ∼70% and only ∼27% in OWMs and hypothesized that the pseudogene fraction can be used as an indicator for olfactory sensory function. As shown in table 1, our analysis revealed that the three great apes (human, chimpanzee, and orangutan) have similar fraction of pseudogenes (∼50%), whereas the pseudogene fraction is smaller for OWM (46%) and NWM (∼40%). The proportion of pseudogenes in NWMs is significantly different from other primates (P < 0.01, Fisher’s exact test), which is consistent with previous reports that NWMs have a relatively lower fraction of pseudogenes (Rouquier et al. 2000; Gilad et al. 2004). However, there are no significant differences among apes and OWMs. Due to the limitation of genome coverage, we only obtained 177 OR pseudogenes in mouse lemurs, therefore, unable to estimate with confidence the pseudogenes in this genome. The DNA sequences of intact OR functional genes from orangutans, marmosets, and mouse lemurs are available at http://zhanglab.ecnu.edu.cn/primateOR.html.
Birth-and-Death Evolution of OR Genes in Primates
Birth-and-death process is one of the most important mechanisms of gene family evolution (Nei and Rooney 2005; Demuth et al. 2006), by which gene families expanded by duplication and contracted by deletion. It is often considered as the result of the animal's adaptation to particular environmental conditions. Niimura and Nei (2005b, 2007) had reported evidence for OR gene gains and losses in vertebrate evolution, and the dynamic evolution of OR repertoires was also reported in Drosophila species (Nozawa and Nei 2007). We are interested to investigate whether the OR gene repertoires in primates had also undergone the birth-and-death process. To better understand the evolutionary dynamics of OR genes in the primate lineage, we estimated the number of OR genes in the common ancestor of present-day primates and ascertained the pattern of gene gains and losses. The results are then compared and reconciled to draw conclusions (see below). Due to the low quality of its genome sequence, the mouse lemur was not included in the following analysis.
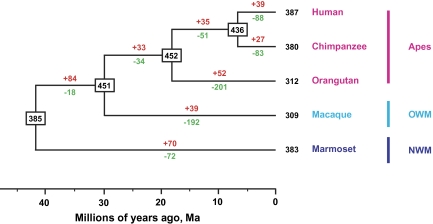
We constructed a phylogenetic tree of all newly identified intact OR genes in orangutans and marmoset, together with previously published intact OR genes in humans, and chimpanzees. Partial OR genes and pseudogenes were not included because they were much shorter than intact genes. The phylogenetic trees and divergence time of these primate groups were calculated according to procedures previously described by Steiper and Young (2006). We then surveyed the evolutionary history of OR genes in primates using a widely used reconciled tree method (Nam and Nei 2005; Nozawa and Nei 2007; Dong et al. 2009). Figure 1 shows the estimated numbers of OR genes in the ancestors of present-day primates and the evolutionary changes in the lineage leading to each species using 70% condensed tree. The result showed the number of ancestral OR gene after the divergence of the Anthropoids (∼42.9 Ma) was almost the same as current species. It is notable that there was no evidence for massive gene contractions at the internal nodes in the phylogenetic tree. We found that the number of OR genes (451) in the ancestor of the apes and OWMs (∼30.5 Ma) is even larger than that in the ancestor of the Anthropoids (385).
FIG. 1.—
Evolutionary changes of the number of OR genes in primates. The numbers within rectangular boxes are the estimated number of OR genes in ancestral species. The red numbers above each branch are those of gained genes, and the green numbers below each branch are those of lost genes.
Although the total number of OR genes stayed mostly stable along the branches in figure 1, we observed evidence for substantial gene turnover throughout the evolution. For example, humans and chimpanzees gained 39 and 27 genes and lost 88 and 83 genes, respectively. There are also more contractions than expansion events in the extant nodes, which caused the modern primates to have fewer functional OR genes than their ancestors. This is especially true for orangutans and macaques as they lost 201 and 192 genes in the branches leading to current species, respectively. The number of OR genes in the ancestral species is only an estimate based on phylogenetic model, such methods have proved to be useful and reasonable in previous analysis of evolution of gene families (Nam and Nei 2005; Niimura and Nei 2007; Nozawa and Nei 2007). To validate our result, we also used 50%, 60%, 80%, and 90% condensed tree, respectively, and got the same results (supplementary fig. S1, Supplementary Material online). Our results clearly showed that OR genes in primates have undergone extensive birth-and-death evolution.
Natural Selection on OR Genes
Previous studies had hypothesized that the difference of the olfaction ability among different primate species might have resulted from the relaxation of selective forces (Rouquier et al. 2000). Gilad, Bustamante, et al. (2003) pointed out that OR genes in humans and chimpanzees were under different selection pressures, some human OR genes are also reported under positive selection. We are interested to know to what extent the natural selection had exerted on the OR genes in primates and how the selective pressure is different in these species and resulted in their respective OR repertoires. To help answer this question, we next investigated the mode and tempo of primate OR genes evolution.
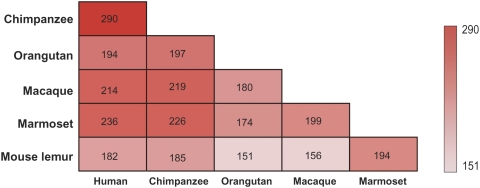
At first, we examined the orthologous relationship of OR genes among primates. Figure 2 shows the number of orthologous pairs between each two species. For example, the figure shows that humans have 290, 194, 214, 236, and 182 intact OR gene orthologous to chimpanzees, orangutans, macaques, marmosets, and mouse lemurs, respectively. It is very intriguing that humans share more orthologous with macaques and marmosets than with orangutans, which further indicated that the OR gene repertoires have experienced extremely dynamic evolution and differ considerably among primates. Again, we excluded mouse lemur from this analysis due to low quality of the genome assembly. At last, 70 orthologous quintets were obtained for further analyses, these are OR genes that are present in all five primates studied.
FIG. 2.—
The number of orthologous OR genes between pairs of primate species. Color intensity (as shown in the color bar) indicates the number of OR gene orthologs.
We further calculated the ratio of synonymous and nonsynonymous rates of substitutions (ω) for the OR genes to estimate the strength of natural selection they are subjected to. We applied three alternative evolutionary models to address this problem. To avoid any bias, we used a maximum likelihood method implemented in the PAML software package and three alternative models (Goldman and Yang 1994; Yang 2007). 1) At first, we selected the “one-ratio model” which assumed the same ω ratio for all lineages (Goldman and Yang 1994); the median ω ratio for individual primate OR genes was calculated to be ranging from 0.09 to 0.6 with a median at 0.32, which suggests that OR genes in primates underwent purifying selection. 2) We then used an alternative “free-ratio model” to calculate independent ω ratios for the different lineages, which can be used to infer different selection pressures between lineages (Yang 1998). We then performed LRTs and found that 24 of the 70 OR genes have significantly different ω ratios among lineages at 5% significance level. Based on these results, we concluded that OR genes in each lineage experienced different degrees of purifying selection. The median ω ratio varied across the five primate lineages with humans having the highest value (table 2). For example, the median ω ratios are 0.79, 0.49, 0.43, 0.22, and 0.24 in humans, chimpanzees, orangutans, macaques, and marmosets, respectively. The macaque and marmoset lineages exhibited a markedly higher degree of purifying selection. Whereas, the ω ratios of OR genes in humans, chimpanzees, and orangutans are significantly higher than in macaques and marmosets (two-sample Kolmogorov–Smirnov tests, P < 0.01). We did not find significant differences for the ω ratios among the three apes. We subsequently concatenated sequences of all the OR orthologous genes and estimated the ω ratio on each lineage. The results were very similar: OR genes evolved under extensively species-specific selective pressures, and apes showed reduced purifying selection (table 2). These results probably reflected an accelerated evolution compared with those of OWMs and NWMs.
Table 2.
Evolutionary Rates of Orthologous Quintets among Primates
| Median | Median (LRT significant) | Concatenated Gene | |
| Human | 0.79 (0.27–2.19) | 0.86 (0.12–2.15) | 0.54 |
| Chimpanzee | 0.49 (0.26–2.45) | 0.64 (0.14–1.58) | 0.43 |
| Orangutan | 0.43 (0.22–0.81) | 0.59 (0.25–1.32) | 0.41 |
| Macaque | 0.22 (0.16–0.47) | 0.23 (0.16–0.59) | 0.31 |
| Marmoset | 0.24 (0.15–0.37) | 0.23 (0.15–0.44) | 0.25 |
NOTE.—Parentheses enclose the number of 20–80 percentile ranges.
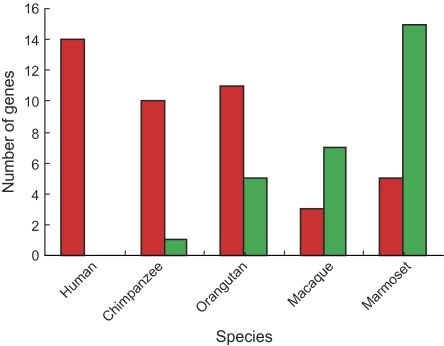
We last used the 3) two-ratio model, which assumed one ω ratio for the foreground branch and another for the background branch. We then conducted the analysis by, respectively, assigning each terminal lineage as foreground branch by using two-ratio model (Yang and Nielsen 1998). The results from this model suggested that apes had a higher number of OR genes showing accelerated evolution than macaques and marmosets, whereas marmosets had the highest number of OR genes under decelerated (fig. 3). By calculating ω ratio from concatenated OR genes, all apes showed significantly accelerated evolution, whereas OR genes in macaques and marmoset are under decelerated evolution (table 3). A high ω ratio suggested a reduced selection constraint on genes, which can be explained by either increased positive selection or relaxed selective pressure. To detect the positively selected coding site in each lineage, the branch site model (Zhang et al. 2005) was used with these species as foreground lineages, respectively. However, we did not find any evidence for positive selection in each species. These results led us to conclude that more OR genes in apes had undergone relaxed natural selection than in OWMs and NWMs.
FIG. 3.—
The number of OR genes under accelerated evolution (red color) and decelerated evolution (green color) in each species.
Table 3.
LRT to Estimate Variable Selection Pressures of Concatenated OR Gene among Primates
| Model | Foreground Species | No. of Parameter | 2*Δℓ | Estimates of Parameters |
| One ratio (ωB = ωF) | 9 | ωB = 0.32 | ||
| Two ratio (ωB ≠ ωF) | Human* | 10 | 33.1 | ωB = 0.31; ωF = 0.57 |
| Chimpanzee* | 10 | 16.2 | ωB = 0.32; ωF = 0.44 | |
| Orangutan* | 10 | 33.2 | ωB = 0.30; ωF = 0.41 | |
| Macaque | 10 | 7.3 | ωB = 0.33; ωF = 0.29 | |
| Marmoset | 10 | 55.4 | ωB = 0.35; ωF = 0.25 |
NOTE.—2 × Δℓ was obtained by 2(lnℓ1 − lnℓ0). lnℓ1 was the likelihood value for two-ratio model. lnℓ0 was the likelihood value for one-ratio model. All tests were compared with one-ratio model. Two-ratio model is significantly better than one-ratio model marked by “*” (P < 0.05). All tests were compared with one-ratio model. The two-ratio model is significantly better than one-ratio model in each lineage. ωB means the ω ratio for the background, and ωF means the ω ratio for the foreground.
Discussion
Generally speaking, primates are always regarded to have a poorly developed sense of smell, thus mainly rely on their visual system (King and Fobes 1974). Based on anatomical characteristics such as surface area of olfactory epithelium, olfactory bulb volume, primates are typical considered as microsmatic species (Stephan et al. 1988). In comparison with the OR gene repertoires in rodents and dogs which are macrosmatic animals, primates are thought to have relatively fewer number of intact OR genes and higher fraction of pseudogenes. It was also hypothesized that the advanced brain functions unique in primates, such as memory and emotion, had evolved in coordination with the reduction of the sense of smell (Laska et al. 2000; Shepherd 2004).
In this study, we identified OR gene repertoires in orangutans, marmosets, and mouse lemurs for the first time based on currently available genome sequences. We found that primates generally have similarly small number of OR functional genes (∼300–400), and there is little variation between different clades (OWM, NWM, and apes). Consistent with previous reports, we observed that the fraction of pseudogenes in NWM (∼40%) is relatively smaller than that in apes (∼50%) and OWMs (∼45%). However, because of the extensive birth-and-death processes, such variation in pseudogenization rates did not cause big difference in the number of intact and functional OR genes in individual primate genomes. In fact, the numbers of functional OR genes in human and chimpanzee are similar to that in NWMs and OWMs. Therefore, the reduced olfaction ability in apes and OWM cannot be explained by the higher pseudogenization rate or by a fewer number of functional OR genes.
Our results provided a complete picture of OR gene evolution, suggesting dramatic variations and dynamic turnover in the course of primate evolution. It is obvious that significant gene losses and gains took place in the branches leading to current species, coinciding with deterioration of olfaction in these species. Birth-and-death evolution is the main mode of evolution in multigene families, and OR gene families offered a good example of such process. Such process was generally considered as an adaptation by animals to their specific surviving environments. For examples, platypuses have limited number of OR genes that might be related to their semiaquatic life (Niimura and Nei 2007); the OR gene repertoire is degenerated in dolphins, likely due to their gained echolocation ability and aquatic lifestyle (Oelschlager and Kemp 1998). The evolution of OR genes is characterized by both adaptation and genomic drift (Nei 2007; Nei et al. 2008), which made the OR gene repertoires change dramatically. Recent observation on the copy number variation of OR genes in human may be the cause of different smelling ability among individuals (Redon et al. 2006; Nozawa et al. 2007; Young et al. 2008).
At the present, it is still far from clear what factors have contributed to the difference in olfactory ability among animals. Both the size of olfactory epithelial surface and the diversity of expressed OR genes were suggested to account for these differences (Ishii et al. 2001). It had been hypothesized that the reduction of sense of smell in humans paralleled with the relaxed selective pressure on human OR genes (Rouquier et al. 2000). Indeed, we found that there are differences in the selective pressures on OR gene orthologous in primates. This result raised two scenarios of OR gene evolution. One is that many OR genes in apes have experienced relaxed natural selection, which coincided with their accumulated OR pseudogenes (Gilad, Man, et al. 2003). Primates may no longer need some of the ORs used by their ancestors due to changes in their surviving external environments. If it is true, it is likely that apes are still in a phase of diminishing olfaction ability. An alternative scenario is that there were positive selections of OR genes in apes. Several previous studies have documented that genes involved in olfaction tend to under positive selection in humans (Clark et al. 2003; Nielsen et al. 2005). However, it is difficult to distinguish between these two opposite scenarios without comparing the genetic variation data between two species. Moreover, the influences of effective population size on natural selection have been well documented (Eyre-Walker and Keightley 1999). The results of present study also correspond to the impact of effective population size exerted on natural selection at the genomic scale. The evolutionary basis for the reduced purifying selection of OR genes in apes is still unknown but may be related to their reduced effective population sizes during the evolution which made natural selection less effective (Li and Tanimura 1987; Chen and Li 2001). It is hard to say that there is a reduced biological importance of olfaction in apes. The ongoing genome variation projects such as 1,000 genomes project promise to shed more light on this question.
Conclusion
In summary, we have identified OR gene repertoires in orangutans, marmosets, and mouse lemurs and demonstrated that extant primate species have the similar number of intact OR genes and the fraction of pseudogenes. In spite of the similar size of OR gene repertoires, there were also dramatic variations during the primate evolution. After comparative analyses of the evolution of OR genes, we found that the OR genes in apes had experiences a lower level of selective constraint.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
Key Construction Program of the National “985” Project and “211” project [to S.Z.]; University of Toronto [to Z.Z.].
Supplementary Material
Acknowledgments
We thank Dr Huabin Zhao and Yang Liu for helpful discussion and the editor and anonymous referees for helpful comments on previous versions of the manuscript. D.D., G.H., S.Z., and Z.Z. conceived this study. D.D. performed the work and the statistical analyses. D.D. and Z.Z. discussed the results and wrote the manuscript. All authors commented on it and approved the final version of the manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baron G, Frahm HD, Bhatnagar KP, Stephan H. Comparison of brain structure volumes in Insectivora and Primates. III. Main olfactory bulb (MOB) J Hirnforsch. 1983;24:551–568. [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. High genomic deleterious mutation rates in hominids. Nature. 1999;397:344–347. doi: 10.1038/16915. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Paabo S. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 2003;73:489–501. doi: 10.1086/378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Man O, Paabo S, Lancet D. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci USA. 2003;100:3324–3327. doi: 10.1073/pnas.0535697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant AA, Skaletsky H, Chess A. Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc Natl Acad Sci USA. 2004;101:9019–9022. doi: 10.1073/pnas.0401566101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y, Niimura Y. Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol Biol Evol. 2008;25:1897–1907. doi: 10.1093/molbev/msn135. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Ishii T, et al. Monoallelic expression of the odourant receptor gene and axonal projection of olfactory sensory neurones. Genes Cells. 2001;6:71–78. doi: 10.1046/j.1365-2443.2001.00398.x. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JE, Fobes JL. Evolutionary changes in primate sensory capacities. J Hum Evol. 1974;3:435–443. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laska M, Seibt A, Weber A. ‘Microsmatic’ primates revisited: olfactory sensitivity in the squirrel monkey. Chem Senses. 2000;25:47–53. doi: 10.1093/chemse/25.1.47. [DOI] [PubMed] [Google Scholar]
- Li WH, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004a;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004b;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005a;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA. 2005b;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Nei M. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci USA. 2007;104:7122–7127. doi: 10.1073/pnas.0702133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlager HH, Kemp B. Ontogenesis of the sperm whale brain. J Comp Neurol. 1998;399:210–228. doi: 10.1002/(sici)1096-9861(19980921)399:2<210::aid-cne5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Quignon P, et al. Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol. 2003;4:R80. doi: 10.1186/gb-2003-4-12-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier S, Blancher A, Giorgi D. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci USA. 2000;97:2870–2874. doi: 10.1073/pnas.040580197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier S, et al. A gene recently inactivated in human defines a new olfactory receptor family in mammals. Hum Mol Genet. 1998;7:1337–1345. doi: 10.1093/hmg/7.9.1337. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. The human sense of smell: are we better than we think? PLoS Biol. 2004;2:E146. doi: 10.1371/journal.pbio.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiper ME, Young NM. Primate molecular divergence dates. Mol Phylogenet Evol. 2006;41:384–394. doi: 10.1016/j.ympev.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Stephan H, Baron G, Frahm HD. Comparative size of brains and brain structures. Comp Primate Biol Neurosci. 1988;4:1–38. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Young JM, et al. Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet. 2008;83:228–242. doi: 10.1016/j.ajhg.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.