Abstract
We determined the complete mitochondrial genome sequences of two species of gall midges (Diptera: Cecidomyiidae), as well as partial sequence from a third cecidomyiid and a species from a related family, the Sciaridae. The sciarid sequence has a number of rearrangements of tRNA genes, relative to other dipterans, but is otherwise unremarkable. In contrast, the cecidomyiid genomes possess a number of very unusual features. First, the two complete sequences are very small compared with other dipteran mitochondrial genomes. The genome of Mayetiola destructor is only 14,759 bp while that of Rhopalomyia pomum is only 14,503 bp, comparable with genome sizes observed in some arachnids. Second, all three cecidomyiid species have very high A + T content—more than 83% for the coding region. Third, all three cecidomyiid species possess a number of rearrangements of tRNA genes, including variations within the family. Fourth, the most extraordinary feature of cecidomyiids examined in this study is an extreme truncation of all tRNA genes, including the loss of TΨC arms and apparent absence of the 3′ part of the aminoacyl stems.
The truncated tRNA genes of cecidomyiids are very similar to those previously reported for spiders and appear to represent a second, independent origin of these structural features. It is likely that they are made functional through RNA editing, perhaps using the 5′ end of the aminoacyl stem as a template for the construction of the required 3′ end.
Keywords: Mayetiola destructor, truncated tRNA genes, RNA editing
Introduction
Animal mitochondrial genomes encode about 37 essential genes, including 12–13 genes that code for components of the electron transport system, and a minimal translation system, which includes 22 transfer RNA (tRNA) and two ribosomal RNA (rRNA) genes (Wolstenholme 1992; Boore 1999). The genomes are very compact, usually without introns, and with few noncoding residues outside of a single control region. Despite the apparent simplicity of animal mitochondrial genomes, their structure and function often exhibit a number of unusual molecular features, including alternative genetic codes, RNA editing, and a diverse array of tRNA structures (Burger et al. 2003).
Typically, tRNAs fold into a cloverleaf secondary structure, with an aminoacyl (or acceptor) stem, a DHU stem and loop, an anticodon stem and loop, a TΨC stem and loop, and a smaller variable loop separating the anticodon stem from the TΨC stem. Although the majority of mitochondrial tRNAs have this standard structure, variations on the basic structure have been found throughout the animal kingdom. Examples have been encountered missing the DHU stem, or the TΨC stem, or both (Masta and Boore 2008). Nematodes, such as Caenorhabditis elegans, have what appears to be the minimal functional tRNA structure, lacking both the DHU and TΨC stems (Wolstenholme 1992). But the most extraordinary modifications of tRNA genes have been found in spiders (Masta 2000; Masta and Boore 2004, 2008; Qiu et al. 2005). Not only are the TΨC stem and loop missing from all tRNA genes but also the 3′ portion of the aminoacyl stem evidently is not coded in the DNA. Because tRNAs cannot function without a paired aminoacyl stem, it is evident that some form of RNA editing must exist to construct the 3′ end de novo. Lavrov et al. (2000) showed that mismatches in the aminoacyl stem of a centipede are corrected by RNA editing using the 5′ end as a template. Masta and Boore (2004) noted that this mechanism could be used to reconstruct the entire 3′ end of the aminoacyl stem of spider tRNAs, using the 5′ end as template. They provided a number of predictions concerning the evolution of this editing capability. These predictions include the possible relaxation of constraints on the sequence of the aminoacyl stems and a general evolutionary trend toward the loss of the 3′ end from all tRNA genes in the genome.
During preliminary studies of the mitochondrial genome of a gall midge (the Hessian fly, Mayetiola destructor), we encountered tRNA-like structures similar to those of spiders. We therefore undertook the sequencing of the complete genome of this species, along with a more extensive examination of flies from this interesting family.
Cecidomyiid flies (Arthropoda: Diptera: Nematocera: Bibionomorpha: Sciaridae: Cecidomyiidae) are an ancient lineage of flies known to exist for more than 150 My (Yukawa and Rohfritsch 2005). Cecidomyiidae underwent explosive diversification in the Cretaceous period coincident with the appearance of angiosperm plants and are now a hyperdiverse family encompassing more than 5,700 described species. The predominant life history mode, as the name gall midge implies, involves the induction of gall structures on various plant tissues followed by larval feeding and development within the galls. They are known to diversify both through host plant shifts (Price 2005) and through ecological partitioning of a single plant (Joy and Crespi 2007; Stireman et al. 2008).
Cecidomyiid flies are known to have some unusual genetic features. Quantification of the size of the nuclear genome of M. destructor showed it to be the smallest known nuclear genome of any insect at 0.09 pg (Gregory et al. 2007). The most unusual features of cecidomyiids involve the behavior of chromosomes during meiosis and early development (White 1973). Although the details vary widely across the family, features include the absence of homologous pairing of chromosomes and the formation of a highly asymmetrical spindle during spermatogenesis, as well as the elimination of chromosomes from somatic tissues (but not the germ line) during early development. In M. destructor, for example, the germ line carries about 40 chromosomes, but as a result of chromosome elimination during cleavage, the somatic tissues of both sexes have only about eight chromosomes. Sex determination occurs by the differential elimination of X chromosomes from somatic tissues (White 1949). Some of these features, including unipolar spindle formation during spermatogenesis, the elimination of chromosomes during early cleavage, and sex determination by elimination of X chromosomes, are shared by the Dipteran family Sciaridae. These genetic features, together with morphological synapomorphies, support a sister relationship between the two families (White 1949; Wood and Borkent 1989; Oosterbroek and Courtney 1995).
In this study we examine the evolutionary extent of truncated tRNA genes within the family Cecidomyiidae by identifying tRNA genes in three species in different genera. We also compare the number and type of gene rearrangements among these cecidomyiid mitochondrial genomes. To place the evolution of the truncation in tRNA genes into a broader context within the Cecidomyiidae, we infer phylogenetic relationships of the taxa under study and among the family more generally using mitochondrial cox1 sequences. All family- or subfamily-level phylogenies for Cecidomyiidae to date have been based exclusively on morphological characters (Gagné 1989; Roskam 2005). No single previous phylogeny of any sort has encompassed all the genera under study here. We also include partial sequencing of the mitochondrial genome of Bradysia amoena, of the related family Sciaridae.
Materials and Methods
Source and Collection of Specimens
Adults of the Hessian fly M. destructor (Cecidomyiidae) were obtained from a laboratory culture of R. Shukle, Purdue University. Adults of B. amoena (Sciaridae) were obtained from a laboratory culture maintained by S. Gerbi, Brown University. Both species were preserved in 95% EtOH and provided through the Dipteran Tree of Life Project. We collected specimens of the stem galling Asphondylia rosetta (Cecidomyiidae) from Larrea tridentata (creosote bush) near Tucson, AZ, and specimens of the leaf galling Rhopalomyia pomum (Cecidomyiidae) were collected from Artemesia tridentata (sagebrush) near Kamloops, BC, Canada. We obtained the sequences of the tRNALeu(UUR) gene from GenBank for Asteromyia carbonifera and A. euthamiae (accession numbers EU439835 and EU439782) for comparison with the homologous sequences obtained in this study. No other identified tRNA sequences were available for representatives of this family.
Genome Sequencing
Individual specimens were ground in the presence of protease K, and total genomic DNA was extracted using a standard phenol–chloroform extraction protocol. After ethanol precipitation, extracts were dried and dissolved in 100–200 μl of distilled water. The general strategy for amplification and sequencing was to amplify fragments of 500–1,500 bp using standard primers (Simon et al. 2006). Details of the amplification conditions and purification of templates are given in Beckenbach and Stewart (2009). Initial attempts with standard primer pairs yielded only a few fragments, scattered about the genomes. In particular, primers based in tRNA genes invariably failed to amplify. Additional sequence was obtained by primer walking using taxon-specific primers based on preliminary sequence, paired with standard primers, or with other taxon-specific primers. Two regions proved most challenging: the region between the nad3 and nad5 genes and the control region, between the small ribosomal subunit and the nad2 gene. We were successful in amplifying across these regions in Mayetiola and Rhopalomyia and completed these sequences using primer walks. Primer sequences and locations are available from the authors. Repeated attempts to amplify across these regions in Asphondylia and Bradysia were unsuccessful.
Annotation of the Sequences
Sequences were assembled manually, based on regions of overlap and on the locations of amplification and sequencing primers. Protein-coding genes were identified as open reading frames, and by alignment with homologous sequences of other Diptera. The rRNA genes were identified by alignment with sequences of other arthropods. Identification of tRNA genes posed the greatest challenges. Gene junctions having unassigned sequence were scanned online using tRNAscan-SE (Lowe and Eddy 1997), with a cove score cutoff of 1. This process found tRNA genes for Bradysia but generally failed for cecidomyiid sequences. Where putative tRNA genes were located in the cecidomyiids, the sequences overlapped downstream genes and showed mismatches in the aminoacyl stem. Examination of these regions suggested that cecidomyiids possessed truncated tRNA sequences, similar to those observed in some arachnids (Masta 2000; Masta and Boore 2008). We used an approach described in Masta and Boore (2004) for locating the genes. We used the following criteria: 1) a well-formed and well-paired anticodon stem and loop, with an appropriate anticodon; 2) a well-formed and well-paired DHU stem and loop, with 3–4 bp in the stem; and 3) at least nine residues upstream of the DHU stem that cannot be assigned to an upstream gene coded on the same strand. This last criterion assures that the 5′ end of the aminoacyl stem is present after processing of the primary transcripts. The most crucial criterion, however, was the conservation of the putative DHU stems and of the entire anticodon stem–loop sequence across the three cecidomyiid species examined here.
Phylogeny Reconstruction and Character Evolution
To characterize the evolution of tRNA truncation within Cecidomyiidae, we inferred a phylogenetic tree and delineated instances of tRNA truncation at the tips. The tree was inferred using combined mitochondrial sequence data from our sequenced genomes, and a 444-bp fragment of the cox1 gene obtained from GenBank for species in all available genera; adequate coverage was not available for any other genes. Sequences were aligned using Clustal (Thompson et al. 1994) and adjusted by eye using Se-Al (Rambaut 1996). The best fitting model of sequence evolution was determined using Modeltest (Posada and Crandall 1998). We also employed MrModeltest 2.2 (Nylander 2004) to determine the best model for use in Bayesian phylogeny estimation. We reconstructed phylogenetic relationships among cecidomyiid species under maximum likelihood (ML) and maximum parsimony (MP) using PAUP* 4.0b10 (Swofford 2002). We also reconstructed phylogenetic relationships using Bayesian methods as implemented in MrBayes 3.12 (Ronquist and Huelsenbeck 2003). Two parallel runs utilizing default priors, four heated chains, while sampling trees from one cold chain every 1,000 generation were run for 10 million generations.
To test the hypothesis that all species displaying the tRNA truncation were monophyletic, we employed the Shimodaira–Hasegawa (SH) test and the Templeton test as implemented in PAUP* (Swofford 2002) to compare the best tree with a constraint tree which forces monophyly of cecidomyiid species known to have truncated tRNA genes.
Data Deposition
Sequences have been deposited in GenBank under the following accession numbers: Mayetiola destructor, GQ387648; Rhopalomyia pomum, GQ387649; Asphondylia rosetta, GQ387650; and Bradysia amoena, GQ387651.
Results
We determined complete mitochondrial genome sequences for two species of gall midges, M. destructor and R. pomum, as well as partial sequences for a third cecidomyiid, A. rosetta, and a sciarid, B. amoena. In all four species, the protein-coding and rRNA genes are in the typical arthropod positions and orientation, but rearrangements involving tRNA genes are evident in all four genomes. The cecidomyiid sequences exhibit a number of unusual features. The most interesting is a severe truncation of all tRNA gene sequences, which is described in detail below.
tRNA Gene Rearrangements
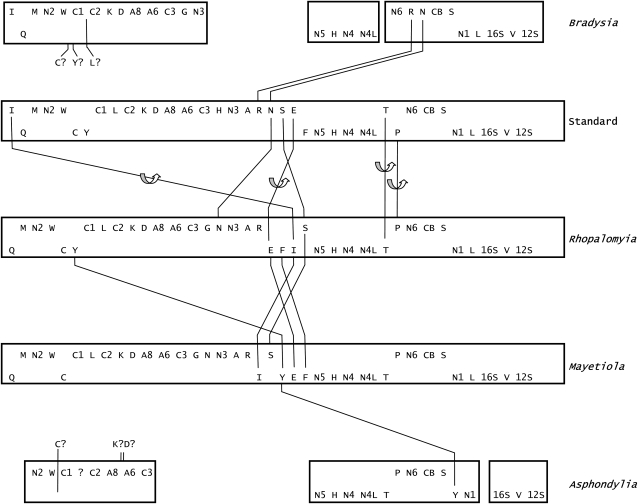
The two complete cecidomyiid sequences show a number of rearrangements involving tRNA genes (fig. 1). The tRNAIle gene has been inverted and transposed in both Mayetiola and Rhopalomyia from the typical arthropod position between the control region and the nad2 gene to the block of tRNA genes between nad3 and nad5. The tRNAAsn gene has been moved from the nad3–nad5 block to a position between the tRNAGly gene and nad3 in both species. The tRNAGlu gene, located within the nad3–nad5 region, has been inverted in both species, relative to the typical arthropod gene arrangement. As these rearrangements involve regions of the genome not determined in Asphondylia, we cannot say whether they are shared by this species.
FIG. 1.—
Organization of the mitochondrial genomes of representatives of the families Cecidomyiidae and Sciaridae. The genome structures are linearized to place the control region at the end. The “typical” gene order is that found in Drosophila and is widespread in insects. The upper line of gene names are coded on the majority (J) strand; the lower line gives those coded on the minority (N) strand. Protein-coding genes: A6, A8 are ATPase subunits 6 and 8; C1–C3 are cytochrome oxidase subunits; CB is cytochrome B; and N1–N6 and N4L are NADH dehydrogenase subunits. Ribosomal genes: 16s and 12s are the large and small subunits. The tRNA genes are indicated by their single-letter amino acid designations. Lines indicate transpositions; curved arrows indicate that an inversion is involved. Question marks indicate tRNA genes not present in their typical positions and assumed to have transposed into regions not sequenced.
Both the tRNAThr and tRNAPro genes, located between the nad4l and nad6 genes, are inverted in all three cecidomyiid species. This observation is particularly interesting, as it requires a minimum of two separate steps because they remain in the same position relative to the typical arthropod genome arrangement. Either they both underwent inversions independently or there was a single inversion involving both genes, followed by a transposition. The two genes are transcribed from different strands, both in the typical gene arrangement and in the inverted arrangement found in cecidomyiids. In Rhopalomyia, the genes are separated by a noncoding block consisting of a tandem repeat of 11 copies of an 18-bp sequence. The genes overlap by 16 bp in Asphondylia and 26 bp in Mayetiola.
The tRNATyr gene appears in three different places in the three cecidomyiid species examined here. In Rhopalomyia it is retained in the typical arthropod position, between the tRNACys gene and cox1. In Mayetiola it has been transposed to the nad3–nad5 tRNA block, whereas in Asphondylia it has been transposed to a position between tRNASer(UCN) and nad1. It is coded on the minority strand in all three genomes. This gene is also moved from the typical arthropod position in the sciarid Bradysia, although it is evidently not located within the regions sequenced in this study.
A number of other rearrangements are evident in the partial Asphondylia sequence. Genes not present in their typical positions include tRNACys, tRNALeu(UUR), tRNALys, and tRNAAsp. Rearrangements evident in the partial Bradysia sequence include tRNACys, tRNATyr, and tRNALeu(UUR), which are not present in their typical positions. We assume they have been moved to positions within the regions not sequenced in this study. In addition, tRNAArg and tRNAAsn, usually located within the nad3–nad5 sequence block, are identifiable between the nad6 and cytb genes in Bradysia. In the typical arthropod mitochondrial gene arrangement the nad6 and cytb genes abut.
Features of Cecidomyiid Mitochondrial Genomes
The two complete cecidomyiid sequences show a number of unusual features. Both genomes are smaller than most insect mitochondrial genomes and comparable to those observed in some spiders (Masta and Boore 2004, 2008). The genome of Mayetiola is 14,759 bp, including a control region of about 600 bp, whereas the genome of Rhopalomyia is only 14,503 bp, with a control region of about 360 bp. Notably, they represent the smallest known dipteran mitochondrial genomes and are comparable to those found among many arachnids (Masta and Boore 2008).
Part of the reduction in overall genome size can be attributed to a reduction in length of most of the protein-coding genes (Table 1). Nearly all the protein-coding genes are shorter than those of other dipterans, especially the NADH dehydrogenase complex genes. This reduction cannot be ascribed to differing views of the annotation of these genes, as all protein-coding genes in Mayetiola and all but two in Rhopalomyia have DNA-encoded terminators, and in most cases the start codons are unambiguous.
Table 1.
Characteristics of protein-coding genes of cecidomyiids and other Diptera
| atp6 | atp8 | cox1 | cox2 | cox3 | cytb | nad1 | nad2 | nad3 | nad4 | nad4l | nad5 | nad6 | |
| Mayetiola | |||||||||||||
| Length | 223 | 51 | 511 | 225 | 260 | 375 | 301 | 324 | 116 | 436 | 90 | 558 | 160 |
| Initiator | ATG | ATT | ATT | ATA | ATA | ATT | ATG | ATA | ATT | ATT | TTT | ATT | ATG |
| Terminator | TAA | (TAA) | TAA | TAA | TAA | TAA | TAA | TAA | TAA | TAG | TAA | TA(A) | TAA |
| A + T (%) | 82.5 | 94.1 | 74.3 | 80.2 | 79.2 | 79.3 | 82.0 | 89.8 | 86.0 | 82.3 | 89.4 | 83.2 | 91.1 |
| Rhopalomyia | |||||||||||||
| Length | 223 | 51 | 512 | 224 | 260 | 372 | 301 | 325 | 116 | 437 | 90 | 554 | 155 |
| Initiator | ATG | ATT | TTT | ATA | TTA | ATA | ATA | ATA | ATA | ATA | ATA | ATT | ATA |
| Terminator | TAA | (TAA) | TA(A) | TAA | TA(A) | T | TAA | TAA | TAA | T | TAA | TAA | TAA |
| A + T (%) | 82.8 | 92.8 | 76.6 | 81.5 | 80.3 | 80.3 | 83.3 | 91.3 | 85.8 | 83.1 | 89.0 | 85.2 | 92.1 |
| Asphondylia | |||||||||||||
| Length | 223 | 52 | 511 | 219 | n/a | 378 | n/a | n/a | n/a | 437 | 90 | n/a | 154 |
| Initiator | ATG | ATT | ATT | ATT | ATT | ATA | ATA | n/a | n/a | ATG | TTT | ATA | ATT |
| Terminator | TAA | (TAA) | TAA | TAA | n/a | TAA | TAG | TAA | n/a | TAA | TAA | n/a | TAA |
| A + T (%) | 83.0 | 91.7 | 74.8 | 81.1 | 80.8 | 81.1 | n/a | n/a | n/a | 85.2 | 88.6 | 84.8 | 92.9 |
| Bradysia | |||||||||||||
| Length | 224 | 55 | 512 | 227 | 262 | 378 | 313 | 344 | n/a | 446 | n/a | 574 | n/a |
| Initiator | ATG | ATC | ATG | ATT | ATG | ATA | ATA | ATA | ATT | ATG | n/a | ATA | n/a |
| Terminator | TAA | (TAA) | TAA | TAA | TAA | TAA | TAA | TAG | TAA | TAA | (TAA) | n/a | TAA |
| A + T (%) | 75.0 | 88.5 | 69.0 | 74.3 | 71.4 | 73.0 | 76.7 | 81.9 | 74.7 | 78.4 | n/a | 78.7 | n/a |
| Anopheles | |||||||||||||
| Length | 226 | 53 | 512 | 228 | 262 | 378 | 314 | 341 | 117 | 448 | 99 | 574 | 173 |
| Initiator | ATG | ATC | TCG | ATG | ATG | ATG | ATA | ATC | ATA | ATG | ATG | TAT | ATT |
| Terminator | TA(A) | (TAA) | T | T | TA | TAA | TAA | T(AA) | TAA | T | TAA | TAA | TAA |
| A + T (%) | 74.3 | 81.8 | 68.6 | 73.1 | 70.4 | 72.4 | 76.6 | 83.0 | 79.4 | 77.7 | 82.7 | 78.2 | 84.9 |
| Drosophila | |||||||||||||
| Length | 224 | 53 | 511 | 228 | 262 | 378 | 315 | 341 | 117 | 447 | 96 | 573 | 174 |
| Initiator | ATG | ATT | TCG | ATG | ATG | ATG | ATA | ATT | ATT | ATG | ATG | ATT | ATT |
| Terminator | TA(A) | (TAA) | TAA | T | TAA | TAA | T | T(AA) | TAA | T | TAA | T | TAA |
| A + T (%) | 75.8 | 82.4 | 69.6 | 73.8 | 71.1 | 73.8 | 78.6 | 81.4 | 79.4 | 79.6 | 83.8 | 77.7 | 84.8 |
Note.—The length of each gene is given in codons. Initiators and terminators are shown as coded in the DNA sequences. Overlaps of one or more residues in the terminator with the downstream gene are indicated with parentheses. n/a, not available.
The three cecidomyiid genomes examined in this study have extremely high A + T contents, ranging from just over 74% in cox1 to over 90% in the atp8 and nad6 genes (Table 1). These values are considerably higher than those that have been observed in other dipterans and are comparable to those of the honeybee, Apis mellifera (Crozier RH and Crozier YC 1993). Overall A + T content of Mayetiola is 84.1%, including 83.6% for the coding region and 90.5% for the control region. The values for Rhopalomyia are 85.2% overall, 84.6% coding and 94.2% for the control region. Regions sequenced from Asphondylia show comparable A + T content, whereas those in the sciarid Bradysia have A + T content more typical of other dipterans (Table 1).
Structural Characteristics of Cecidomyiid tRNAs
The tRNAs coded in all three species evidently lack TΨC stem–loop structures, as well as the 3′ end the aminoacyl (acceptor) stem. Structures similar to these have been previously observed in spiders (Masta and Boore 2004, 2008; Qiu et al. 2005). Evidence that some arthropods have evolved a mechanism to reconstruct the 3′ end of the aminoacyl stem through RNA editing, presumably using the 5′ end as a template, was provided by Lavrov et al. (2000).
The sequences of the tRNAs identified in this study are given in fig. 2 and supplementary fig. S1 (Supplementary Material online); examples of our interpretation of the folding structures are given in fig. 3. Folded structures of all tRNAs are given in supplementary figs. S2–S5 (Supplementary Material online). The regions including the entire anticodon stem and loops are very well conserved across the cecidomyiid species in all 22 tRNAs. In nearly all instances where a nucleotide substitution is observed in the aminoacyl stem, a compensatory substitution retains pairing capability. The DHU stems are also well conserved across each species, again with compensatory substitutions observed in some cases. Most of the variations observable in the DHU stem–loop regions are within the loops, including both nucleotide substitutions and indels (fig. 2 and supplementary fig. S1).
FIG. 2.—
Comparison of tRNA gene sequences in cecidomyiids. The Mayetiola sequence is given for each group, while sequences of Rhopalomyia and Asphondylia are compared below, with dots indicating identical residues and dashes indicating indels. In the top line of each grouping, dots indicate pairing regions; the first two groups of dots are the DHU stem, whereas the third and fourth are the anticodon stem. The anticodon is given, along with the start or end of an adjacent downstream gene where they fall within the region shown. The arrow associated with the downstream gene shows the coding direction. Where the arrow is to the left, it indicates the end of that gene.
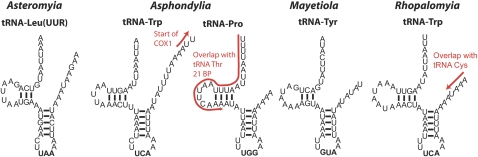
FIG. 3.—
Examples of inferred tRNA secondary structure from each cecidomyiid genome. Note consistent lack of a TΨC arm across genera; the overlap of the tRNAPro gene with tRNAThr on opposing strands in Asphondylia; the overlap of tRNATrp with the cox1 gene in Asphondylia; and tRNATyr shown here in Mayetiola is found in a different genomic positions in each genus, between nad2 and cox1 in Rhopalomyia, between nad3 and nad5 in Mayetiola, and between cytb and nad1 in Asphondylia.
In contrast to the conservation of the anticodon stem–loops and DHU stems, there is little conservation either upstream or downstream from these structures. We assume that nine residues are required upstream of the DHU stem, including two unpaired residues and seven residues needed to form the 5′ end of the aminoacyl stem. In no case do these nine residues overlap an upstream gene coded on the same strand, and in many instances this putative 5′ stem region follows immediately after the terminator codon of an upstream protein-coding gene. We take these observations as evidence of a functional role for these nine residues.
Phylogenetic Results
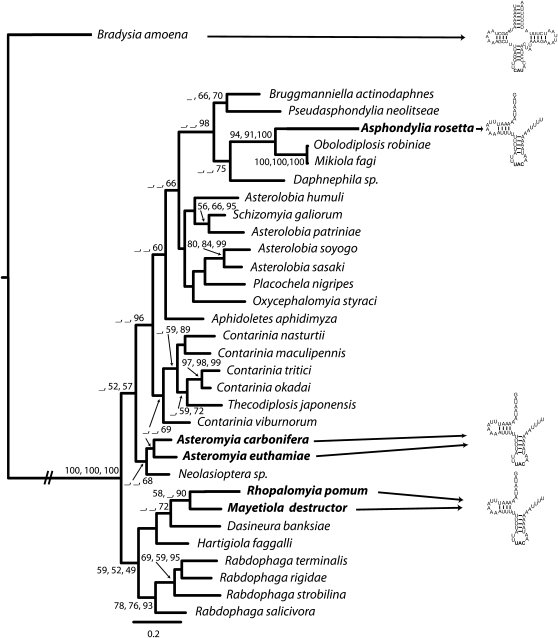
Figure 4 illustrates the reconstructed phylogenetic relationships among genera in the subfamily Cecidomyiinae (family Cecidomyiidae). All available data for Cecidomyiidae are from this subfamily. The MP, ML, and Bayesian analyses yielded trees of very similar topology. The support for the recovered nodes was generally robust toward the tips and declined with depth in the tree, apparently due to mutational saturation of the cox1 locus at this level of divergence.
FIG. 4.—
Phylogeny of cecidomyiid genera and outgroup based on sequence data from cytochrome oxidase subunit I gene. Numbers above branches are MP bootstrap, ML bootstrap, and Bayesian posterior probabilities. Taxa displaying incomplete inferred secondary tRNA structure are denoted at the tips.
SH test (difference in −ln L = 79.95058, P < 0.001) and Templeton test (difference in length = 47, P < 0.001) strongly reject the hypothesis that taxa sharing the character state of truncated tRNA genes form a monophyletic group relative to other genera.
Discussion
Complete sequences of the mitochondrial genomes from two representatives of gall midges (family Cecidomyiidae), as well as partial sequence from a third, show that the genomes are highly modified in several ways. They have a very high A + T content, a general reduction of overall length compared with most other animal mitochondrial genomes, rearrangement of some of the tRNA genes, and, most notably, truncation of all tRNA genes. The overall length reduction is partly due to a shortening of most of the major genes and to a severe truncation of the tRNA genes. Phylogenetic tests support the placement of four genera displaying truncated tRNA genes in disparate parts of the cecidomyiid phylogeny, indicating either repeated evolution within the family or, more likely, origin of the truncation mechanism in a common ancestor shared by all cecidomyiine species. The presence of fully coded tRNA genes in Bradysia (family Sciaridae) implies that truncated tRNA structures arose after the separation of these two families. Further investigation of the structure of tRNA genes in the Cecidomyiidae will provide insights into the evolutionary origins of truncated tRNA genes.
Rearrangement of tRNA Genes
All the taxa examined in this study have tRNA rearrangements relative to the typical arthropod mitochondrial genome organization. These rearrangements include inversions and transpositions. All three cecidomyiid genomes share inversions of both the tRNAThr and tRNAPro genes. These changes appear to simplify transcription and processing of genes in this region, by bringing together the N-strand–encoded genes from tRNAThr to nad5 and the J-strand genes from tRNAPro to tRNASer(UCN) into continuous, uninterrupted blocks. Mapping these changes onto the phylogeny of the subfamily Cecidomyiinae (fig. 4) shows that these changes occurred early in the diversification of this subfamily. The inversion and transposition of tRNAIle gene, the inversion of the tRNAGlu gene, and the transposition of the tRNAAsn gene in both Mayetiola and Rhopalomyia indicate that these rearrangements occurred prior to the separation of these two genera. These genes were not located within the regions sequenced in Asphondylia, so we cannot pinpoint the origin of these changes on this phylogeny.
Several tRNA gene rearrangements have occurred since the separation of Mayetiola and Rhoplalomyia. These changes include transposition of tRNATyr in Mayetiola from its typical position, which is retained in Rhopalomyia, and transposition of a small block, which includes the tRNAGlu and tRNAPhe genes. These two genera are placed in the same Tribe by morphology and appear as sisters in our cox1 molecular phylogeny. The number of changes that we observe in tRNA gene organization, however, suggests that they are not close on an absolute timescale.
The observation of multiple shared, as well as taxon-specific, tRNA gene rearrangements indicates that tRNA gene arrangements may provide useful markers for more detailed phylogenetic reconstruction in the Cecidomyiidae. The sciarid examined in this study, Bradysia, also possesses tRNA gene rearrangements, suggesting that gene arrangements may be generally useful for phylogenetic reconstruction in this section of the superfamily Sciaridae.
Truncated tRNA Genes
Sequences interpreted here as tRNA genes are well conserved in the anticodon stems and loops, and in the DHU stems. This conservation is evident even where the tRNA genes appear in different places in the genome, as we observe for the tRNATyr gene. Despite strong conservation of two of the arms of the standard cloverleaf structure, there is little or no evidence of TΨC stems or loops, or sequence corresponding to the 3′ end of the aminoacyl stem. Nonetheless, the presence of well-formed anticodon stems and loops corresponding to all 22 expected tRNA genes in both Mayetiola and Rhopalomyia strongly suggests a coding role for these sequences. As they occupy regions between the protein-coding and rRNA genes and no other coding role is evident for these sequences, we conclude they are the functional tRNA genes. Further, the sequence conservation evident in the anticodon loops and DHU stems would degrade rapidly in the mutation-prone (Lynch et al. 2006) mitochondrial genome. Intergenic residues where no coding role is evident are not conserved among these species. Thus, selection likely maintains the conserved tRNA sequences in cecidomyiid mitochondrial genomes, further supporting a coding role for them. Certainly, the conservation would not persist over the millions of years of evolution, which separates the cecidomyiid genera displaying truncated tRNA genes if they were not functional.
The truncation of these genes poses problems in locating and annotating the tRNA genes (Masta and Boore 2004). In the absence of a well-paired acceptor stem, the 3′ end is not clearly defined. The region downstream from the anticodon stem is extremely variable in sequence and length. In some cases possible stem structures can be found downstream, but such structures are not consistent and often overlap downstream genes. For example, the region downstream of the anticodon stem in tRNAAsp in Rhopalomyia could be folded in several ways, but the region is absent in Mayetiola (supplementary fig. S1, Supplementary Material online). Similarly, a region that could be folded in tRNAMet from Mayetiola is missing from Rhopalomyia. Rather than hypothesize a variety of structural differences among the tRNA genes and between the same tRNA gene in different cecidomyiid species, it is more reasonable to assume that all the tRNA genes function in a similar manner.
The absence of well-paired aminoacyl stems poses interesting questions regarding the processing of primary transcripts in cecidomyiids. In all Metazoa where transcription has been documented, the primary transcripts are polycistronic and must be processed to yield the necessary messenger (mRNA), tRNA, and rRNA transcripts. The tRNA punctuation model was proposed to account for the processing in the human mitochondrial genome (Ojala et al. 1981). In this model, the tRNA sequences are removed from the primary transcripts, producing the mature mRNA and rRNA transcripts in the process. There is evidence supporting this model in Drosophila melanogaster (Stewart and Beckenbach 2009). It is generally assumed that the secondary structure of the tRNA sequences provides the required signals for processing of the primary transcripts (Ojala et al. 1981; Clary and Wolstenholme 1985). In the absence of standard cloverleaf tRNA structures, some modification of this model appears necessary. It is possible, of course, that the anticodon stem alone provides the appropriate signals.
A second implication of the tRNA punctuation model is that genes coded on the same strand cannot overlap because transcripts for adjacent genes are derived from the same processing events. The only exceptions are the atp8/atp6 and nad4l/nad4 genes, which are translated from bicistronic transcripts (Berthier et al. 1986). In the cecidomyiid sequences examined here, 11 of the 22 tRNA genes have an adjacent downstream gene coded on the same strand (figs. 1 and 2 and supplementary fig. S1, Supplementary Material online). If the tRNA punctuation model holds for cecidomyiids, the processed transcripts for these tRNAs must be severely truncated at the 3′ end prior to any editing steps.
The 5′ ends of the aminoacyl stems are poorly conserved (fig. 2). Most are extremely A + T rich, so it is often possible to find an A + T–rich sequence downstream that will pair as many as five of the seven residues we have assigned to the 5′ end of the aminoacyl stem. In some cases potential matching regions are within downstream genes. Rather than postulate different structures for some of the tRNA genes and for the same gene in different species, it seems more reasonable to hypothesize that all genes function in a common manner.
The most likely mechanism for proper functioning of truncated tRNA genes is RNA editing, using the 5′ end of the aminoacyl stem as a template for the construction of a well-paired stem (Lavrov et al. 2000; Masta and Boore 2004). Editing of the 3′ end of the aminoacyl stem has been demonstrated in snail mitochondrial tRNA sequences, as well (Yokobori and Pääbo 1995). The tRNA genes in snails, where the most extensive editing is required, are those that overlap with a downstream gene. It is likely that they are actually truncated at the start of the downstream gene and reconstructed using the 5′ end as a template. If this mechanism is used, it assures a fully matched stem regardless of the 5′ end sequence and would appear to relax the constraints on the actual sequence of those stems (Masta and Boore 2004). Our observations support this prediction (fig. 2).
There is, however, another potential source of constraint on the aminoacyl stem sequence: the requirement for proper recognition of the tRNA by amino acid–charging enzymes. In both prokaryotes and nuclear-encoded eukaryote systems, tRNA-synthetase recognition is based on determinants (specific residues) in both the aminoacyl stem and anticodon loop (Giege et al. 1998; Beuning and Musier-Forsyth 1999). The lack of conservation of the 5′ end of the tRNAs in cecidomyiids would seem to require either that proper recognition of the tRNA during the charging process relies solely on determinants in the anticodon stem and loop or a rapid co-evolution of aminoacyl determinants with the aminoacyl synthase genes.
Parallel Evolution of Truncated tRNA Genes
The finding of truncated tRNA genes in cecidomyiids implies that this feature has arisen at least twice within the Arthropoda. Masta and Boore (2008) provided evidence of multiple losses of TΨC arms within arachnids, but loss of both the TΨC arm and 3′ portion of the aminoacyl stem from all tRNA genes may have a single origin in spiders. A second independent origin within the cecidomyiids supports the prediction that development of mechanisms to edit the 3′ end of tRNAs and the ability of the ribosome to accommodate tRNAs lacking the TΨC arms can lead to the pattern of truncation of tRNA genes observed in spiders and cecidomyiids (Masta and Boore 2004).
The observation that all, not just a subset, the tRNA genes in both spiders and gall midges have become truncated suggests that natural selection may favor truncation, once a mechanism evolves which allows the proper functioning of these genes. Several hypotheses might favor the development of such a mechanism. One hypothesis is that the mechanism evolves as a result of selection for a smaller genome size (Dufresne et al. 2005; Giovannoni et al. 2005, 2008). Selection may favor smaller genomes to enhance replication speed in organisms, which must develop rapidly to take advantage of ephemeral conditions. The small size of both the nuclear and mitochondrial genomes of cecidomyiids is consistent with this idea. A second hypothesis is that the RNA-editing mechanism may evolve as a way to ameliorate the mutational consequences of asexuality in asexual organellar genomes (Lynch 1997; Lynch et al. 2006). A third hypothesis is that marked population subdivision can result in extremely small local effective population size, allowing a variety of deleterious mutations to be fixed. A final hypothesis states that modification of the ribosome in a common ancestor may result in the relaxation of rules governing interactions between ribosome and tRNA molecules.
Phylogenetic Origin of Truncated tRNA Genes in Cecidomyiids
Our phylogeny and results of SH and Templeton tests for the cox1 gene support the hypothesis that taxa displaying reduced genome size and truncated tRNA genes do not form a compact monophyletic group relative to other available genera in the family. Monophyly of the taxa under study here would also be unsupported by phylogenies inferred from morphological characters (Roskam 2005). Inferences about cecidomyiid phylogeny based on morphological characters place Asphondylia and Mayetiola species in highly divergent clades (Roskam 2005). Further, Mayetiola and Asphondylia are morphologically very different and display dramatic differences in life history. Thus, multiple lines of evidence (genetic, morphological, and life history) support the premise that cecidomyiid taxa, which have evolved tRNA truncation, are widely dispersed across the subfamily. These results imply either that the mechanism which allows truncation of the tRNA genes evolved once in a common ancestor, or that it has evolved multiple times, convergently, within the family. We favor a single origin in the Cecidomyiidae.
The truncated and rearranged tRNA genes shown in this study illustrate the dynamic nature of cecidomyiid mitochondrial genomes and extend the taxonomic breadth of the observation that in some lineages tRNA genes are severely truncated to Diptera. As more data become available on both the distribution of truncated tRNA genes and the mechanisms that allow them to function, it will become more tractable to test hypotheses about the roles of various evolutionary forces favoring the development of the mechanism.
Supplementary Material
Supplementary figures S1–S5 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada [to A.T.B.] and National Science Foundation (US) grant [EF-0334949, Building the Dipteran Tree of Life, Brian Wiegmann, principal investigator].
Supplementary Material
Acknowledgments
We thank the SFU FAB* laboratory for helpful discussion and comments, and two anonymous reviewers for suggestions which improved the manuscript.
References
- Beckenbach AT, Stewart JB. Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera), a giant lacewing and an owlfly (order Neuroptera) Genome. 2009;52:31–38. doi: 10.1139/G08-098. [DOI] [PubMed] [Google Scholar]
- Berthier F, Renaud M, Alziari S, Durand R. RNA mapping on Drosophila mitochondrial DNA: precursors and template strands. Nucleic Acids Res. 1986;14:4519–4533. doi: 10.1093/nar/14.11.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymer. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné RJ. The plant-feeding gall midges of North America. Ithaca (NY): Cornell University Press; 1989. [Google Scholar]
- Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–1782. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- Gregory TR, et al. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–D338. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy JB, Crespi B. Adaptive radiation of gall-inducing insects within a single host-plant species. Evolution. 2007;61:784–795. doi: 10.1111/j.1558-5646.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TD, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Masta SE. Mitochondrial sequence evolution in spiders: intraspecific variation in tRNAs lacking the TΨC arm. Mol Biol Evol. 2000;17:1091–1100. doi: 10.1093/oxfordjournals.molbev.a026390. [DOI] [PubMed] [Google Scholar]
- Masta SE, Boore JL. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 2004;21:893–902. doi: 10.1093/molbev/msh096. [DOI] [PubMed] [Google Scholar]
- Masta SE, Boore JL. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol Biol Evol. 2008;25:949–959. doi: 10.1093/molbev/msn051. [DOI] [PubMed] [Google Scholar]
- Nylander JA. MrModeltest v2. Program distributed by the author. Uppsala (Sweden): Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Oosterbroek P, Courtney G. Phylogeny of the nematocerous families of Diptera (Insecta) Zool J Linn Soc. 1995;115:267–311. [Google Scholar]
- Posada D, Crandall KA. MODELEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Price PW. Adaptive radiation of gall-inducing insects. Basic Appl Ecol. 2005;6:413–421. [Google Scholar]
- Qiu Y, Song D, Zhou K, Sun H. The mitochondrial sequences of Heptathela hangzhouensis and Ornithoctonus huwena reveal unique gene arrangements and atypical tRNAs. J Mol Evol. 2005;60:57–71. doi: 10.1007/s00239-004-0010-2. [DOI] [PubMed] [Google Scholar]
- Rambaut A. 1996. Se-Al: sequence alignment editor, version 2.0a11 [cited 2009 January] Available from: http://tree.bio.ed.ac.uk/software/seal/ [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roskam H. Phylogeny of gall midges (Cecidomyiidae) In: Raman A, Schaefer CW, Withers TM, editors. Biology, ecology, and evolution of gall-inducing arthropods. Enfield (NH): Science Publishers Inc; 2005. pp. 205–320. [Google Scholar]
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2006;37:545–579. [Google Scholar]
- Stewart JB, Beckenbach AT. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model. Gene. 2009;445:49–57. doi: 10.1016/j.gene.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Stireman JO, Janson EM, Carr TG, Devlin H, Abbot P. Evolutionary radiation of Asteromyia carbonifera (Diptera: Cecidomyiidae) gall morphotypes on the goldenrod Solidago altissima (Asteraceae) Biol J Linn Soc Lond. 2008;95:840–858. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Ver. 4. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. Cytological evidence on the phylogeny and classification of the Diptera. Evolution. 1949;3:252–261. doi: 10.1111/j.1558-5646.1949.tb00025.x. [DOI] [PubMed] [Google Scholar]
- White MJD. Animal cytology and evolution. 3rd ed. New York: Cambridge University Press; 1973. [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. In: Jeon KW, Wolstenholme DR, editors. Mitochondrial genomes. International review of cytology. Vol. 141. Elsevier Press; 1992. pp. 173–216. [DOI] [PubMed] [Google Scholar]
- Wood DM, Borkent A. Phylogeny and classification of the Nematocera. In: McAlpine JF, editor. Manual of nearctic Diptera. Volume 3. Agriculture Canada monograph 32. 1989. Hull Quebec Canada: Agriculture Canada. ISBN: 0-660-12961-2.1333–1370. [Google Scholar]
- Yokobori S-I, Pääbo S. Transfer editing in land snail mitochondria. Proc Natl Acad Sci USA. 1995;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa J, Rohfritsch O. 2005. Biology and ecology of gall inducing Cecidomyiidae (Diptera). Raman A, Schaefer CW, Withers TM Biology, ecology, and evolution of gall-inducing arthropodsScience Publishers Inc., Enfield (NH)273–304. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.