Abstract
Analysis of gene architecture and expression levels of four organisms, Homo sapiens, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana, reveals a surprising, nonmonotonic, universal relationship between expression level and gene compactness. With increasing expression level, the genes tend at first to become longer but, from a certain level of expression, they become more and more compact, resulting in an approximate bell-shaped dependence. There are two leading hypotheses to explain the compactness of highly expressed genes. The selection hypothesis predicts that gene compactness is predominantly driven by the level of expression, whereas the genomic design hypothesis predicts that expression breadth across tissues is the driving force. We observed the connection between gene expression breadth in humans and gene compactness to be significantly weaker than the connection between expression level and compactness, a result that is compatible with the selection hypothesis but not the genome design hypothesis. The initial gene elongation with increasing expression level could be explained, at least in part, by accumulation of regulatory elements enhancing expression, in particular, in introns. This explanation is compatible with the observed positive correlation between intron density and expression level of a gene. Conversely, the trend toward increasing compactness for highly expressed genes could be caused by selection for minimization of energy and time expenditure during transcription and splicing and for increased fidelity of transcription, splicing, and/or translation that is likely to be particularly critical for highly expressed genes. Regardless of the exact nature of the forces that shape the gene architecture, we present evidence that, at least, in animals, coding and noncoding parts of genes show similar architectonic trends.
Keywords: eukaryotic gene structure, eukaryotic gene architecture, selection on gene compactness, genomic design, intron functionality, intron density
Introduction
One of the hallmarks of the architecture of eukaryotic genes is their fragmented structure (genes in pieces), where exons encoding parts of a protein are separated by noncoding introns. The fraction of intron-containing genes widely differs among eukaryotes; genes of many unicellular forms are intron poor but in multicellular eukaryotes (plants and animals), a substantial majority of the genes contain multiple introns (Rodriguez-Trelles, Tarrio, and Ayala 2006; Roy and Gilbert 2006). Generally, introns are considered to be nonfunctional, although there are many anecdotal reports on the involvement of intronic sequences in various cellular functions (Maniatis and Reed 2002; Ast 2004; Hong, Scofield, and Lynch 2006; Ying and Lin 2006; Zhao and Hamilton 2007).
It has been reported that introns in highly expressed genes in human and worm tend to be shorter than those in genes expressed at lower levels (Castillo-Davis et al. 2002). Subsequent research supported and expanded these findings by revealing the positive correlation between gene compactness and expression level in humans (Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Comeron 2004; Vinogradov 2004; Chen et al. 2005; Li, Feng, and Niu 2007), mouse (Li, Feng, and Niu 2007), worm (Vinogradov 2004; Fahey and Higgins 2007), fly (Vinogradov 2004; Fahey and Higgins 2007), the plant Arabidopsis thaliana (Seoighe, Gehring, and Hurst 2005), and the moss Physcomitrella patens (Stenoien 2007). This pattern, hereinafter denoted the C↑E (compactness increasing with expression) trend to indicate the positive association between compactness and expression, was observed when different technologies were used to measure expression levels, namely, expressed sequence tag libraries (Castillo-Davis et al. 2002; Fahey and Higgins 2007; Stenoien 2007), serial analysis of gene expression (Urrutia and Hurst 2003; Chen et al. 2005; Seoighe, Gehring, and Hurst 2005), and microarrays (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Comeron 2004; Vinogradov 2004; Seoighe, Gehring, and Hurst 2005; Li, Feng, and Niu 2007).
Why highly expressed genes are more compact? The cell invests considerable time and energy in transcription: transcription of a single nucleotide requires at least two adenine triphosphate (ATP) molecules (Lehninger, Nelson, and Cox 1982) and about 0.05 s (Ucker and Yamamoto 1984; Izban and Luse 1992). Thus, the compactness of highly expressed genes was attributed to selective forces that act to minimize the expenditure of energy and/or time on transcription (Castillo-Davis et al. 2002). This “selection hypothesis” received wide support from many authors (Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Chen et al. 2005; Seoighe, Gehring, and Hurst 2005; Li, Feng, and Niu 2007).
However, an important observation that is not readily explained by the selection hypothesis is that intergenic regions also tend to get shorter near highly expressed genes (Urrutia and Hurst 2003; Vinogradov 2004). Although Urrutia and Hurst (2003) showed that the C↑E trend remains highly significant even after controlling for this effect, Vinogradov (2004) reached the opposite conclusion. These findings led Vinogradov to propose an alternative concept, the “genomic design” hypothesis, according to which broadly expressed genes need simpler regulation, and therefore fewer regulatory elements, and less space to accommodate them than genes that are expressed only in specific conditions or tissues. The latter, narrowly expressed genes are thought to require more complex regulation and accordingly more space (both within the gene and in the flanking regions) to accommodate regulatory elements. Given that expression breadth and expression level are positively correlated, broadly expressed genes are, on average, also more highly expressed, thus highly expressed genes are more compact.
The presence of short intergenic regions near highly expressed genes correlates with the tendency of highly expressed genes to cluster along the genomic DNA (Caron et al. 2001; Lercher, Urrutia, and Hurst 2002), suggesting that local mutational bias might also contribute to the C↑E trend. This complication notwithstanding, controlling for various local characteristics like GC content, several authors demonstrated that the C↑E trend remains highly significant (Castillo-Davis et al. 2002; Urrutia and Hurst 2003; Comeron 2004; Seoighe, Gehring, and Hurst 2005).
Surprisingly, given the consistent support of the positive correlation between gene expression and compactness in diverse organisms, the opposite trend, hereinafter denoted the C↓E trend, has been reported in flowering plants, the monocot Oryza sativa (rice), and the dicot A. thaliana (Ren et al. 2006).
These opposing observations require explanation. One obvious possibility is that the forces that shape the gene architecture in plants and animals are different (Ren et al. 2006). Else, there could be a substantial methodological difference between the work of Ren et al. (2006) and the studies that report the C↑E trend, especially considering that the study of Ren et al. is unique in using the massively parallel signature sequencing (MPSS) technology (Brenner, Johnson, et al. 2000; Brenner, Williams, et al. 2000) to measure expression levels.
Given the reported opposite trends and the uncertainty with regard to the evolutionary forces that shape the dependence between gene compactness and expression, we sought to analyze this dependence in multicellular eukaryotes within a more comprehensive framework. We examined compactness in terms of multiple-length variables, such as the length of the entire transcript, the length of the protein-coding sequence, the combined lengths of all coding and noncoding exons, and the combined lengths of the introns. In addition, we investigated a distinct characteristic of genes related to compactness, the intron density, in connection with expression. We show that, in both animals and plants, highly expressed genes are more compact than lowly expressed genes and explain how the analysis of Ren et al. (2006) might give rise to an appearance of a negative trend. The relationship between expression level and gene compactness in both plants and animals turned out to be nonmonotonic. We demonstrate, particularly for introns, a tendency to become longer (hence the genes to become less compact) with increasing expression levels, for lower expression levels. This nonmonotonic trend might result from the combined effect of opposing selective forces that make genes more compact for high levels of expression but make them less compact for lower levels of expression owing, at least, in part, to accumulation of regulatory elements in introns.
Methods
Genome Annotation
The following genome annotations were used:
Homo sapiens: RefSeq GenBank flat files build 36 (14 September 2006), downloaded from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens;
Caenorhabditis elegans: RefSeq GenBank flat files (28 November 2005), downloaded from ftp://ftp.ncbi.nih.gov/genomes/Caenorhabditis_elegans;
Drosophila melanogaster: FlyBase version 3.1, downloaded from ftp://ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r3.1/fasta;
Arabidopsis thaliana: RefSeq GenBank flat files (downloaded on 15 April 2008), downloaded from ftp://ftp.ncbi.nih.gov/genomes/Arabidopsis_thaliana.
Genome files were parsed using in-house Perl and Matlab scripts. Only genes for which messenger RNA (mRNA) record as well as coding sequence (CDS) record could be identified were included in the analysis, as lengths of introns, exons, and so on, were computed by comparing the mRNA structure with the CDS structure. Genes annotated as pseudogenes were removed. Genes annotated as having alternative isoforms were excluded from the analysis, unless all the isoforms differed only in their untranslated regions (UTRs). In that case, introns and exons within the coding region were analyzed, and the UTRs’ information was discarded.
Expression Level
Expression-level measurements were from the following sources:
Homo sapiens: The data of Su et al. (2004) obtained with the Affymetrix GeneChip Human Genome U133 Array Set HG-U133A (see http://www.affymetrix.com/products/arrays/specific/hgu133.affx) were employed. In this study, the microarrays were used to measure expression in 79 different tissues, each in two repetitions. Some tissues, such as brain, blood, and bone marrow, are overrepresented in Su's data set. Other tissues are cancerous. We have therefore selected 32 nonredundant, normal tissues for the analyses in the current study (supplementary table S5, Supplementary Material online).
Caenorhabditis elegans: Combined measurements from several studies, all employing the Affymetrix GeneChip C. elegans Genome Array (see http://www.affymetrix.com/products/arrays/specific/celegans.affx) (Baugh et al. 2005; Fox et al. 2007; Falk et al. 2008), were used. Only expression in wild-type specimens was included. The final data set included 34 samples from Baugh et al. (2005), 7 samples from Fox et al. (2007), and 7 samples from Falk et al. (2008), 48 samples in total.
Drosophila melanogaster: Measurements from two studies using the Affymetrix GeneChip Drosophila Genome 2.0 Array (see http://www.affymetrix.com/products/arrays/specific/fly_2.affx) (Chintapalli, Wang, and Dow 2007; Kadener et al. 2007) were analyzed. Only expression in wild-type specimens was included. The final data set included 10 samples (control only) from Kadener et al. (2007) and 44 samples from Chintapalli, Wang, and Dow (2007).
Arabidopsis thaliana: The results of Meyers et al. (2004), which comprise one of the plant MPSS databases (Nakano et al. 2006), were analyzed. Expression was measured using the 17-bp signature MPSS technology. Only untreated samples were used, summing up to 10 samples in total (AP1, AP3, AGM, INS, ROS, SAP, LES, GSE, CAS, SIS [see “Library information” in http://mpss.udel.edu/at]).
Probes/target sequences that had matches in multiple genes were removed from the analysis. When multiple probes/target sequences matched a single gene, their average was taken. Only genes for which expression measurements were available were included in the analysis. The final data set consisted of 9,355 human genes, 10,071 fly genes, 15,438 nematode genes, and 14,184 Arabidopsis genes (see supplementary table S6, Supplementary Material online, for more statistics on the data set). Therefore, for a given organism, each gene in the data set has one expression-level measurement per sample. In order to put expression-level measurements from different experiments and different tissues on the same scale, we followed a technique proposed by Ren et al. (2006): For each organism, the expression data are a matrix ED with ng rows (number of genes) and ns columns (number of samples). Let C be a predefined number of categories. Then, each column (sample data) is ranked into C categories, such that the genes with the lowest expression levels have the value 1 and the genes with the highest expression levels have the value C. The number of genes in each category is kept approximately equal (as closely as possible). At the end of this process, we obtain another ng × ns matrix, ER, with ranks replacing the original expression values. In order to obtain a single expression-level value per gene, the ranks in every row were averaged, and the result was rounded to the nearest integer. Formally, the expression of a gene g is given by
In this work we always use C = 30, but the results are robust to the C value (data not shown). Such definition of the expression of a gene allows combining many expression data sources but comes at the expense of using ranks. This approach allows one to analyze the general trends in the relationship between expression and compactness, as we do in this work, but masks the specific details of this relationship. We verified that adopting another strategy, based on the popular log2 expression values instead of ranks over samples, yielded qualitatively the same results (supplementary fig. S1, Supplementary Material online).
Expression Breadth
It is customary to define a threshold for making a binary decision whether a gene is present or absent in a particular tissue/condition. Such binary decisions are, by definition, somewhat arbitrary, so in this study we employed six different threshold values. These threshold values were taken as the expression levels that correspond to a certain percentile of the expression levels, ranging from 10% to 60% (see supplementary table S7, Supplementary Material online). Expression breadth was computed only for human genes and was defined as the number of tissues where the gene is called expressed under the corresponding threshold.
Segmented Regression
Segmented regression is the process of fitting data to possibly more than one linear segment (Oosterbaan 1994). We used the SegReg (http://www.waterlog.info/segreg.htm) software, which selects the most statistically significant linear model that consists of up to two linear segments. When a two-segment model is the most appropriate one, the program computes the bend point and estimates its standard error.
Results
The Relationship between Gene Compactness and Expression Level Is Universal and Nonmonotonic
We measured gene compactness using several length variables. Total lengths variables are the total transcript length, the length of the protein-coding (CDS) region, the combined lengths of all coding and noncoding exons, and the total length of introns (supplementary fig. S2, Supplementary Material online). Whenever available, we also analyzed lengths of the 3′ untranslated regions (UTRs) and the 5′UTRs. Per-gene length variables are the mean and median lengths of the exons and the introns. Expression levels were binned into 30 classes (see Methods), and the average value of each of the length variables in each of the 30 expression levels was computed across all the genes.
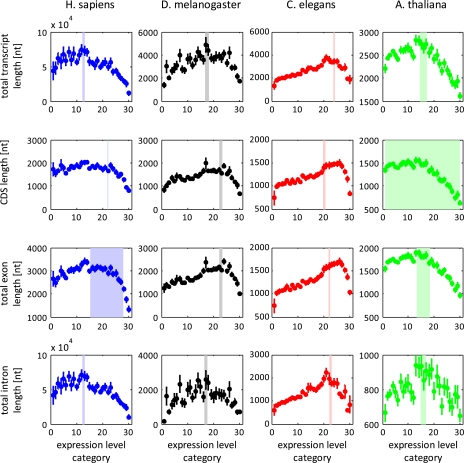
Qualitatively, all the organisms showed the same nonmonotonic dependence between expression level and each of the total length variables (fig. 1, supplementary fig. S3, Supplementary Material online). The compactness of genes decreases with the increasing expression level for low expression levels (the C↓E trend) and then increases with the increasing expression level for the high expression levels (the C↑E trend). To statistically validate the trend of monotonic increase of the gene length variable values followed by the monotonic decrease (hereinafter the Λ-shape), we employed segmented regression using the SegReg software (see Methods). The Λ-shape was statistically significant in all four organisms and for all the four total length variables (table 1). Notably, the Λ-shape is more significant than any monotonic trend alone. Specifically, the Λ-shape was highly significant in A. thaliana, and we did not obtain any support for the claim of Ren et al. (2006) that highly expressed genes were less compact than lowly expressed genes in this plant. The lack of agreement with Ren et al. appears to stem, largely, from a series of methodological differences. Among these are the differences in selecting the tissues and genes that comprised the database and differences in the way that highly expressed genes were compared with lowly expressed ones. A detailed discussion of the possible reasons for the failure to detect the shortening of highly expressed genes by Ren et al. (2006) is available in the Supplementary Text (Supplementary Material online).
FIG. 1.—
Total length variables as functions of expression-level category. All lengths are measured as number of nucleotides. Expression levels are binned into 30 categories, with higher categories matching higher expression levels. Each dot is the mean value for all genes in the given expression category, and the error bar indicates the standard deviation of the mean. Dark areas depict the area (standard error) of the bend point where the trend changes from increasing to decreasing, according to SegReg.
Table 1.
The Nonmonotonic Dependence between Gene Length Variables and Expression
| Bend Point | Left Fraction | Left Slope | Right Slope | ||
| Total transcript length | HS | 12.6 ± 0.5 | 0.37 | 1,170 ± 487 | -2,440 ± 243 |
| DM | 17.5 ± 0.7 | 0.59 | 118 ± 24 | -119 ± 19 | |
| CE | 23.9 ± 0.4 | 0.80 | 84.4 ± 4.7 | -258 ± 18 | |
| AT | 15.8 ± 1.3 | 0.80 | 22.5 ± 4.2 | -68.6 ± 3.0 | |
| CDS length | HS | 21.9 ± 0.2 | 0.76 | 6.17 ± 3.4 | -109 ± 12 |
| DM | 22.8 ± 0.5 | 0.79 | 30.9 ± 3.4 | -102 ± 7 | |
| CEa | 20.1 | 0.71 | 17.2 ± 1.8 | -38.8 ± 6.5 | |
| AT | 15.8 ± 16.6 | 0.80 | 6.09 ± 2.5 | -55.5 ± 1.7 | |
| Total exon length | HS | 21.6 ± 6.3 | 0.76 | 18.3 ± 4.8 | -171 ± 7 |
| DM | 22.8 ± 0.5 | 0.79 | 42.4 ± 3.6 | -81.4 ± 8.0 | |
| CEa | 22.2 | 0.77 | 25.7 ± 1.7 | -57.9 ± 9.2 | |
| AT | 15.8 ± 2.7 | 0.80 | 12.7 ± 2.5 | -55.8 ± 1.8 | |
| Total intron length | HS | 12.6 ± 0.5 | 0.37 | 1,070 ± 484 | -2,370 ± 241 |
| DM | 16.9 ± 0.6 | 0.56 | 76.1 ± 17.4 | -101 ± 25 | |
| CE | 22.5 ± 0.4 | 0.77 | 60.7 ± 4.3 | -161 ± 14 | |
| AT | 15.8 ± 1.0 | 0.80 | 9.8 ± 2.3 | -12.8 ± 1.7 | |
| Mean exon length | HS | -5.58 ± 1.31 | -13.40 ± 1.76 | ||
| DMb | -2.09 ± 1.32 | -22.2 ± 2.26 | |||
| CE | 1.69 ± 0.34 | 4.62 ± 0.29 | |||
| AT | 9.4 ± 0.4 | 0.64 | 4.25 ± 2.14 | -11.10 ± 1.22 | |
| Mean intron length | HS | -93 ± 29 | -237 ± 37 | ||
| DM | 14 ± 2.3 | 0.48 | 9.48 ± 6.45 | -10.2 ± 2.9 | |
| CE | 21.9 ± 0.6 | 0.74 | 7.77 ± 0.92 | -17.2 ± 2.2 | |
| ATc |
The results of segmented regression applied to the data in figure 1. Bend point: The expression-level category that is the border between the increasing and the decreasing parts, as decided by SegReg. The ± symbol indicates standard error (SE). Whenever the curve does not show Λ-shape, the bend point is not reported. Left fraction: The fraction of genes in the increasing part of the curve. Left slope: The slope of the left part of the curve, as computed by SegReg. The ± symbol indicates SE. Right slope: The slope of the right part of the curve, as computed by SegReg. The ± symbol indicates SE. If not otherwise indicated, SegReg found statistical support in favor of two joint linear segments.
SegReg found statistical support for two disjoint linear segments.
Computation was made on median values.
Computation failed in SegReg (software crashed).
In all organisms and for all total length variables, the slope of the increasing part has a smaller absolute value than the slope of the decreasing part. In other words, the Λ-shape is always nonsymmetrical: The C↓E trend seen for the weakly to moderately expressed genes is relatively weak and gradual, whereas the C↑E trend that is characteristic of more highly expressed genes is considerably steeper (fig. 1).
Comparing the lengths of UTRs to expression levels yielded no significant trend (supplementary figs. S4 and S5, table S1, Supplementary Material online). It remains uncertain whether this is an indication of a genuine lack of dependence or a reflection of the poor annotation of the UTRs.
Individual Introns Show Λ-Shape Dependence on Expression Level, whereas Individual Exons Show Monotonicity
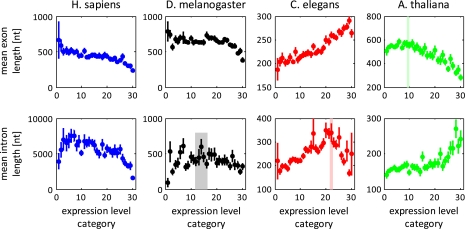
Mean exon lengths do not display a Λ-shape dependence on the expression level except for a possible weak effect in Arabidopsis (fig. 2 and supplementary fig. S6 [Supplementary Material online], table 1). Human and fly show a clear monotonic decrease (a trend that is seen also in Arabidopsis except for the slight increase for lowly expressed genes), whereas, in a striking contrast, C. elegans shows a monotonic increase. As mean exon lengths reflect the combined effect of intron density and total protein lengths, the increase in average exon length with increasing expression level in C. elegans is likely to be linked to the high rate of intron loss in nematodes (Carmel, Wolf, et al. 2007).
FIG. 2.—
Mean lengths of exons and introns as functions of expression-level category. All designations are as in figure 1.
By contrast, length–expression curves for the mean (fig. 2, table 1) and median (supplementary figs. S7 and S8, table S2, Supplementary Material online) intron length show a Λ-shape in all organisms. Thus, the nonmonotonic Λ-trend that is observed for the total length measures seems to be primarily associated with the changes in the length of introns.
Similar Dependence on Expression Level for Coding and Noncoding Regions
Both the selection and the genomic design hypotheses imply dependence between the expression level of a gene and the total length of the introns or the exons rather than the corresponding means. Our present findings reveal qualitatively the same, Λ-shape of the length–expression curve for all employed total length variables. In itself, this universal form of the dependence does not, of course, indicate that the details of this dependence, in particular, for exons and introns, are the same.
To further compare the dependences of the coding and noncoding regions on expression level, we computed for all the genes in each organism the correlation between the CDS length and the total intron length (the raw correlation) and found it to be positive and significant in all cases (table 2). If this correlation ensues from the two variables being similarly dependent on the expression level, the correlation is expected to increase when computed for the mean values in each expression-level class (binned correlation). Indeed, such an increase in the correlation coefficient was invariably seen (table 2). Such an increase in correlation is expected with any binning that is identical for both variables due to noise reduction. Therefore, we tested the significance of the specific increase due to binning by expression level by randomly assigning the expression-level classes to the genes 1,000 times, and computing the correlation coefficient between the CDS length and the total intron length for the resulting random data sets. The increase in the correlation above the normal increase due to binning was found to be significant in all species except A. thaliana (table 2). Thus, at least, in animals, coding and noncoding parts of the gene seem to show similar dependence on the expression level.
Table 2.
Raw versus Binned Correlation Coefficients
| CDS Length - Total Intron Lengtha | Intron Density - Expression Levelb | |||||
| Raw Correlation | Binned Correlation | P Value | Raw Correlation | Binned Correlation | P Value | |
| HS | 0.30 | 0.85 | 0.01 | 0.11 | 0.93 | <0.001 |
| DM | 0.20 | 0.69 | 0.01 | 0.13 | 0.73 | <0.001 |
| CE | 0.28 | 0.81 | 0.01 | -0.20 | -0.93 | <0.001 |
| AT | 0.52 | 0.58 | 0.40 | 0.06 | 0.84 | <0.001 |
Raw correlation: The correlation was computed using all the genes. Binned correlation: The correlation was computed between the mean values across the expression-level categories. P value: The significance of the correlation change due to the binning was computed using 1,000 bootstrap repetitions.
Pearson (linear) correlation.
Spearman correlation.
However, although qualitatively similar, these dependencies are clearly distinct as indicated by the different shapes of the dependence for the mean lengths of exons and introns (fig. 2). We further explored these differences by comparing the locations of the bend point, that is, the point where the trend changes from C↓E to C↑E, for the exons and the introns. Comparing expression-level categories between organisms involves considerable uncertainty. Nevertheless, assuming similar expression-level distributions in all species, the relative positions of the bend point can be compared by counting the fraction of genes that are in the increasing (C↓E) part (table 1). Of course, such a comparison is meaningful only when the behavior of the entire ensemble of genes is considered: Identical bend points in different organisms are likely to represent different absolute expression levels. The positions of the bend point differed between organisms and, at least for human and fly, between the introns and the exons. In general, bending occurs at lower expression levels in introns compared with exons. Together with the observations of the preceding section, these findings indicate that the Λ-shape is more prominent in introns; in particular, the tendency of introns to be elongated with increasing expression (for lower expression levels) is more pronounced than the similar tendency for exons. Furthermore, the bend point in the plots for total transcript length and the total intron length but not the CDS or total exon length is shifted to the left in humans compared with all other organisms (fig. 1, table 1). The difference in the position of the bend point suggests that in humans the selective forces that underlie the C↑E trend become prevalent at a relatively lower expression level than in other organisms, in all likelihood, because humans on average have much longer introns than invertebrates or plants.
Expression Level Correlates with Gene Compactness Stronger than Expression Breadth
In agreement with previous observations (Eisenberg and Levanon 2003), gene expression breadth (i.e., the number of tissues in which a gene is expressed above a threshold level) in human is positively and highly significantly correlated with expression level (supplementary table S3, Supplementary Material online). However, for all total length variables and for all threshold values, the segmented regression against breadth explained less variance than the segmented regression against expression level (supplementary fig. S9, table S3, Supplementary Material online).
Differences in GC Content Cannot Explain the Observed Relationship between Gene Compactness and Expression Level
To determine whether our findings could be affected by local mutational biases that are known to depend on the base pair composition of DNA (Duret, Mouchiroud, and Gautier 1995), we computed average GC values for all exons, all introns, and the protein-coding sequences in human. As with expression breadth, GC content did not show any significant trend, neither concave nor monotonic, with the expression level (supplementary fig. S10, table S4, Supplementary Material online).
Intron Density Tends to Increase with Expression Level
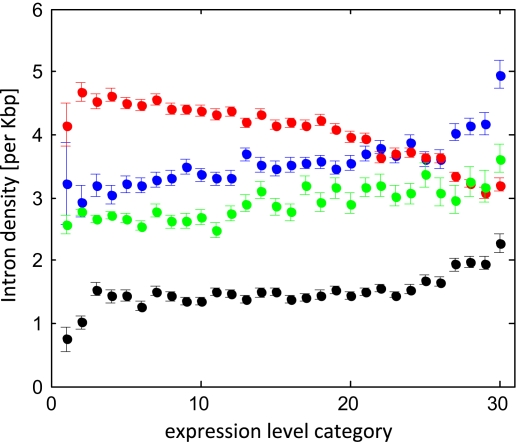
We observed previously that introns appear to be gained at higher rates in evolutionarily highly conserved genes than in faster evolving genes, with the implication that the intron density of a gene might be functionally relevant (Carmel, Rogozin, et al. 2007). Similarly, it was shown that ancient eukaryotic genes, on average, have a higher intron density than genes of apparent more recent origin (Wolf et al. 2009). We computed intron density for the four analyzed organisms as a function of expression-level category for all four organisms (fig. 3). In accord with the previous findings (Carmel, Rogozin, et al. 2007), there was a significant positive correlation between expression level and intron density in three of the four analyzed organisms (table 2). Only C. elegans showed the opposite trend, as previously reported by Fahey and Higgins (2007), possibly, owing to its atypically high intron loss rate (Carmel, Wolf, et al. 2007). Our results agree with those of Fahey and Higgins for the nematode but not for D. melanogaster for which they report the same trend as in C. elegans, whereas we observe a trend consistent with that seen in human and Arabidopsis (fig. 3).
FIG. 3.—
Intron density of genes as a function of expression-level category. The intron density is measured as the number of introns per kilobase of the CDS. Color codes: blue = human, black = Drosophila, red = nematode, and green = Arabidopsis. All other designations are as in figure 1.
The increase in intron density in parallel with increased expression is likely to reflect widespread functional importance of introns in eukaryotic genomes. This relationship is further supported by the substantial increase of the correlation coefficient when computed using the mean value of the intron density in each expression-level class (table 2).
Discussion
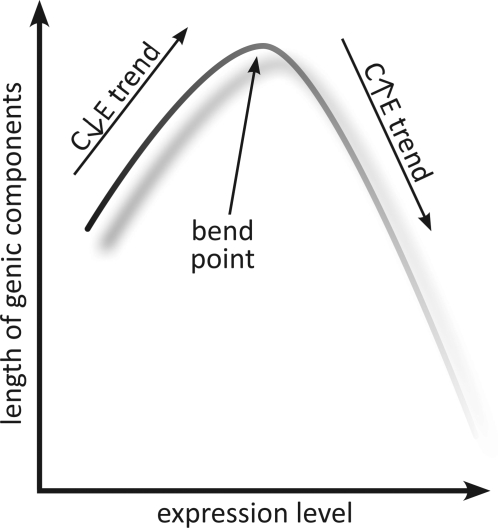
Comparative analysis of the connections between gene architecture and expression levels in four multicellular eukaryotes, H. sapiens, C. elegans, D. melanogaster, and A. thaliana, revealed a universal qualitative relationship between expression level and compactness (fig. 4). Unexpectedly, this trend is nonmonotonic whereby, up to a certain limit, genes become less compact with the increase of the expression level (the C↓E trend), but with further increase in expression, become more compact again (the C↑E trend). Thus, somewhat paradoxically, eukaryotic genes with the lowest expression level tend to be about as compact as the most highly expressed genes, whereas moderately expressed genes are the least compact ones.
FIG. 4.—
The universal nonmonotonic relationship between the expression level of a gene and its compactness: a schematic depiction.
Numerous results of comparative genomics and evolutionary systems biology indicate that gene evolution is shaped by complex interaction of many factors, some of which are general, whereas others are gene-specific, and some are selective, whereas others are neutral (Nei 2005; Lynch 2007; Ellegren 2008; Koonin 2009). The existence of a universal, nonmonotonic dependence between gene compactness and expression level seems to suggest that gene architecture depends on the interplay of several factors at least some of which are selective. The bending of all the expression–length curves at the transition from moderate to high expression levels (fig. 1) seems to be incompatible with a single underlying mechanistic cause. Furthermore, we identified a significant positive correlation between intron density and expression level, in all analyzed organisms except for C. elegans. Combined with the previous observations on the higher intron density and increased intron gain rate in evolutionarily conserved genes (Carmel, Rogozin, et al. 2007; Wolf et al. 2009) and the strong positive correlation between expression level and evolutionary conservation (Pal, Papp, and Hurst 2001; Koonin and Wolf 2006; Drummond and Wilke 2008; Wolf et al. 2009), this link is compatible with a functional significance of introns in expression, perhaps, leading to selection for intron accumulation. Indeed, multiple pieces of evidence in support of the involvement of introns in the regulation of expression of individual genes have been reported (Le Hir, Nott, and Moore 2003; Nott, Meislin, and Moore 2003; Moore 2005).
In contrast to the highly significant (even if complex) link between expression level and gene compactness, we found a much weaker dependence between compactness and expression breadth across animal tissues. This observation directly falsifies the principal prediction of the genome design hypothesis (Vinogradov 2004).
Thus, the results of the present analysis favor the selection hypothesis as the principal explanation why highly expressed genes are more compact than genes expressed at a lower level (the C↑E trend). What factors underpin this selection remains an open question. The original explanation implicated the minimization of ATP expenditure and time spent on the expression (transcription combined with transcript maturation) of the given gene (Castillo-Davis et al. 2002). These remain potentially relevant factors, but their ultimate importance could be questioned considering that the expenditure of both energy and time during transcription is small compared with that during translation, whereas the connection between intron length and splicing rate is uncertain. An alternative selective factor could be the fidelity of transcription, splicing, and in the case of exon sequences, translation. Evolution of both nonsynonymous and synonymous sites in protein-coding sequences, in particular, those of highly expressed genes, appears to be substantially constrained by selection for robustness to mistranslation-induced protein misfolding (Drummond and Wilke 2008; Wolf, Wolf, and Koonin 2008). Selection for short exons in highly expressed genes could be part of the same general trend. Similarly, selection for short introns might have to do with the minimization of splicing errors that result in the formation of partially spliced transcripts which could be not only a waste of time and energy but also, perhaps, more importantly, a source of potentially toxic, misfolded proteins.
Explaining the negative trend (C↓E) between gene compactness and expression that is observed for genes that are expressed at lower levels seems to be more difficult than interpreting the positive trend. Highly expressed genes, on average, encode more evolutionarily conserved proteins than lowly expressed genes (Koonin and Wolf 2006; Drummond and Wilke 2008). This trend is a key aspect of the “status” of a gene in an organism, a proxy of its biological importance, broadly understood (Koonin and Wolf 2006). The increased total length of introns in moderately expressed genes compared with lowly expressed genes, in part, is due to the increased intron density. However, the (C↓E) trend is clearly seen even in C. elegans, where intron density drops with the increase in expression level. The most relevant phenomenon to account for the (C↓E) trend might be the accumulation, in introns, of regulatory elements contributing to expression (Le Hir, Nott, and Moore 2003; Nott, Meislin, and Moore 2003; Moore 2005). Thus, perhaps paradoxically, a version of the genomic design hypothesis could be relevant to explain the negative correlation between expression and gene compactness that is seen on the low end of the expression scale.
The most surprising finding of this analysis seems to be the universality of the dependence between expression level and gene compactness in all analyzed organisms (figs. 1 and 4), in spite of the major differences in their gene architectures. As a case in point, the trend between the total gene length and expression level is essentially the same in humans and Arabidopsis despite the fact that, in humans, the total length of introns is much greater than that of exons, whereas the opposite holds for Arabidopsis. Thus, the evolutionary factors that shape the dependence between gene compactness and expression appear to be more fundamental than those that govern the evolution of gene architecture (determined, primarily, by intron lengths), in agreement with the growing evidence that expression is one of the major determinants of gene evolution (Drummond and Wilke 2008; Wolf, Wolf, and Koonin 2008). The effects of gene compactness on expression are certainly amenable to further comparative-genomic and experimental analyses, which should lead to insights into the nature of the universal relationship described here.
Supplementary Material
Supplementary figures S1–S10, tables S1–S7, and supplementary text are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/)
Supplementary Material
Acknowledgments
The authors would like to thank Miklós Csűrös and Igor Rogozin for helpful discussions. The authors’ research is supported by the intramural funds of the US Department of Health and Human Services (National Library of Medicine).
References
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Baugh LR, et al. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132:1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- Brenner S, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brenner S, et al. In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci U S A. 2000;97:1665–1670. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Rogozin IB, Wolf YI, Koonin EV. Evolutionarily conserved genes preferentially accumulate introns. Genome Res. 2007;17:1045–1050. doi: 10.1101/gr.5978207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet. 2005;21:203–207. doi: 10.1016/j.tig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Comeron JM. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J Mol Evol. 1995;40:308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 2008;17:4586–4596. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- Fahey ME, Higgins DG. Gene expression, intron density, and splice site strength in Drosophila and Caenorhabditis. J Mol Evol. 2007;65:349–357. doi: 10.1007/s00239-007-9015-y. [DOI] [PubMed] [Google Scholar]
- Falk MJ, et al. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008;93:388–397. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, et al. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol. 2007;8:R188. doi: 10.1186/gb-2007-8-9-r188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Evolutionary systems biology: links between gene evolution and function. Curr Opin Biotechnol. 2006;17:481–487. doi: 10.1016/j.copbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. New York: Worth; 1982. pp. 615–644. [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- Li SW, Feng L, Niu DK. Selection for the miniaturization of highly expressed genes. Biochem Biophys Res Commun. 2007;360:586–592. doi: 10.1016/j.bbrc.2007.06.085. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Meyers BC, et al. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nakano M, et al. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Selectionism and neutralism in molecular evolution. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterbaan RJ. Frequency and regression analysis of hydrologic data. In: Ritzema HP, editor. Drainage principles and applications. Wageningen (the Netherlands): ILRI Publication; 1994. pp. 175–224. [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X-Y, Vorst O, Fiers MWEJ, Stiekema WJ, Nap J-P. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22:528–532. doi: 10.1016/j.tig.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C, Hurst LD. Gametophytic selection in Arabidopsis thaliana supports the selective model of intron length reduction. PLoS Genet. 2005;1:e13. doi: 10.1371/journal.pgen.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien HK. Compact genes are highly expressed in the moss Physcomitrella patens. J Evol Biol. 2007;20:1223–1229. doi: 10.1111/j.1420-9101.2007.01301.x. [DOI] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucker DS, Yamamoto KR. Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J Biol Chem. 1984;259:7416–7420. [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wolf MY, Wolf YI, Koonin EV. Comparable contributions of structural-functional constraints and expression level to the rate of protein sequence evolution. Biol Direct. 2008;3:40. doi: 10.1186/1745-6150-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 2009;106:7273–7280. doi: 10.1073/pnas.0901808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J Biomed Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- Zhao C, Hamilton T. Introns regulate the rate of unstable mRNA decay. J Biol Chem. 2007;282:20230–20237. doi: 10.1074/jbc.M700180200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.