Abstract
Next-generation sequencing is revolutionizing the identification of transcription factor binding sites throughout the human genome. However, the bioinformatics analysis of large datasets collected using chromatin immunoprecipitation and high-throughput sequencing is often a roadblock that impedes researchers in their attempts to gain biological insights from their experiments. We have developed integrated peak-calling and analysis software (Sole-Search) which is available through a user-friendly interface and (i) converts raw data into a format for visualization on a genome browser, (ii) outputs ranked peak locations using a statistically based method that overcomes the significant problem of false positives, (iii) identifies the gene nearest to each peak, (iv) classifies the location of each peak relative to gene structure, (v) provides information such as the number of binding sites per chromosome and per gene and (vi) allows the user to determine overlap between two different experiments. In addition, the program performs an analysis of amplified and deleted regions of the input genome. This software is web-based and automated, allowing easy and immediate access to all investigators. We demonstrate the utility of our software by collecting, analyzing and comparing ChIP-seq data for six different human transcription factors/cell line combinations.
INTRODUCTION
Although chromatin immunoprecipitation (ChIP) was first adapted for use with mammalian cells <10 years ago (1,2), it is now the gold standard experiment for the identification of a target gene of a particular transcription factor. Recent advances allow investigators to use the ChIP assay to identify and characterize the entire set of binding sites for a given factor. Such large-scale studies of transcription factor binding began using promoter-specific microarrays, a technique called ChIP-chip (3–7). However, many binding sites will be completely missed on such arrays because some factors localize mainly to regions outside of the tiled core promoters (8,9). ChIP-chip has now been extended to the entire human genome using a series of microarrays that contain oligonucleotides spaced ∼35–100 nt apart (10,11). This gapped spacing is necessary due to the large number of arrays (and thus the large cost) that would be required if overlapping oligomers were used. However, the gapped spacing results in the genome-scale ChIP-chip experiments being less precise in mapping the exact location of a binding site than if overlapping oligomers were used. The latest development, ChIP-seq, which uses the immunoprecipitated sample to create a library that is analyzed using high-throughput next generation sequencers, also provides genome-scale analysis of binding sites (12–15). Because ChIP-seq is not limited to a specific tiled region but can sample the entire genome, this technique can provide a very precise mapping of a peak location (16). A comparison of an E2F4 binding site identified in the GMNN promoter using both ChIP-chip and ChIP-seq is shown in Supplementary Figure S1. Although both technologies correctly identify the GMNN promoter as a target for E2F4, ChIP-seq provides a more accurate location of the binding site. Since the ChIP-seq technology provides a genome-scale analysis that is less costly than genome-wide ChIP-chip and because it allows for more precise mapping of binding site locations, most investigators are moving to this technology as the method of choice for identifying transcription factor binding sites. However, as described below, like any other technology, ChIP-seq also has issues that must be considered.
The first step in the analysis of ChIP-seq data is to identify all sequenced tags that map uniquely to the genome of interest. For many ChIP-seq experiments, investigators analyze very short reads (e.g. 27 nt). This short-read length can sometimes result in a sequenced tag mapping to more than one place in the genome. If this occurs, the tag will be discarded and not included in peak analyses. In most cases, this is not a problem because the region surrounding the ‘non-unique’ tag contains many unique 27-mers and a peak can still be identified. However, if a peak lies within a large region that is not unique within the genome, it will be completely missed (i.e. it will be a false negative). This is especially problematic for genes that have been duplicated over evolutionary time and thus have several identical (or almost identical) copies that reside in different genomic locations (Supplementary Figure S2). Another reason that false negatives can arise in ChIP-seq analyses is due to effects of chromatin structure on the fragmentation step. Investigators use either sonication or micrococcal nuclease to digest the chromatin before using it in a ChIP assay. However, the sonication and/or digestion step does not always provide a representative population of fragments in the right size range; this is especially problematic for heterochromatic regions. These regions will be underrepresented in the sequencing library and peak identification can be adversely affected using ChIP-seq (Supplementary Figure S3). Conversely, just as heterochromatic regions are lost during sample preparation, promoter regions are sometimes artificially enriched. Promoters appear to be more easily fragmented into small chromatin than other regions of the genome and often show up as a small peak in an input sample (17). However, proper analysis using appropriate input libraries can improve the accuracy of binding site identification (see below for more details). Another problem that must be considered when analyzing ChIP-seq data is that certain regions always appear as peaks in a given cell type, independent of the factor being tested (Supplementary Figure S4). These false positives can be due to repetitive regions being mis-annotated as unique. This is especially problematic when studying cancer cell lines and tissues, which have many amplified genomic regions. It is critical that these false positives are removed from the set of called peaks. As described below, we have addressed many of these problems by identifying binding sites as regions that are significant over background, independent of sequence density. We present a software package, called Sole-Search, to analyze ChIP-Seq data and determine statistically significant peaks, with minimal false positives and false negatives. We demonstrate the utility of our software by collecting, analyzing and comparing ChIP-seq data for six different human transcription factors/cell line combinations; E2F4, E2F6 and YY1 in K562 cells; YY1 in Ntera2 cells, TCF7L2 (called in this article by its other name TCF4) in HCT116 cells, and TFAP2A (called in this article by its other name AP2α) in HeLa cells; the analyses of these datasets are provided in Supplementary Data S1–S21, whereas the sgr visualization files and sequenced tag files are available on the UCSC browser.
MATERIALS AND METHODS
ChIP
E2F4, E2F6 and YY1 ChIP samples were prepared from human chronic myelogenous leukemia cells (K562, ATCC #CCL-243), which were grown in RPMI supplemented with 10% FBS, 2 mM l-Glutamine, 100 U/ml Pen-Strep and harvested at a density of 106 cells/ml cells. YY1 samples were also prepared from Ntera2 embryonal carcinoma cells (ATCC #CRL-1973) which were grown in DMEM (GIBCO #11960) with 10% FBS, 2 mM l-Glutamine and 100 U/ml Pen-Strep and harvested at 90% confluency. TCF4 ChIP samples were prepared from HCT116 cells (ATCC #CCL-247) which were grown in McCoy’s 5A Medium supplemented with 10% FBS and 1% Penicillin/Streptomycin until 80% confluency. AP2α ChIP samples were prepared from HeLa cells (ATCC #CCL-2.2) which were grown in 5% BCS DMEM, 2 mM l-Glutamine and harvested at 50–55% confluency. All cell cultures were cross-linked for 10 min by adding formaldehyde to the growth media to a final concentration of 1%. Cross-linking was stopped by the addition of glycine to 125 mM final concentration, and cells were washed twice with ice cold PBS. Chromatin was fragmented using the Bioruptor sonicator (Diagenode) for 30 min (30 s pulses, 1.5 min pauses in between) to produce fragments ∼500 nt in size. ChIP assays were performed using ∼5 × 107 to 1 × 108 cells for each ChIP as described at http://www.genomecenter.ucdavis.edu/farnham/protocol.html. Antibodies used were: anti-TCF4 antibody (Cell Signaling Technology, #9751), anti-E2F4 antibody (SantaCruz, #sc-866x), anti-E2F6 (SantaCruz, #sc-22823x), anti-AP2α (Santa Cruz, #sc-8975), and anti-YY1 SantaCruz, #sc-1703x). Immunoprecipitates were collected using either Staph A or protein G magnetic beads (Cell Signaling Technology); further details are available upon request.
ChIP-seq library construction and quantitation
ChIP samples were tested by PCR using positive and negative control primer sets prior to making the library. ChIP libraries were created according to Robertson et al. (12), using 15 cycles of amplification. Libraries were run on a 2% agarose gel and the 150–450 bp or 400–600 fraction of the library was extracted and purified (except as noted in Supplementary Figure S3, which compares libraries made from 150 to 400 bp and 400 to 1 kb samples); the library with the highest enrichment, as monitored by qPCR was used for sequencing. The libraries were initially quantitated using a Nanodrop. However, we noted that the number of clusters obtained from sequencing the libraries was often much lower than expected from the concentration determined by the Nanodrop. Therefore, to estimate relative amplification potential, the DNA was quantitated using serial dilutions and compared to a reference library by real-time PCR using primers complementary to the library adapters. The amplification value relative to the reference library was then used to estimate the flow-cell loading concentration. For example, to produce the desired number of clusters for TCF4, 10 times more sample was loaded onto the flow cell than would have been used if the concentration was determined using the Nanodrop. The ChIP-seq libraries were run on an Illumina GA2 by the DNA Technologies Core Faciity at the University of California-Davis (http://genomecenter.ucdavis.edu/dna_technologies/). The tag files for the E2F4, E2F6, YY1, AP2α and TCF4 ChIP-seq experiments are publicly available on the UCSC browser, as part of the ENCODE Consortium (http://www.genome.ucsc.edu/ENCODE/); see also Supplementary Table S1.
Statistical basis of peak calling by Sole-Search
Sole-Search employs several different analysis steps in the peak calling, each step enabling elimination of different types of false positives or false negatives in the final peak list. Below, we describe each step in the program and the consequences of removal of any individual step.
Step 1: Identification and compensation for amplified and deleted regions of the genome
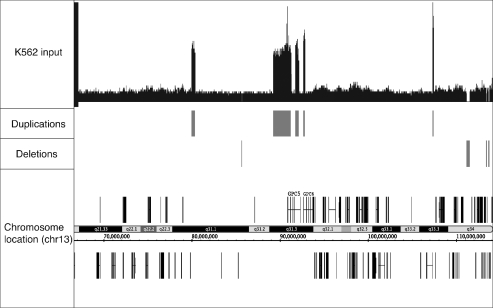
Human cell lines are well known to have genomic instability and often contain duplications and deletions of regions of the genome. Since the regions that are duplicated in a cell line are found in a single copy in the reference genome, they are identified as unique in the initial stages of tag binning. However, since these regions are actually found in multiple copies, the entire region is overrepresented and can appear to be composed of many binding sites; thus, false positive peaks are called in these regions. The plethora of false positives found in these regions may skew downstream analysis of the set of peaks (such as target gene identification, location analysis, etc.). Therefore, it is beneficial to identify such regions as being non-unique and estimate copy number, so that peak calling in these regions may be adjusted accordingly. To identify the duplicated and deleted regions, the input data are first smoothed. Using a sliding window, all input data sequenced for that cell type is binned into 200 bp bins and smoothed using a sliding average (the values of ten bins on either side of a center bin are averaged to create the center bin’s smoothed value). For our analyses, regions that have no sequence at all, and are greater than 10 000 bp (and less than 2 900 000, to eliminate calling centromeres as deletions) are considered deletion events; these regions are identified in the deletions.gff file. Similarly, we define large duplicated regions as being at least 10 000 bp long: 1/3 of the bins must contain at least 3-fold more tags than the average bin tag count in the genome. Small non-unique regions are defined as being at least 2000 bp long: 1/3 of the bins within the region must contain at least 4-fold more tags than average. Copy number of the duplicated region can be roughly estimated as fold increase of average bin count within the duplicated region over average bin count within the genome. Regions that comply with either the small or large duplication parameters are listed with fold increase values in the duplications.gff file. Since the method for calling peaks, as described below, assumes that there is only one copy of every region of the genome, Sole-Search adjusts duplicated regions so that they are represented by only one copy; to do so, tag counts, in both background data and ChIP data, are divided by the fold increase of the duplicated region.
Consequences of elimination of Step 1
If amplification is not taken into consideration by the peak-calling program, a large number of false positive peaks will be called in these regions (Figures 1, 5C and Supplementary Figure S9, Panel A). Furthermore, we note that peaks within the duplicated regions will be ranked differently by programs that do versus do not take into account the extent of genomic amplification. Specifically, these peaks will appear much higher on the ranked list of peaks called by programs that do not take duplications into account.
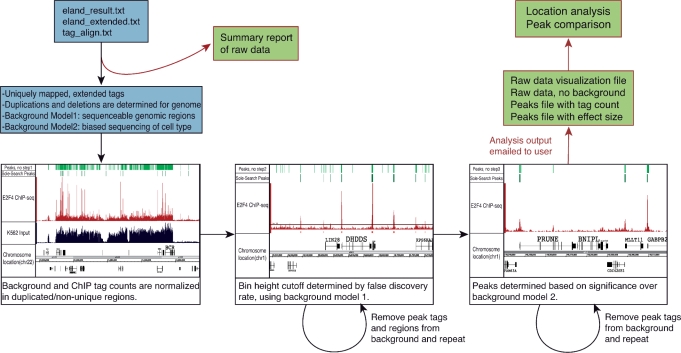
Figure 1.
Sole-Search schema. A user can upload one Solexa raw data file into the online program or can upload several files and have the data merged into one file for analysis. The program parses the data and gives the user a summary report which details the number of reads, the number of reads that match the human genome and the number of unique reads. A message is also provided indicating that the remainder of the analysis results will be provided via email when the analysis is completed. Next, two background models are created to reflect the test sample submitted. The first background model reflects all regions of the genome that are both unique and sequenceable. The second model reflects the biased sequencing of input from that cell type. In the first step of the program, the duplicated and deleted regions of the genome are determined and background and ChIP tag counts are normalized to reflect a single copy. In the second step, a peak height threshold is determined based on background model 1 and a false discovery rate (0.0001 or 0.001 is recommended). The most significant peaks determined in the first pass are removed from the background model and this step is repeated for an accurate height cutoff. In the third step, peaks are determined significant over background model 2. Again, the most significant peaks determined in the first pass of this step are removed from the second background model, and this step is repeated, resulting in a final peaks list. After the second pass is complete, output is sent to the user via email. The files produced can be used as visualization tools (Figures 2 and 3) or for further analysis with additional online software (Figure 4). Shown on the top of the browser shots in each panel are the peaks called using Sole-Search and the peaks called if step 1, step 2 or step 3 of the program is removed (see also Supplementary Figure S9 for a larger view of each panel).
Step 2: Background estimation using the sequenceable tags (Background Model 1)
The first pass of peak calling is to determine an accurate, statistically significant height cutoff for peaks. This cutoff is often determined by means of a false discovery rate (fdr): the basic concept is to divide the number of false positive peaks determined with a specific height cutoff in a simulated background, by the number of peaks determined at that same height cutoff in the ChIP-seq sample. These backgrounds are meant to represent the ChIP tags randomly distributed among unique genomic regions (12,18,19). However, certain regions of the genome will have a higher abundance of sequenced reads because binding sites are present in that region, and thus there are many more tags in that location. Likewise, certain regions are sequenced more than the rest of the genome because they are misannotated as unique (such a pericentromeric region or other duplication events; Supplementary Figure S4). If the tag count represented in these densely covered regions was randomly distributed among the potential unique sequences, the background model produced will be denser than the actual background noise of the experiment. To account for this problem, the fdr can be calculated after eliminating reads estimated to be in the binding sites (18). Unique background models, however, do not take into account the fact that a region is not necessarily going to be sequenced even if it is unique (e.g. because certain regions are not easily fragmented and are less enriched in the sequencing library; Supplementary Figure S3). Also, individual libraries can show biased sequencing; reads are not randomly distributed among unique regions, but cluster possibly due to the amplification step. In these cases, distributing sequences randomly across an estimated unique genome will spread the tags too thin because the experimental method does not allow for random unbiased sequencing of the entire unique genome. In the program that we present, we account for these factors, and create a background model that more accurately reflects biology and experimental manipulation. The first Sole-Search background model uses combined reads from several different input libraries, representing not only unique reads, but sequenceable reads. In the first round of analysis, the same number of unique tags that were available from the ChIP sample is chosen randomly from the merged input dataset. A sliding window of 30 bp is used to determine how many 30 bp bins would represent all sequenced reads for a typical run. The tags within these bins are then scrambled and peak height cutoff is determined using a fdr of 0.0001 or a user chosen value. We define fdr as false peaks (i.e. the number of peaks determined in the background model at a given a height cutoff) over total peaks (i.e. all false and true-positive peaks determined within the ChIP data with the same height cutoff). To eliminate the problem of distributing reads found within peaks and false positive regions, these reads are removed and the process is then repeated: in the second round, the number of tags used to produce the background model is reduced by the number of tags found under the peaks determined from the first round. Also, the tags found in these regions are removed from the possible sequenceable reads. This results in a representative number of background reads/bins in the background regions of the genome. The peak height cutoff is then determined using this more accurate model. Peaks that pass this step are further analyzed in Model 2.
Consequences of elimination of Step 2
Sequence tags may cluster by chance into small peak-like structures. Step 2 eliminates such regions of the genome from being considered peaks by placing a height restriction on peaks. Therefore, if this step were removed, peaks called would contain an abundance of small false positive peaks. For example, ∼90 000 peaks are called for E2F4 in the absence of step 2, in comparison to ∼17 000 peaks called if Step 2 is left in place (Figure 1 and Supplementary Figure S9, Panel B). This peak set had an average peak height of 13.45 and a median height of 8, compared to using all three steps, which produced a peak set with an average peak height of 37 and a median of 27. While these additional ‘peaks’ are considered significant over a scarce background (for example, zero, one or two tags), as determined by step 3, they have occurred by chance and are normally removed by step 2.
Step 3: Peak elimination using specific inputs (Background Model 2)
Because some larger peaks with heights surpassing the significant height cutoff in Model 1 are not necessarily binding sites, but have accumulated tags because a region is more easy to sequence, either because it is a promoter region, which fragments easily to the most desirable size, or because the region is misannotated as unique, fdr cannot be used as the only means to determine peaks. Therefore, to distinguish true positives from false positives, sequenced ChIP data must be compared directly to sequenced input data. To obtain the most accurate results, the user can upload and analyze their data using their own input file(s) prepared from the same cells as their ChIP sample. However, our website also offers several options of input from different cell types and an option for ‘generic’ cell type (created using tags from multiple, different human cell lines) which can be used if an input library is not available for a particular cell type. However, we stress that having an input library from the same cell type is very important for final analyses, due to amplifications and deletions in the genomes of human cell lines. As indicated above, Sole-Search will eliminate false positives due to amplified regions. However, it will also identify deleted genomic regions, which is very useful information when characterizing the functional elements in a cell line (e.g. this can provide insight into why a ‘known’ binding site is not identified in a particular ChIP-seq experiment). ChIP-seq tags and input tags (the same number of tags as in the uploaded ChIP sample data set) are binned using a sliding window of adjacent 30 bp. Each ChIP bin that had passed height cutoff is compared to input using a one sample t-test. Using a user-defined significance cutoff, ChIP bins are retained if they are significant over background. As described above, tags represented in peaks should not be used to determine background, so the tags in the peaks determined in round 1 of Model 2 are removed and a second round of analysis is performed. Peaks that pass both height cutoff (from Model 1) and significance cutoff (from Model 2) are kept as potential peaks. Then, any potential peak whose length is greater than the user-specified length of chromatin fragment is kept to form the final peak list.
Consequences of elimination of Step 3
Step 3 essentially removes insignificant ‘peaks’ from consideration and narrows binding sites to their essential elements. Therefore, removal of this step will produce a peak set that includes small false positive peaks and extended true positive peaks. For example, removing the third step produced 20 352 E2F4 peaks (instead of ∼17 000); the extra peaks that were called were smaller than the average and median heights of the entire peak set and were often in shoulder regions of larger peaks (Figure 1 and Supplementary Figure S9, Panel C).
Sole-Search implementation
The Sole-Search tool set employs a client-server architecture. Users initially access the application through a web browser interface. A graphical client-side Java application for data entry and analysis request submission is automatically downloaded and executed on the users local computer via Java Web Start Technology (http://java.sun.com/developer/technicalArticles/WebServices/JWS_2/-JWS_White_Paper.pdf). This client application is downloaded again for future invocations only if an upgraded version of the application has been installed on the server. Executing the rich client interface on the users’ local computer system allows Sole-Search to transparently format and compress data files for upload to the server, relieving users of the need to perform these steps manually and reducing the bandwidth and time required for data set upload. The client-side application runs on any computer system (including those running Windows, OS X or Linux) configured with Java JRE version 1.4 or higher. Analysis requests are posted to the application server using the CGI protocol. In response, the application server invokes a set of Perl and shell scripts installed on the web server. The server installation depends on Linux, Perl and Apache web server technologies. Users are notified of analysis completion via email; results may be downloaded using URLs included in this e-mail message within two days of completion.
The Sole-Search system architecture is designed specially to handle the typically very large ChIP-seq data files (i.e. often several gigabytes in size). To avoid http time-outs, the client-side application transparently compresses very large files and splits them into multiple HTTP request. The system automatically retries failed data transfers. Because each analysis run requires a large amount of disk space, system memory and CPU resources, the Sole-Search server queues analysis requests. The maximum number of concurrent analysis jobs can be configured according to the computing resources available on the server. Ease of use is a primary concern since many users will lack programming skills. The client interface behaves like a locally installed application. Sole-Search assists the user in handling the very large data files by providing users with two modes of data transfer (based on a fast or slow network); in the high speed network mode, large files are only coarsely compressed before being uploaded, greatly speeding up request submission and dramatically reducing local resource consumption. A comprehensive help system is built into the client application and system administrators are provided information concerning the system logs status and error information related to each job submission, execution and completion (administrators are notified by email automatically when errors occur).
RESULTS AND DISCUSSION
Sole-Search: a web-accessed ChIP-seq data analysis program
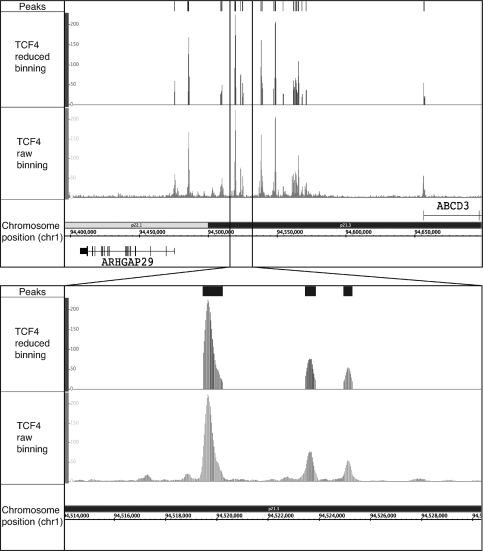
Below, we describe the Sole-Search software package, which is available in combination with additional analysis tools. Importantly, we have made this software available online so that experimentalists with minimal bioinformatics expertise can easily analyze their ChIP-seq data. A schematic representing the input files, the analysis steps used to call peaks and the output files is shown in Figure 1. The program is entered via a public web interface (http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi). This web interface allows the user to upload one or several lanes of ChIP-seq data (e.g. if more than one lane of sequencing data are available for a sample, the program can merge the reads into a single file for analysis) from the Illumina pipeline, as well as to upload one to several lanes of sequenced input. As detailed above, the user also has the option of choosing from a set of available input sequence files. The user indicates the size cut-off to be used for peak width (e.g. the smallest size of the chromatin), the number of permutations of the background tags (2 to 10 is allowed), the false discovery rate to be used for peak identification (0.0001 is recommended for factors that bind to >10 000 sites and 0.001 for factors that bind to <10 000 sites in the genome), and the α-value, which determines significance over background, to be used (0.001 is recommended). The program then combines and parses the ChIP-seq raw data and provides the user with input statistics, including the number of sequenced tags, the number of tags matching the reference genome, and the number of tags mapping to only one location of the reference genome (this set of ‘unique’ tags is used for all further analyses). Links to the remaining output files (Supplementary Figure S5) are provided via an email that is sent to the user after the analysis is completed. These output files include a job summary file (Summary_Job.txt) that includes run statistics and the parameters chosen by the user for this particular analysis (see below for more details) and visualization files of the data (rawbinning.sgr), developed using a sliding window of adjacent 30 bp (Figure 2). For easier manipulation of this large quantity of data, and so that multiple datasets can be visualized at once in an appropriate browser, these data are divided into files separated by chromosome. The program next produces several files related to peaks. One file is a summary (Summary_Job_signifpeaks.txt) that details the number of peaks, average peak height, median peak height, highest peak, lowest peak and average peak width (Table 1 for the information provided by the summary file for each dataset analyzed in this article). A visualization file (redbin.sgr) that includes tags under the called peaks, but removes all of the background is also provided (Figure 2); this file allows the user to visualize the shape of each peak. Because the redbin.sgr file is much smaller than the rawbinning.sgr files (due to the fact that most of the tags sequenced in any ChIP-seq experiment correspond to background and do not contribute to the peaks), it can include all peaks in the genome and does not need to be separated into individual chromosomes. A user can upload several different redbin.sgr files into a browser (such as the Affymetrix Integrated Genome Browser) at once, allowing easy visualization of the peaks of multiple factors without crashing the program due to large file sizes. For further analysis, two statistically significant peaks files are produced: one peaks file (Figure 2) is characterized by the number of sequenced tags per peak (signifpeaks.gff) and the other is characterized by effect size of each peak (effectsize.gff), which measures how significant the peak is over background. The effect size and significant peaks files list the same peaks; the only difference is the rank order of the peaks (strict tag count versus significance over background); smaller peaks in a region of extremely low background may be ranked higher in the effectsize file than in the signifpeaks file. Because many investigators are studying cancer cells and tissues, there is an issue with amplifications and deletions of large regions of the genome. Sole-Search identifies the amplified and deleted regions (and approximates copy number) and provides visualization files (smear.sgr) identifying these regions (Figure 3), as well as files listing the amplified (duplications.gff) and deleted (deletions.gff) regions of the genome of the cell line or tissue used.
Figure 2.
Visualization of ChIP-seq data using Sole-Search output files. ChIP-seq data for the merged TCF4 replicate dataset were analyzed using Sole-Search (Table 1). Shown for a region of chromosome 1 is: (Peaks) the visualization file (signifpeaks.gff) indicating the called peaks; (TCF4 reduced binning) the visualization file (redbin.sgr) of the tags that correspond only to regions called as peaks, and (TCF4 raw binning) the visualization file (rawbinning.sgr) of all binned and mapped tags. The inset shows the same files, but with an expanded view of a region of chromosome 1. The rawbinning.sgr files are provided for each individual chromosome, due to their large size (e.g. the size of the TCF4 chromosome 1 rawbinning.sgr file is 64.1 MB). However, the redbin.sgr file and the signifpeaks.gff file are much smaller (e.g. the size of the TCF4 redbin.sgr file, which shows TCF4 peaks for all chromosomes, is only 4.5 Mb) and are provided as single files for the entire genome, for ease in comparing different datasets.
Table 1.
Shown is the information provided by Sole-search after analysis of the indicated ChIP-seq datasets
| E2F4/K562 | E2F6/K562 | TCF4/HCT116 | YY1/K562 | YY1/Ntera2 | |
|---|---|---|---|---|---|
| No. of peaks | 17 673 | 26 043 | 21 102 | 2408 | 4443 |
| Average peak height | 37 | 48 | 60 | 44 | 51 |
| Median peak height | 27 | 32 | 43 | 33 | 33 |
| Highest peak | 224 | 229 | 229 | 210 | 229 |
| Lowest peak | 13 | 13 | 14 | 13 | 13 |
| Average peak width | 451 | 520 | 378 | 354 | 353 |
| No. of uniquely mapped reads | 12 917 986 | 10 383 166 | 14 559 371 | 6 926 438 | 6 311 210 |
Figure 3.
Identification of amplified and deleted regions of the genome. Shown for a region of chromosome 13 is the visualization file (smear.sgr) that allows easy detection of the amplified and deleted regions of the genome being analyzed, a file showing the regions called as duplicated (duplications.gff), and a file showing the deleted regions of the genome (deletions.gff).
Use of the Sole-Search ChIP-seq Tool set to analyze E2F4 ChIP-seq data from K562 cells
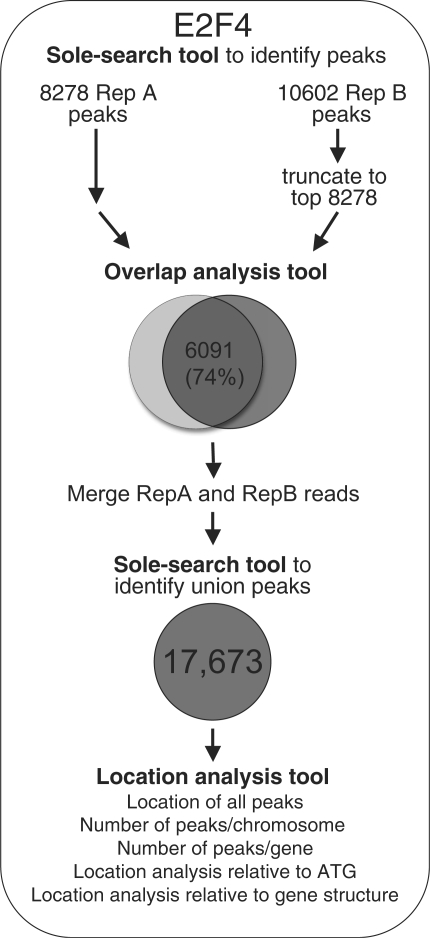
To demonstrate the utility of the Sole-Search ChIP-seq Tool set, we performed two ChIP assays for E2F4 in K562 cells. For the first replicate, 4.7 million unique reads were analyzed and for the second replicate, 8.8 million unique reads were analyzed (see Supplementary Table S1 for a summary of the number of unique reads used for all factors analyzed in this study). As described below, each replicate was independently analyzed, the experiments were compared using the Gff-Overlap Tool to determine replicate quality, the replicate samples were merged and reanalyzed using Sole-Search to produce a final peaks file, the final peaks file obtained using Sole-Search was compared to that obtained using two different peak calling programs, and the Location-Analysis Tool was used to characterize the E2F4 dataset (Figure 4).
Figure 4.
Step-wise analysis of ChIP-seq data. Shown are the steps taken to analyze the E2F4 ChIP-seq data. First, each replicate was analyzed separately (Supplementary Folders S1 and S2) using Sole-Search, then the replicate peak lists were sorted by peak height and truncated to the same length. Then, the two peak lists were compared using the Overlap analysis Tool (Supplementary Folder S7). The overlap was determined to be 74% and then Sole-Search was repeated, inputting both replicates using the option to merge the replicate files (Supplementary Folder S3). The peaks were then characterized using the Location-Analysis tool (Supplementary Folder S9 and Figure S6).
It is important to perform at least two independent ChIP experiments (for the same factor and same cell type, but with cells grown on separate days) and to prepare and sequence libraries from each of the samples. A comparison of the two datasets will then provide information as to the reproducibility of the ChIP-seq results for that particular factor and cell line combination. The two E2F4 ChIP samples were first analyzed independently using Sole-Search; the output files from the Sole-Search analysis of each of the E2F4 replicates can be found in Supplementary Data S1 and S2. To determine reproducibility of the datasets, we next used the Gff-Overlap Tool. The Gff-Overlap tool allows an automated comparison for replicate datasets using as input files the peaks lists that are obtained as output files from the Sole-Search program (signifpeaks.gff). We also note that any other peaks files, such as those from other ChIP-seq or ChIP-chip peak calling programs, can be used as input for the Gff-Overlap program, as long as they are in gff format. It is important to keep in mind that the number of peaks increases with the number of tags sequenced (until saturation has occurred) and that replicate datasets may identify different numbers of peaks. Therefore, for comparison of peaks files, we recommend truncating the longer list to the length of the shorter list. This is illustrated in Figure 4 for replicate datasets for E2F4. The peaks files provided by the Sole-Search program for each replicate (Supplementary Data S1 and S2) were sorted in descending tag height and then the longer replicate list was truncated to the length of the shorter list. The two peak files were then uploaded into the GFF-Overlap Tool for comparison. Output from this program, which is sent to the user via email, includes a summary file which indicates the total number of peaks and the number of overlapping peaks for each dataset, a file listing the overlapping peaks, a file listing the non-overlapping peaks, and a final file listing the union of both peak sets (see Supplementary Data S7 for the overlap analysis of the E2F4 replicates). These files are in gff format and can be loaded into a genome browser, similar to the peak files from which they are derived. If the peaks from the two biological replicates show a high degree of overlap, we recommend that the union set of all sequenced reads for a given factor and cell type be used for further analysis of the factor. This allows peaks to be called using the largest number of reads, providing a very robust set of binding sites. Therefore, at this point, the user can rerun Sole-Search, uploading all lanes for both replicates into the program to produce a final peaks file of the merged replicate dataset (see Supplementary Data S3 for the Sole-Search analysis of the merged E2F4 datasets). The final peak file from the merged replicates should be considered the list of peaks for a factor and is thus used for further analyses, as described below.
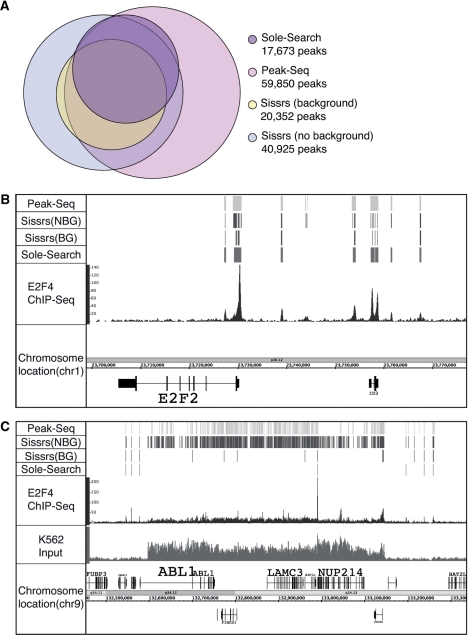
Several other ChIP-seq peak calling programs for site-specific DNA binding transcription factors have recently been published, such PeakSeq (18) and Sissrs (16). Therefore, we have compared the E2F4 peak file obtained using Sole-Search to the peak files obtained using the same merged E2F4 dataset analyzed with PeakSeq or Sissrs. Default parameters were used to call peaks with all three programs. The E2F4/K562 dataset constitutes 12 917 986 uniquely mapped reads and was produced by merging two independently derived E2F4 datasets. Sole-Search identified 17 673 peaks (Table 1), Peakseq identified 59 850 peaks (using the recommended fdr of 0.05) and Sissrs identified either 40 925 peaks (using the no background option) or 20 352 peaks (using the background option). Since running Sissrs with too many input tags exceeds memory, we only used three out of the five possible K562 input lanes for analysis with this program; all five input lanes were used as background for the other two programs. As shown in Figure 5A, the peaks identified by Sole-Search are contained within the larger peak sets identified by the other two programs: 80% of peaks called by Sole-Search are found in all three other peak sets; of the remaining 20%, 19% are found in two other peak sets and <1% of peaks identified by Sole-Search are only found in one other set. A direct comparison of the characteristics of the peaks identified by the three programs revealed that the average height of the Sole-Search, PeakSeq and Sissrs (with background) were comparable: 37, 38 and 34, respectively. Importantly, the median peak height of the PeakSeq set was much lower (15) than for Sole-Search (28) or Sissrs (25) indicating that the additional ∼40 000 peaks called by PeakSeq were very small. We suggest that the fdr recommended for use with PeakSeq is too lenient and that the peak height average resembles the averages of the other two programs because the large number of small peaks are balanced by very large false positive peaks (i.e. peaks at centromeric regions). Likewise, using Sissrs without a background identifies a large number of peaks, suggesting that using a background model is very important for the elimination of false positives. Another reason why Sissrs calls so many more peaks is shown in Figure 5B; a region which is called as one or two peaks by Sole-Search and PeakSeq may be called as many peaks by Sissrs (with or without background). It is important to account for this fact if using Sissrs peaks to find de novo motifs, as this may skew results. Also illustrated in Figure 5B are additional very small peaks that are called by PeakSeq when using the recommended fdr and Sissrs if a background model is not used. Finally, many of the ‘extra’ peaks called by PeakSeq and by Sissrs are in the amplified region of the K562 genome (Figure 5C). In the region shown, Sole-Search identifies one peak, Sissrs (with background model) identifies six peaks, and both PeakSeq and Sissrs (no background model) identify a very large number of peaks that are due to the over-represented tags in the amplified region.
Figure 5.
Comparison of peak calling by different programs. (A) Peaks were called using the 12 917 986 uniquely mapped tags from the E2F4 merged dataset using Sole-Search, PeakSeq and Sissrs (either with or without using a background; note that only 3 of the 5 lanes of input could be used with Sissrs because the program could not handle the larger number of reads but all 5 lanes were used with the other two programs). The Venn diagram shows the number of peaks called by each program and the relative overlap of the different datasets. (B) Shown for a region of chromosome 1 is the sgr visualization file for the K562 input sample, the E2F4 ChIP sample and the peaks called by each program. (C) Shown for a region of chromosome 9 is the sgr visualization file for the K562 input sample, the E2F4 ChIP sample, and the peaks called by each program; note the very large number of peaks called by PeakSeq and Sissrs in the amplified genomic region.
Analyzing ChIP-seq data is more complicated than analyzing ChIP-chip data and a major problem in the field is that many experimentalists lack the necessary bioinformatics skill sets. For example, when using promoter arrays, the ‘nearest’ gene is already known for every binding site and so a list of target genes is quite easy to obtain. In contrast, when using ChIP-seq, binding sites can be located at a large distance from a gene. Therefore, we have included in our software package, several tools to allow certain follow-up characterizations that most experimentalists would like to perform. For example, detailed characterizations of binding patterns are possible with the Location-Analysis Tool of the Sole-Search software package (see Supplementary Data S9 for the Location-Analysis output for E2F4). The user simply uploads the final peaks file (from the merged replicates if two high quality replicates are available) into the program. Again, we note that any peaks file, independent of origin, can be uploaded into this program, as long as it is in gff format and the coordinates are human hg18, human hg19 or mouse mm9 (the user specifies which genome build to use for the location analysis). The analysis is automatically performed and the output is sent to the user via email. The output includes: (i) an analysis of the number of hits per chromosome (chrom_count) that can be visualized graphically, (ii) a file (loc_analysis) listing the chromosomal location of each peak, the name and chromosomal location of the gene nearest to each binding site (the nearest gene can be located 5′ or 3′ of the binding site), the distance between the binding site and the start site of transcription of the nearest gene, and a classification as to whether the binding site is located close or distal upstream, close or distal downstream or within the target gene, (iii) an analysis of whether the factor binds in one or multiple places near a target gene (gene_count); to derive this information, the loc_analysis list is collapsed such that each gene is listed only once and the number of sites that match to each gene is tallied, (iv) a file (dist_analysis) that allows a graphical analysis of the location of all binding sites for a factor with respect to the start site of transcription of the nearest gene, (v) a file listing the number of binding sites located upstream, downstream or within a gene (pos_info) and (vi) a file listing the breakdown of the intragenic binding sites into different exons and introns (intron_information). Graphical representations of the chrom_count, gene_count, dist_analysis, pos_info, and intron_information files for the E2F4 dataset is shown in Supplementary Figure S6; the large loc_analysis excel tables for the E2F4 dataset can be found in Supplementary Data S9. As we expected, based on previous ChIP-chip analysis of 1% of the human genome (20,21), most E2F4 binding sites in K562 cells identified using Sole-Search fall within 1 kb upstream or downstream of the start site (Supplementary Figure S6). As a comparison, we also performed a location analysis of the peaks identified using PeakSeq and Sissrs (with and without background correction). As shown in Supplementary Figure S8, Sissrs (with background correction) identified a similar number of binding sites as did Sole-Search and the set of sites identified by Sissrs had an almost identical location pattern with respect to the start site of transcription as did the sites identified by Sole-Search (Supplementary Figure S8A and C). However, the larger number of sites identified by PeakSeq and Sissrs with no background correction had very different location profiles (Supplementary Figure S8B and D), suggesting that many of the extra peaks were not bona fide E2F4 binding sites.
Using the Sole-Search ChIP-seq Tool set to gain biological insight into transcriptional regulation
In addition to allowing a rapid, facile, user-friendly and statistically based identification of the binding sites for a particular factor, the Sole-Search ChIP-seq Tool set can be used to (i) compare binding patterns of two different factors, to determine if they tend to bind in similar regions (with respect to the start site of transcription), (ii) compare binding sites of two different members of a family of transcription factors, (iii) compare binding sites of a single factor in two different cell types, (iv) identify binding modules (enhanceosomes) and (v) assist in performing motif analyses.
(a) Comparison of binding patterns of different transcription factors
As noted above, most E2F4 binding sites are located in core promoter regions, very near to the start site of transcription. However, most transcription factors do not show this type of location analysis pattern (12,16). To illustrate this point, a complete analysis (using two replicates, as outlined in Figure 4) was performed for TCF4. A previous analysis of TCF4 binding using ChIP-chip identified ∼6800 sites and found that most of the sites were located outside of core promoter regions (22). However, because of possible differences between experimental platforms, we needed to identify the set of TCF4 sites using ChIP-seq. Therefore, two independent replicate TCF4 datasets were produced and analyzed using Sole-Search, the resultant individual peak files were compared using the Overlap-Tool, the individual datasets were then merged and re-analyzed using Sole-Search, and then the merged peak file (21 102 peaks) was analyzed using the Location-Analysis Tool; see Supplementary Data S4–S6 for the Sole-Search output files for each single ChIP-seq replicate of TCF4 and the merged dataset; Supplementary Data S8 for the Overlap-Analysis of the TCF4 replicates, Supplementary Data S10 for the output files from the Location-Analysis of TCF4 peaks, and Supplementary Figure S7 for the graphical representations of the chrom_count, gene_count, dist_analysis, pos_info, and intron_information files for the TCF4 dataset. Although both E2F4 and TCF4 bind to a similar number of places in the human genome (Table 1), their binding patterns are very different (compare Supplementary Figures S6C and S7C). In particular, regions corresponding to 10–100 kb upstream or downstream from the start site of genes (i.e. regions considered typical of where enhancers are located) are highly enriched in the TCF4 set of binding sites (see Figure 2 for an example of TCF4 binding in between two genes). Thus, E2F4 binding sites are promoter-specific, but TCF4 binding sites are found at both promoters and enhancers.
(b) Comparison of different members of a family of transcription factors
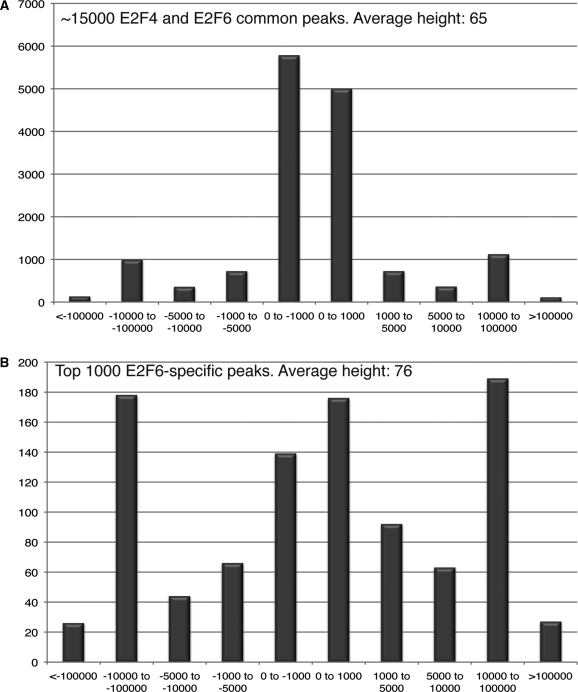
Most site-specific DNA-binding transcription factors are members of multi-gene families, with each member having a very similar DNA binding domain. A common question that is asked about the different family members is whether they bind to the same genomic locations (and therefore regulate the same set of target genes) or if they bind to different locations (and therefore regulate distinct sets of genes). The E2F family of transcription factors consists of eight genes, each having a highly conserved DNA binding domain (23). In particular, E2F4 and E2F6 have very similar DNA binding and hetero-dimerization domains (both proteins require dimerization with DP1 to bind to DNA in vitro). However, the C-terminal regions of E2F4 and E2F6 are very different, with E2F4 having a pocket protein binding domain [that mediates interaction with the retinoblastoma (Rb) protein family members] and a transactivation domain, both of which are absent in E2F6. Comparisons of E2F4 and E2F6 binding has been performed previously using ChIP-chip and promoter arrays (21). However, a genome-wide comparison of the binding patterns of these two E2F family members has not been performed. Therefore, we performed ChIP-seq using two biological replicates of K562 cells for both E2F4 and E2F6. Sole-Search and the Overlap-Analysis Tool, as illustrated in Figure 4, were used to analyze each dataset. Briefly, each replicate was sequenced and peaks were called using Sole-Search (Supplementary Data S1, S2 for E2F4 and S11, S12 for E2F6) then the Gff-Overlap Tool was used to confirm a high degree of overlap for the E2F4 replicates and for the E2F6 replicates. The merged E2F4 and merged E2F6 replicates were then re-analyzed using Sole-Search (Supplementary Data S3 for E2F4 and S13 for E2F6); the information from the Summary Text output file for each dataset can be found in Table 1. To determine if E2F4 and E2F6 bind to the same locations, the signifpeaks.gff files for the merged E2F4 and merged E2F6 datasets were then uploaded into the Gff-Overlap Tool, using 0 nt distance, such that the peaks must overlap by at least 1 nt (Supplementary Data S15). In this case, we did not truncate the lists to the same length because we are not comparing replicates but rather are comparing two different family members (and the number of uniquely mapped reads was similar for each dataset). When we compared the 17 611 E2F4 and 25 944 E2F6 peaks, we found that 14 700 of the E2F4 peaks were in the E2F6 dataset (83%), but because there are so many more E2F6 sites, only 54% of the E2F6 sites were in the E2F4 dataset. To determine if the characteristics of the E2F4 versus E2F6 overlapping versus non-overlapping sites were similar, these two sets of peaks were analyzed using the Location-Analysis Tool (Supplementary Data S16 and S17). As shown in Figure 6A, the ∼15 000 peaks in common for E2F4 and E2F6 are all localized to core promoter regions. In contrast, when the ∼12 000 peaks that were unique to E2F6 were analyzed, they were found to be enriched in promoters and enhancer regions (data not shown). One caveat to this analysis could be that if many of the E2F6-specific peaks were small, false positives, this could skew the results. Therefore, the ∼12 000 E2F6-specific peaks were ranked and the top 1000 were selected and analyzed for their location pattern. As shown in Figure 6B, these peaks are all very strong (having an average peak height higher than that of the common peaks). However, the binding pattern of these E2F6-specific peaks is quite different than the E2F4 and E2F6 overlapping peaks. Thus, although both E2F4 and E2F6 bind to the same ∼15 000 promoters in the genome, E2F6 also a set unique set of binding sites that differ in location relative to the start site of transcription from sites bound by both family members. The common and unique peaks files identified for the genome wide ChIP-seq for E2F4 and E2F6 are provided in Supplementary Data S15.
Figure 6.
Comparison of E2F4 and E2F6 binding sites. The signifpeaks.gff files for the merged E2F4 and merged E2F6 datasets were uploaded into the Gff-Overlap Tool, using 0 nt distance (Supplementary Folder S15). (A) The ∼15 000 binding sites that were identified (using the overlapping peaks file from the Overlap Analysis Tool) to be bound by both E2F4 and E2F6 in K562 cells were analyzed using the Location-Analysis Tool (Supplementary Folder S16). A graphical representation of the dist_analysis file is shown. (B) The top 1000 of the ∼12 000 E2F6-specific binding sites that were identified (using the non-overlapping peaks file from the Overlap Analysis Tool) were analyzed using the Location-Analysis Tool (Supplementary folder S17). A graphical representation of the dist_analysis file is shown.
(c) Comparison of binding patterns of one factor in different cell types
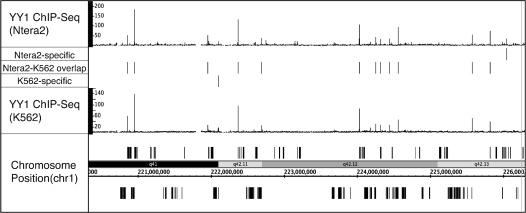
Many site-specific factors are expressed in many different tissues (24). However, very few studies have examined binding of a factor in more than one cell type. YY1 is a site-specific factor that is expressed in most cell types. To determine if YY1 binds to the same locations and regulates the same target genes in different cell types, we analyzed YY1 binding in K562 chronic myeloid leukemia cells and Ntera2 embryonal carcinoma cells. ChIP-seq data for YY1 in both cell types was analyzed using Sole-Search (Table 1); 4443 sites were identified in Ntera2 cells and 2408 sites were identified in K562 cells (Supplementary Data S18–19). The Ntera2 list was truncated and both sets of 2408 peaks were compared using the Gff-Overlap Tool. In general, the binding sites are very similar in the two cell types; an overlap of 74% was obtained, which is similar to the overlap obtained when two replicates in the same cell type are analyzed. Examples from the overlapping and non-overlapping peaks files (Supplementary Data S20), along with visualization of the Ntera2 and the K562 YY1 ChIP-seq data, are shown for a region of chromosome 1 in Figure 7.
Figure 7.
Comparison of YY1 binding sites in two different cell types. The YY1 binding sites (signifpeaks.gff) identified using Sole-Search for K562 cells (Supplementary Folder S18) and for Ntera2 cells (Supplementary Folder S19) were compared using the Overlap Analysis Tool (Supplementary Folder S20). Shown for a region of chromosome 1 is the sgr visualization file for the YY1 ChIP-seq data from K562 and from Ntera2. Also shown for this region are the peaks that are common to both cell types and the peaks that are specific for each cell type.
(d) Identification of binding modules (enhanceosomes)
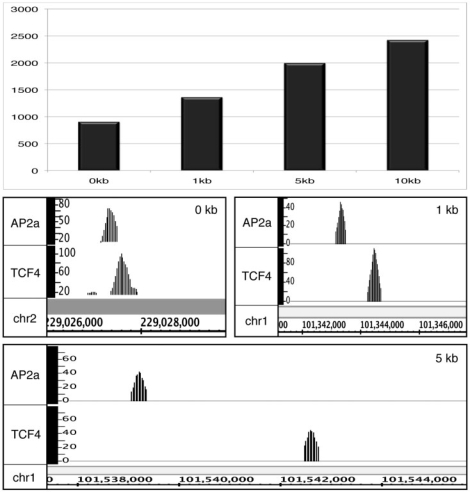
Although the GFF-Overlap Tool has an option to extend the region that you consider to represent overlapping peaks, for replicates of the same factor we recommend that the value be 0 (i.e. at least 1 nt of the peaks should overlap). However, changing the overlap distance can allow a user to determine co-localization of different factors in the genome. In this case, one would not expect the center of the peaks to be at the exact same location. Rather, one can use this tool to identify possible enhanceosomes, as defined as relatively small regions, located far from a core promoter, that are bound by different factors. To illustrate this use of the Sole-Search Tool set, we wanted to analyze two different factors that have approximately half of their binding sites far from start sites. As shown in Supplementary Figure S7, the TCF4 binding pattern fits this requirement. However, E2F4, E2F6 and YY1 all have the majority of their sites in core promoters. Therefore, we needed to identify another factor that binds far from start sites. We tested several factors and found that the AP2α binding pattern fits this requirement. Therefore, we performed two ChIP-seq replicates of AP2α, called peaks using Sole-Search, determined that the two sets showed a high degree of overlap, merged the two replicates, and called peaks again using Sole-Search, identifying 17 118 AP2α peaks. We then analyzed the AP2α signifpeaks file (17 118 peaks) and the TCF4 signifpeaks file (21 102 peaks) using the Location-Analysis Tool. The 6272 AP2α peaks and the 6682 TCF4 peaks that were identified as being ±10 to 100 kb from the start site of a gene (and thus located in a region that may correspond to an enhancer) were then compared using the Overlap Tool. The number of sites identified when spacing was varied between 0 kb (peaks must overlap) to 10 kb is shown in Figure 8. Also shown are examples of AP2α and TCF4 peaks, all of them far from start sites of genes, that overlap or are separated by less than 1 kb or less than 5 kb. The several thousand regions identified to contain both AP2α and TCF4 binding sites are good candidates for enhanceosomes. We anticipate that as more and more factors are analyzed using ChIP-seq, the GFF-Overlap Tool will be very useful for identifying additional enhanceosomes.
Figure 8.
Using the Sole-Search Tool set to identify enhanceosomes. A location analysis of the 17 118 AP2α peaks and 21 102 TCF4 peaks was performed using the Location-Analysis Tool. The 6272 AP2α and 6682 TCF4 enhancer sites (defined as ±10 to 100 kb from the start site of a gene) were then compared using the Overlap Tool. The number of sites identified when the spacing between TCF4 and AP2α sites was varied between 0 kb (peaks must overlap) to 10 kb is shown in the top panel. Also shown are examples of AP2α and TCF4 peaks, all of them far from start sites of genes, that overlap by 0, <1 kb or <5 kb.
(e) Motif analysis
Although some factors, such as members of the E2F family, appear to lack a requirement for a specific motif for binding in vivo (25), other factors appear to be recruited to a majority of their binding sites via a common motif. For example, each of the sets of binding sites for p63, STAT1 and REST (also known as NRSF) show a high enrichment for a specific motif (12,26,27). However, these previous studies did not examine whether the identified motif was similarly enriched in all categories of binding sites for a particular factor. Specifically, they did not determine if a particular factor was recruited to promoter regions versus enhancer regions using the same motif. To address this question, we have used our ChIP-seq data for TCF4. We began by developing a position weight matrix for the TCF4 motif using our ChIP-seq data and the de novo motif search program Meme; the position weight matrix that we derived was similar to the TCF4 consensus motif identified previously (22). We then determined that 46% of the top 500 binding sites identified by Sole-Search contained a good match to the TCF4 position weight matrix. We next used PeakSeq and Sissrs (with and without background correction) to identify TCF4 binding sites and then determined the percentage of those binding sites that contained a match to the TCF4 position weight matrix (Supplementary Table S2). We found that TCF4 motifs were found at about the same percentage in Sole-Search and PeakSeq but that Sissrs peak sets had a much lower percentage of sites that contained a match to the TCF4 consensus. It should be noted that the average peak width called by each of the programs is different. In particular, PeakSeq calls wider peaks and Sissrs calls narrower peaks than does Sole-Search. However, even after correction for the width of the peak, a greater percentage of top 500 binding sites identified by Solesearch or by PeakSeq contained the TCF4 consensus motif than did the top 500 binding sites identified by Sissrs. These results suggest that the peaks identified by Sissrs are likely to be too narrow for optimal motif analyses.
In our ChIP-seq experiments, we identified 21 102 TCF4 binding sites in the human colon cancer cell line HCT116. Using our Location-Analysis Tool, we determined that 6682 sites are located in enhancer regions (±10 to 100 kb from the start site) and 9341 sites are located in promoter sites (±2 kb from start site); we note that the location analysis for our large ChIP-seq dataset is similar to that reported for a smaller number of sites previously identified by ChIP-chip (22). We determined that the 6682 TCF4 enhancer sites have an average peak height of 66 tags and the 9341 TCF4 promoter sites have an average peak height of 54 tags. Thus, TCF4 appears to be recruited to these two subsets of sites with equal efficiency. To determine if the same motif is used for TCF4 recruitment in the two subsets of sites, we performed motif analysis of the top 500 TCF4 promoter binding sites and the top 500 TCF4 enhancer binding sites using the de novo motif search Meme (28). The top three motifs with a length of seven were selected; a length of seven was chosen because it is the length of a previously determined TCF4 motif (22). In the promoter subset, only the known TCF4 consensus motif was identified. However, in the enhancer subset, motifs for three different transcription factors were identified: the AP1 motif (consensus TGAGTCA): E-value = 1.1e-097; the TCF4 motif (consensus ATCAAAG): E-value = 7.9e-078; and the ETS1 motif (consensus CAGGAAG): E-value = 3.1e-032; see Figure 9A.
Figure 9.
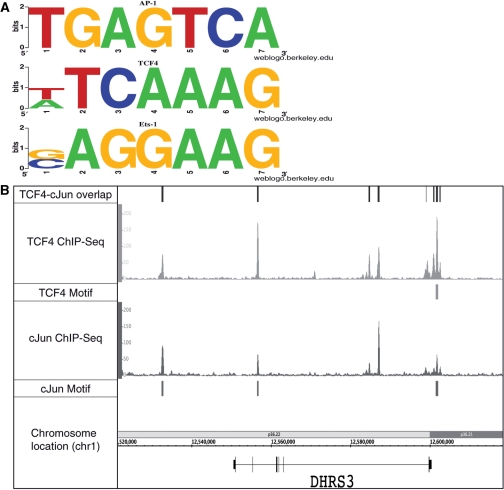
Motif analysis of TCF4 ChIP-seq data. The signifpeaks.gff file for the merged TCF4 dataset was analyzed using the Location Analysis Tool (Supplementary Folder S10). Then, the sites corresponding to promoters (±2 kb from a transcription start site) or enhancers (±10 to 100 kb from a start site) were selected. Each set was then analyzed using Meme for de novo motif identification. As shown in (A), the three motifs identified in the enhancer binding sites correspond to known motifs for AP-1 (TGAGTCA), TCF4 (T/ATCAAAG) and ETS-1 (G/CAGGAAG). (B) The similar binding patterns of TCF4 (HCT116 cells) and JUN (K562 cells) across a region of chromosome 1. Also shown for that region of chromosome 1 are the overlapping sites identified when the Gff-Overlap Tool was used to compare the TCF4 and JUN peak files and the TCF4 and JUN motifs identified in the TCF4 peaks.
Our results suggested that different motifs may be important for recruitment of TCF4 to enhancers versus promoter regions. However, it was possible that the AP1 and ETS1 motifs are present in the promoter regions bound by TCF4, but were just not identified using Meme. Therefore, we more directly examined the prevalence of TCF4, AP1, and ETS1 motifs in various subsets of TCF4 binding sites. Using the weight matrix for the TCF4, AP1 and ETS1 motifs (as determined by Meme in the first analysis), four different sets of binding sites were analyzed; these included all 9341 TCF4 ChIP-seq promoter binding sites, the top 500 ChIP-seq TCF4 promoter binding sites, all 6682 TCF4 ChIP-seq enhancer binding sites, and the top 500 TCF4 ChIP-seq enhancer binding sites. Potential motifs in the four different peak sets were scored using a log-odds score (and a strict threshold of 8) that takes into account the background nucleotide frequencies of the sequences being scanned. We found that all three motifs were enriched in the TCF4 enhancer binding sites, when compared to a set of random sequences (Table 2). For example, the set of 500 highest-ranking TCF4 enhancer sites showed a very strong enrichment of the identified motifs (45% contained a TCF4 motif, 38% contained an AP-1 motif, and 34% contained an ETS1 motif, as compared to 500 random regions of which only 3%, 4% and 15% contained a TCF4, AP-1 or ETS1 motif, respectively). Interestingly, the TCF4, AP-1 and ETS1 motifs were all found at about the same frequency in the TCF4 enhancer regions. To determine if the AP-1 and ETS1 motifs were enriched in a set of binding sites of another transcription factor, we also analyzed the promoter and enhancer binding sites from AP2α ChIP-seq data (Table 2). We found that the TCF4 motif was not significantly enriched in the AP2α promoter or enhancer peaks, that the AP-1 motif was somewhat enriched in the AP2α enhancer, but not promoter, peak set, and that the ETS1 motif was somewhat enriched in the AP2α promoter, but not the enhancer, peak set.Our motif analysis results suggest that TCF4, AP1 and ETS family members may bind to many of the same genomic locations. To test our bioinformatics predictions, additional ChIP-seq data was required. Fortunately, ChIP-seq data for JUN (one of the heterodimeric components of AP1) in K562 cells was available from the Snyder laboratory’s contributions to the ENCODE consortium (http://genome.ucsc.edu/ENCODE/). Although this is a different cell type than was used for the TCF4 ChIP-seq experiments, it was possible that many of the JUN targets would be the same in K562 and HCT116 cells. Therefore, we downloaded the JUN ChIP-seq data from the UCSC browser, called peaks using Sole-Search, and then used the Gff-Overlap Tool to determine overlaps between the TCF4 and JUN binding sites. We found ∼7400 locations that were bound by both proteins. A comparison of TCF4 and JUN binding across the DHRS3 gene on chromosome 1 is shown in Figure 9B. Clearly, the binding patterns of these two proteins are very similar, even though different cell lines were used for the ChIP-seq experiments. Interestingly, TCF4 has previously been shown to physically interact with JUN (29). It has been previously suggested that the JUN and TCF4 interaction is a molecular mechanism that integrates the activation of the TCF/ CTNNB1 (β-catenin) pathway by the JNK pathway. Using genome-wide ChIP-seq of TCF4 and motif analysis of the TCF4-bound enhancer regions, our studies suggest that interaction with JUN may be a major mechanism for recruitment of TCF4 to the genome. Also, JUN has been shown to physically interact with ETS family members (30). Future ChIP-seq experiments are required to determine if ETS binding is coincident with TCF4 and JUN throughout the genome.
Table 2.
| No. of Peaks | TCF4 motifs (%) | AP-1 motifs (%) | Ets motifs (%) | |
|---|---|---|---|---|
| TCF4 binding sites | ||||
| Promoters | 9341 | 14 | 4 | 23 |
| top 500 | 45 | 10 | 48 | |
| Enhancers | 6682 | 23 | 24 | 19 |
| top 500 | 45 | 38 | 34 | |
| AP2α binding sites | ||||
| Promoters | 4548 | 6 | 6 | 19 |
| top 500 | 11 | 12 | 35 | |
| Enhancers | 4850 | 5 | 18 | 11 |
| top 500 | 8 | 31 | 20 | |
| Random regions | 9341 | 4 | 4 | 17 |
| 500 | 3 | 4 | 15 |
CONCLUSIONS
As described above, we have developed statistically based, integrated analysis software for ChIP-seq data that uses reads from the Illumina pipeline to create visualization files, calls peaks (taking into account background due to input characteristics), provides different types of peak files, and automatically provides information concerning critical characteristics of the binding patterns. We demonstrate the utility of this analysis approach by experimentally collecting and analyzing 10 different ChIP-seq datasets (two replicates each of E2F4, E2F6, TCF4, YY1 and AP2α). We use the Sole-Search Tool set to identify and compare binding patterns of different E2F family members and binding patterns of YY1 in different cell types, to identify potential enhanceosomes, and to provide insight into the mechanism by which TCF4 regulates transcription. All analysis files and tag files for each dataset are provided as supplementary data or are publicly available on the UCSC browser as a resource for the scientific community and so that they can be used by investigators who wish to familiarize themselves with the various tools of the Sole-Search program prior to analyzing their own ChIP-seq data.
We note that Sole-Search has both similarities and differences in comparison to several other recently described ChIP-seq peak-calling programs. The earliest ChIP-seq peak calling programs did not take into account peaks that also occur in input sequences (12,15) and thus peak lists derived using these early methods include many false positives. However, similar to Sole-Search, more recently developed programs, such as PeakSeq, Sissrs, MACS, CisGenome and GLITR (16,18,31–33), allow the user to take into account sequencing biases in the input samples; a comparison of the ways in which several different programs use input data to identify bona fide binding sites can be found in Tuteja et al. (33). Of these published programs, Sole-Search is the only program that specifically controls for amplified regions of the genome and automatically provides files listing the amplified and deleted regions of the genome in the cell line used for the ChIP-seq study, making Sole-Search especially well-suited for studying transcription factor binding sites and chromatin profiles of cancer genomes. We showed that programs that do not take into account the amplified regions call far too many peaks in certain regions of the genome. We compared binding patterns of E2F4 peaks and motif analyses of TCF4 peaks using Solesearch, PeakSeq and Sissrs. We showed that the smaller peak sets called by Sole-Search and Sissrs (with background correction) provide more accurate E2F4 location analyses than do the larger sets of peaks called by PeakSeq and Sissrs (without a background correction) and that the TCF4 peaks called by Sole-Search and PeakSeq contain more consensus TCF4 motifs than the peaks called by Sissrs. We note that unlike many of the previous programs, both Sole-Search and CisGenome (32) provide an integrated set of downstream analysis programs. Finally, we emphasize that one unique aspect of Sole-Search is that it is web-based (http://chipseq.genomecenter.ucdavis.edu/cgi-bin/chipseq.cgi) so that experimentalists with minimal bioinformatics expertise can quickly analyze their ChIP-seq data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (CA45250, 1U54HG004558, GM007377); the National Science Foundation (IIS- 0612326). Funding for open access charge: 1U54HG004558.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Farnham, Ludäscher, and Korf labs for helpful discussions.
REFERENCES
- 1.Boyd KE, Farnham PJ. Myc versus USF: Discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 3.Squazzo SL, Komashko VM, O’Geen H, Krig S, Jin VX, Jang S-W, Green R, Margueron R, Reinberg D, Farnham PJ. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberley MJ, Inman D, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J. Biol. Chem. 2003;278:42466–42476. doi: 10.1074/jbc.M307733200. [DOI] [PubMed] [Google Scholar]
- 5.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 lysine 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinmann AS, Yan PS, Oberley MJ, Huang THM, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sties during S phase. Oncogene. 2003;22:1445–1460. doi: 10.1038/sj.onc.1206264. [DOI] [PubMed] [Google Scholar]
- 8.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 10.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'G;een H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ. Genome-Wide Analysis of KAP1 Binding Suggests Autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:1–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 13.Wederell ED, Bilenky M, Cullum R, Thiessen N, Daqpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 16.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrançois P, Struhl K, Gerstein M, Snyder M. Proc. Natl Acad. Sci. USA. 2009;106:14926–14931. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZD, Rozowsky J, Snyder M, Chang J, Gerstein M. Modeling ChIP sequencing in silico with applications. PLoS Comput. Biol. 2008;4:e1000158. doi: 10.1371/journal.pcbi.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bieda M, Xu X, Singer M, Green R, Farnham PJ. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation, and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 24.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 25.Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus vs. non-consensus binding sites. Genome Res. 2008;18:1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 29.Nateri A, Spencer-Dene B, Behrens A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature. 2005;437:281–285. doi: 10.1038/nature03914. [DOI] [PubMed] [Google Scholar]
- 30.Bassuk AG, Leiden JM. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H, Jian H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing CHIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp536. [June 24, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.