Abstract
6-Phenylpyrrolocytidine (PhpC), a structurally conservative and highly fluorescent cytidine analog, was incorporated into oligoribonucleotides. The PhpC-containing RNA formed native-like duplex structures with complementary DNA or RNA. The PhpC-modification was found to act as a sensitive reporter group being non-disruptive to structure and the enzymatic activity of RNase H. A RNA/DNA hybrid possessing a single PhpC insert was an excellent substrate for HIV-1 RT Ribonuclease H and rapidly reported cleavage of the RNA strand with a 14-fold increase in fluorescence intensity. The PhpC-based assay for RNase H was superior to the traditional molecular beacon approach in terms of responsiveness, rapidity and ease (single label versus dual). Furthermore, the PhpC-based assay is amenable to high-throughput microplate assay format and may form the basis for a new screen for inhibitors of HIV-RT RNase H.
INTRODUCTION
The continuing emergence of drug resistant variants of the human immunodeficiency virus (HIV) calls for new tools to combat this epidemic (1). Current clinical treatment of HIV targets essential enzymatic activities associated with the reverse transcription of the viral genome and the protease activity required for maturation of protein components of the virus. The most effective control of the disease has been achieved with combination therapy targeting these pathways simultaneously. The inclusion of additional targets in a combination drug regimen is highly desirable for potent and long-term disease management.
The ribonuclease H (RNase H) activity associated with HIV-reverse transcriptase (HIV-RT) degrades the viral RNA genome in RNA/DNA hybrids (2), and has been identified as a potential target for antiretroviral therapy as it is required for virus infectivity (3); yet there are no antiRNase H agents in clinical use. Few inhibitors of HIV-1 RNase H were identified until the transition of testing methods from gel-based techniques to fluorescent assays amenable to high-throughput screening (HTS) (4–8). The most widely used assay was developed by Parniak and co-workers (6) and utilizes a two label, molecular beacon strategy (9) in which the RNA strand is labeled with a 3′-terminal fluorophore (fluorescein, F) and a DNA strand with a quencher (dabcyl, Q) at the 5′-terminus (Scheme 1).
Scheme 1.
Representation of fluorescent RNase H assay using a dual label system employing a fluorophore and a quencher.
Fluorescent studies involving nucleic acids most often utilize luminescent tags such as fluorescein or rhodamine sometimes in combination with a quencher such as 4-(dimethylaminoazo)benzene (dabcyl). However, these probes can be perturbing to the processes under investigation due to the steric bulk or non-polar groups they introduce to DNA and RNA. Through our own studies on inhibitors of HIV-1 RNase H, we have observed that the 5′-dabcyl quencher substantially reduces the catalytic efficiency of RNase H for its RNA/DNA substrate (SI).
The use of intrinsically fluorescent nucleotide base analogs offers a more conservative approach to fluorescence labeling of nucleic acids. Using phosphoramidite solid-phase synthesis (10), a fluorescent nucleotide can be incorporated at any position on an oligomer without the use of linkers or postsynthetic modifications. In addition, the fluorescence intensity of many base modified nucleotides are responsive to changes in their microenvironment, making them reporters for nucleic acid structure and dynamics (11). Despite these advantages, few fluorescent nucleobase analogs have found widespread use as a tool for molecular biology, 2-aminopurine (12) and the pteridine (13) base analogs being historically the most important. Recent work from Tor and others have elegantly demonstrated the value and utility in the design and discovery of new fluorescent nucleobases (14–17). Despite these and other recent advances, there remains a paucity of intrinsically fluorescent cytidine analogs that demonstrate responsiveness to their microenvironment and state of hybridization (18) thus motivating this work.
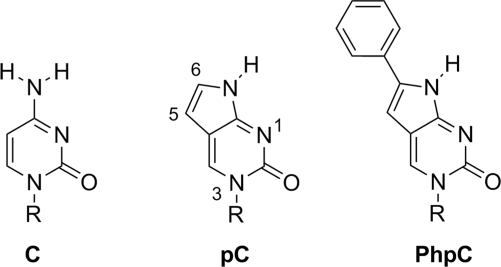
The success of a fluorescent nucleobase analog as a useful reporter resides in its ability to form proper Watson–Crick base pairs, thermostability in double stranded nucleic acids, changes in fluorescence with different hybridization states, recognition of nucleic acid processing/binding enzymes, and its fluorescence intensity. The cytidine analogs pyrrolocytidine (pC, Figure 1) with 6-methyl substitution (MepC) (19–21) satisfies the above criteria as a fluorescent reporter, though its fluorescence intensity, as manifested by a modest quantum yield (Φ), lags behind competing chemistries ultimately making it a less sensitive probe. One of our laboratories has shown that the low quantum yield of MepC can be remedied by substituting the C-6 position with an aromatic group without any penalties on sensitivity or base pairing fidelity (22–24).
Figure 1.
The structure of cytidine (C) and unsubstituted pC, with numbering, compared to PhpC. R = ribose or 2′-deoxyribose.
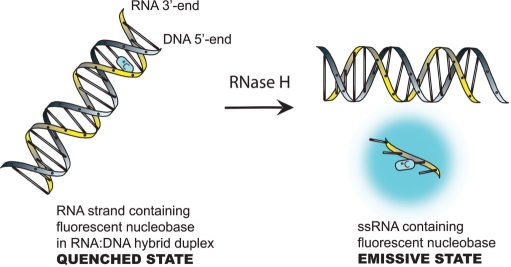
We have now synthesized the ribonucleoside of 6-phenylpyrrolocytidine (PhpC, Figure 1) and found that, like its DNA and PNA homologs, it ranks among the brightest C-analog luminiophores. The fluorescence of PhpC is sensitive to its microenvironment, and displays a λmax excitation (360 nm) and λmax emission (466 nm) significantly red-shifted from the absorption of other biomolecules. The combination of these properties makes PhpC unique among fluorescent nucleobase analogs and has enabled its use as a fluorescent reporter group in an RNase H assay. Maintaining the same RNA sequence as the traditional FQ-based assay (6) (RNA-1, Table 1), we replaced a single rC nucleoside with PhpC close to the 3′-terminus of the RNA/DNA hybrid, PhpC-1, where it exhibits low fluorescence (quenched state) as depicted in Scheme 2. Treatment with HIV-1 RT RNase H generates an RNA tetranucleotide (25) bearing PhpC, and quickly dissociates from its DNA complement with a concomitant dramatic increase in fluorescence emission.
Table 1.
Oligonucleotides used in RNase H assays and Tm data
| Name | Sequence (5′–3′)a |
Tm (°C)b |
|
|---|---|---|---|
| DNA-1 | RNA-2 | ||
| RNA-1 | GAU CUG AGC CUG GGA GCU | 65.1 | 78.5 |
| PhpC-1 | GAU CUG AGC CUG GGA G*CU | 64.7 | 78.8 |
| PhpC-2 | GAU CUG AGC *CUG GGA GCU | 67.5 | 80.8 |
aPhpC indicated by *C.
bTm values represent the average of at least 3 independent experiments within 1°C.
Scheme 2.
Reporting via a single fluorescent nucleotide analogs. *C = 6-phenylpyrrolocytidine (PhpC).
MATERIALS AND METHODS
5′-O-(4,4′-dimethoxytrityl)-5-iodouridine (2)
Uridine (2.44 g, 10 mmol) was added to a mixture of iodine (2.54 g, 10 mmol) and silver sulfate (3.12 mg, 10 mmol) in methanol (200 ml) at room temperature (r.t.). The mixture was stirred for 8 min, after which time the yellow solid was removed by filtration and the filtrate was evaporated under reduced pressure. The residue was co-evaporated with 2 : 1 MeOH/H2O (50 ml) to remove iodine (which sublimed away) and crystallized from water to give pure 5-iodouridine as a white solid (4.44 g, 60%). 5-Iodouridine (2.22 g, 6 mmol) was dissolved in pyridine (30 ml) and 4,4′-dimethoxytritylchloride (2.64 g, 7.8 mmol) was added in four portions during a 4 h period. After an additional 2 h of stirring at r.t. the reaction was quenched by adding 1 ml of 5% NaHCO3 for 15 min. The solution was concentrated under reduced pressure, redissolved in 50 ml of dichloromethane (DCM) and washed with a solution of 5% NaHCO3 (30 ml). The product was further extracted with two 25 ml portions of DCM, and the combined organic layer was dried over Mg2SO4 and evaporated to leave the crude product. The residue was then purified by silica gel column chromatography using an elution gradient with DCM/MeOH/TEA (98 : 1 : 1 to 96 : 3 : 1). Fractions containing 2 were evaporated to give a white foam (3.72 g, 55%). 1H NMR (400 MHz, CDCl3): δ = 8.14 (s, 1 H), 7.21–7.42 (m, 11 H), 6.84 (m, 4 H), 6.31 (dd, 1J = 6.8 Hz, 2J = 5.9 Hz, 1 H), 4.55 (m, 1 H), 4.07 (m, 1 H), 3.79 (s, 6 H), 3.78 (m, 1 H), 3.36–3.44 (m, 2 H), 2.46–2.64 (m, 1 H), 2.30–2.33 (m, 1 H) p.p.m. ESI-TOF (m/z) 695.4 (M + Na).
5′-O-(4,4′-dimethoxytrityl)-5-(phenylethynyl)uridine (3)
In a 50 ml Schlenk flask, 5′-O-(4,4′-dimethoxytrityl)-5-iodouridine (0.67 g, 1 mmol) was mixed with dry THF (15 ml), phenylacetylene (6 mmol), Et3N (4 mmol) and was then deoxygenated with N2 for 5 min. Subsequently, Pd(PPh3)4 (0.1 mmol) and CuI (0.2 mmol) was added and the mixture was stirred in the dark for 36 h at r.t. The solution was then filtered, concentrated to dryness, redissolved in CH2Cl2 (50 ml) and washed sequentially with 5% NaHCO3 (30 ml), 5% EDTA (30 ml) and 5% NaHCO3 (30 ml) and dried over Na2SO4. The solvent was evaporated and the resulting residue was purified by silica gel column chromatography using a gradient of hexanes/acetone/TEA (78 : 20 : 2 to 40 : 58 : 2) to provide product 3 as a pure compound in 60% yield (0.39 g). 1H NMR (400 MHz, CDCl3) δ = 8.15 (s, 1H, 6-H); 7.05–7.42 (m, 14H, ArH), 6.74–6.78 (m, 4H, ArH), 5.80 (d, 3J = 4.0 Hz, 1H, H-1′), 4.44, 4.34, 4.30 (3m, 3H each, H-2′, H-3′, H-4′), 3.66 and 3.65 (s, 6H, 2 OMe), 3.44(d, 1H, 2J = 12.0 Hz, 3J = 4.0 Hz, CHAHB-5′), 3.29 (d, 1H, 2J = 12.0 Hz, CHAHB-5′). 13C NMR(100 MHz, CDCl3) δ = 164.89, 158.71, 152.42, 144.68, 142.24, 135.92, 135.74, 131.84, 130.24, 130.12, 128.21, 128.09, 127.12, 122,99, 113.52, 100.68, 93.56, 90.62, 87.09, 84.99, 81.27, 76.12, 71.28, 71.28, 63.36, 55.33. HRMS (EI) m/z calcd[Na]: 669.2913; found: 669.2924.
5′-O-(4,4′-dimethoxytrityl)-6-phenylfuranouridine (4)
Compound 3 (0.645 g, 1 mmol) was dissolved in deoxygenated acetone (10 ml), then AgNO3 (0.1 mmol) was added. The reaction mixture was stirred at r.t. in the dark for 24 h and monitored by TLC (5–15% MeOH/DCM) until reaction completion. After filtration and concentration of the solution in vacuum, the solid residue was purified by silica gel column chromatography using an elution gradient of hexanes/EtOAc/TEA (50 : 50 : 2 to 15 : 85 : 2) to provide 4 (0.450 g, 70%). 1H NMR (400 MHz, CDCl3) δ 8.90 (s, 1H, 4-H); 7.24–7.64 (m, 14H, ArH), 6.81–6.84 (m, 4H, ArH), 6.50 (d, 1H, 3J = 2.0 Hz, H-1′), 5.77 (s, 1H, 5-H), 5.56 (m, 1H, H-2′), 4.48 (m, 1H, H-3′), 34.39 (m, H, H-4′), 3.72, 3.74 (2s, 6H, 2 OMe), 3.55 (m, 2H, H-5″). 13C NMR (100 MHz, CDCl3): δ 171.38 (C-7a); 158.60(C-2), 158.55(C-6), 155.04 (Carom), 143.91 (Carom), 137.35 (C-4) 135.61, 135.18 (2 Carom), 130.16, 129.99 (2 CHarom), 129.35(Carom), 128.70, 128.34, 128.20, 128.08 (4 CHarom), 126.98 (Carom), 124.57, 113.32 (2 CHarom), 108.40 (C-5), 98.10 (C-4a), 93.23 (C-1′), 87.00 (C–Ph(Ph–OCH3)2) 83.77 (C-4′), 75.67 (C-2′), 68.72 (C-3′), 61.14 (−CH2O), 60,29 (C-5′), 55.45 (2CH3OAr). ESI-TOF (m/z) 669.2 (M + Na).
5′-O-(4,4′-dimethoxytrityl)-6-PhpC (5)
A pressure bottle with compound 4 (0.64 g, 1.0 mmol), concentrated NH3OH (4 ml) and MeOH (6 ml) was stirred at 55°C for 24 h. Progress of the reaction was followed by TLC (Rf 0.38 in 10 : 1 DCM/MeOH). After completion of the reaction (24 h), the yellow solid was filtered and then dissolved in CH2Cl2 and dried over Na2SO4 and the solvent was removed and the residue was further dried under vacuum overnight (0.57 g, 60%). 1H NMR (400 MHz, CDCl3) δ = 11.57 (s, 1H), 8.83 (s, 1H), 7.57 (d, 2H), 7.37 (d, J = 7.5, 3H), 7.33–7.06 (m, 13H), 7.03 (t, J = 7.55, 1H), 6.75 (dd, J = 8.8, 6.9, 4H), 6.13 (s, 1H), 5.52 (s, 1H), 4.48 (s, 1H), 4.46 (d, J = 5.8, 1H), 4.32 (d, J = 5.8, 1H), 3.69, 3.58 (m, 2H), 3.63 (d, J = 10.2, 6H). 13C NMR (101 MHz, CDCl3) δ = 171.40, 159.27, 158.88, 158.82, 155.38, 144.40, 140.89, 136.54, 136.20, 135.71, 130.48, 130.30, 130.26, 128.90, 128.62, 128.38, 127.25, 125.50, 113.65, 113.43, 111.17, 97.22, 93.44, 87.22, 83.90, 76.23, 69.36, 61.73, 60.64, 55.37, 55.34. HRMS (EI) m/z calcd for C38H36N3O7: 645.2475; found [M + H]: 646.2585.
5′-O-(4,4′-Dimethoxytrityl)-2′-O-tert-butyldimethylsilyl-6-PhpC (6)
To a 25 ml round-bottom flask containing 315 mg (0.48 mmol) of compound 5 dissolved in 2 ml of dry DMF, TBDMS-Cl (88 mg, 0.58 mmol) and imidazole (82 mg, 1.2 mmol) was added and stirred overnight. The formation of the 2′-O-silyl and 3′-O-silyl products were monitored by TLC (Rf values of 0.56 and 0.13, respectively in 1 : 3 EtOAc/DCM) until the starting material was consumed. The reaction was worked-up in 5% NaHCO3, filtered over MgSO4 and evaporated to dryness. The 2′-O-silyl regioisomer was obtained by silica gel column chromatography using a gradient of hexanes-ethyl acetate from 1 : 3 to 1 : 1 keeping 1% TEA in the solvents. The remaining crude 2′- and 3′-regioisomer were mixed for 2 h in 5 ml of pyridine and three drops of water to form an equal mix of isomers. From this mix, the 2′-regioisomer was again purified by silica gel and both purified fractions were evaporated over high vacuum to give 213 mg of a yellow powder (58% yield). 1H NMR (500 MHz, DMSO) δ = 11.76 (s, 1H), 8.75 (s, 1H), 7.61 (d, J = 7.6, 2H), 7.47–7.10 (m, 16H), 6.92 (d, J = 4.1, 4H), 5.78 (s, 1H), 5.41 (s, 1H), 5.20 (d, J = 6.1, 1H), 4.37 (s, 1H), 4.15 (s, 1H), 4.11 (d, J = 8.3, 1H), 3.70 (d, J = 6.6, 7H), 3.45 (dd, J = 9.9, 58.4, 2H), 0.89 (s, 9H), 0.15 (s, 3H), 0.10 (s, 3H) p.p.m. 13C NMR (126 MHz, DMSO) δ = 160.38, 158.89, 158.84, 154.53, 144.68, 139.67, 136.79, 136.67, 136.35, 135.72, 130.82, 130.70, 130.54, 130.31, 129.70, 129.53, 128.74, 128.58, 127.53, 125.43, 125.31, 114.10, 114.01, 109.63, 97.04, 92.40, 92.28, 86.88, 82.06, 81.95, 77.58, 77.46, 68.37, 61.76, 55.76, 55.62, 26.45, 26.41, 18.66, −4.12, −4.20 p.p.m. ESI-TOF (m/z) 782.4 (M + Na).
5′-O-(4,4′-Dimethoxytrityl)-2′-O-tert-butyldimethylsilyl-3′-O-(2-cyanoethyldiisopropyl-phosphoramidite)-6-PhpC (7)
Compound 6 (213 mg, 0.28 mmol) was sublimed in 3 ml of distilled benzene over dry ice on high vaccum overnight, purged with argon, and dissolved in freshly distilled THF. Dry diisopropylethylamine (0.2 ml, 1.12 mmol) and diisopropylamino-(2-cyanoethyl)-phosphoramidic chloride (63 µl, 0.28 mmol) were added and the reaction was stirred for 2 h and monitored by TLC (5% methanol in DCM). At the end of this time, the reaction was washed with 5% NaHCO3, dried over sodium sulfate and loaded on a silica gel column neutralized in 2.5% triethylamine in hexane. The purified phosphoramidite diastereomers were eluted in 6 : 4 haxane-ethyl acetate, rotary evaporated and sublimed in 3 ml of distilled benzene overnight to yield a yellow foam (240 mg, 0.25 mmol). 31P NMR (200 MHz, ACN) δ = 151.3 (s, 1P), 149.5 (s, 1P) p.p.m. ESI-TOF (m/z) 982 (M + Na).
Oligonucleotide synthesis
Solid-phase synthesis of oligonucleotides was carried out on an Applied Biosystems 3400 DNA synthesizer using standard protocols (26). Coupling yields for PhpC were comparable to standard 2′-O-TBDMS ribonucleonucleoside phosphoramidites based on analytical denaturing PAGE (SI). Cleavage from the solid support was carried out in a 3 : 1 mixture of NH4OH:EtOH at r.t for 48 h, and removal of the 2′-O-silyl protecting groups was achieved by treatment with distilled triethylammonium trihydrofluoride for 48 h. The crude, deprotected oligonucleotides were precipitated in ice-cold butanol and 3M sodium acetate and quantitated by their UV absorbance at 260 nm on a Cary-300 UV-VIS spectrophotometer (Varian Inc.). Crude products were then analyzed and purified by denaturing PAGE (7 M urea) and visualized by UV shadowing using 254 nm light for non-fluorescent oligonucleotides, and 365 nm light for fluorescent oligonucleotides. Full-length products were excised from the gels using a sterile surgical blade, and eluted in sterile water. The eluted products were desalted by size-exclusion chromatography on G-25 Sephadex (GE Healthcare) and quantitated.
Thermal denaturation and circular dichroism studies
For UV–Vis thermal denaturation experiments oligonucleotides were analyzed in 1 ml solutions at 1 µM concentrations in a buffer of 10 mM sodium phosphate at pH 7.0 and 50 mM NaCl. Double-stranded oligonucleotides were annealed in equimolar amounts by heating to 95°C and then slowly cooling to r.t. on a heating block. Thermal denaturation experiments were performed on a Cary-300 UV–Vis spectrophotometer (Varian Inc.) equipped with a 6 × 6 cell changer and Peltier temperature controller. Samples were heated from 10°C to 95°C at a rate of 0.5°C/min and the change in absorbance was measured every 1.0°C. The melting temperature (Tm) values were obtained by the baseline (alpha) method, and defined as the point when the mole fraction of duplex was equal to 0.5. These values represent the averages of at least three independent experiments. Fluorescent thermal denaturation plots were obtained in a similar fashion on a Cary Eclipse fluorescent spectrophotometer equipped with a multicell Peltier temperature controller and automated polarization accessories. Measurements were carried out in 1 cm × 1 cm quartz cells in 2 ml volumes and 1 µM concentration of oligonucleotides
Circular dichroism (CD) spectra were obtained using a Jasco J-800 spectropolarimeter. Samples were prepared in the same fashion as with Tm experiments. Scans were performed in triplicate at 20°C at a rate of 50 nm/min from 350 to 190 nm. The data were averaged, corrected against a blank and smoothed using the Spectra Manager CD software provided by the manufacturer.
RNase H assays
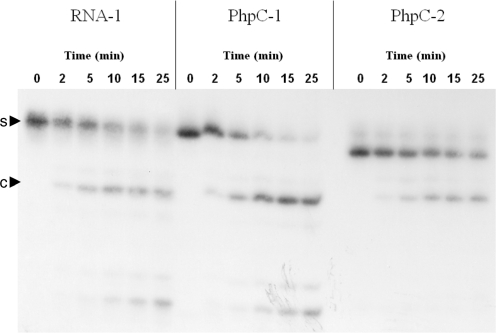
Wild-type p66/p51 HIV-1 RT was a generous gift from Dr M. Götte (McGill), prepared by his group as described earlier (2). RNA substrates were 5′-radiolabeled with γ-32P ATP by T4 polynucleotide kinase (Fermentas) using the manufacturer’s recommended procedure. The DNA and 5′-32P labeled RNA strands were combined in a 1.2 : 1 ratio and annealed by heating to 95°C followed by slow cooling to r.t. HIV-1 RT (2.5 nM final concentration) was incubated for 10 min at 37°C in RNase H reaction buffer (50 mM Tris–HCl, pH 7.8, 60 mM KCl, 5 mM MgCl2, 0.1 mM DTT and 0.01% Tween-20). The reactions were initiated by the addition of duplexed RNA/DNA substrate to a concentration of 50 nM. Aliquots were removed at various times as indicated in Figure 2 and stopped by the addition of an equal volume of loading buffer (98% deionized formamide, 1 mg/ml bromophenol blue and 1 mg/ml xylene cyanol) followed by heat inactivation at 95°C for 5 min. Cleavage products were resolved on a 16% denaturing polyacrylamide gel and visualized by autoradiography.
Figure 2.
5′-32P-labeled PAGE assay for HIV-1 RT RNAase H activity. Arrowheads indicate substrate (s) and major cleavage product (c).
Fluorescent RNase H assays of HIV-1 RT RNase H activity was monitored by the changes in emission of PhpC on a Cary Eclipse fluorescent spectrophotometer equipped with a multicell Peltier temperature controller and automated polarization accessories. These assays were run in 2 ml volumes in identical buffer conditions as the gel-based assays, but were initiated by the addition of MgCl2 (5 mM final concentration) in order to monitor fluorescence prior to cleavage by HIV-1 RT. Substrate concentrations varied from 1 to 0.2 µM and enzyme concentrations were changed to 25 or 5 nM, respectively. The fluorescence intensity was monitored every 15 s. PhpC RNase H assays were monitored at an excitation wavelength of 360 nm (5 nm bandwidth) and emission wavelength of 465 nm (5 nm bandwidth). The fluorescein-RNA/dabcyl-DNA pair was monitored at an excitation wavelength of 485 nm (5 nm bandwidth) and emission wavelength of 520 nm (5 nm bandwidth). Fluorescence polarization (FP) RNase H assays were run under identical conditions as stated above.
Fluorescent RNase H assays on 96-well microplates
RNase H activity of HIV-1 RT was monitored on a Gemini XS and M5 spectrofluorometers by Molecular Devices in 96-half well opaque plates. FP measurements in 96-well microplates were performed on a BioTek Synergy 4. Experiments were run under identical buffer conditions as gel-based assays in a final volume of 100 µl. Reactions were initiated by the addition of a 50 µl solution containing HIV-1 RT and MgCl2. The concentration of substrate was varied from 1 µM to 10 nM, and enzyme concentrations varied from 25 to 5 nM, respectively. Excitation and emission wavelengths for PhpC were 360 and 465 nm, respectively, and 485 and 520 nm for the dabcyl/fluorescein assay. To determine Km, Vmax and kcat, the initial velocity of RNase H cleavage was measured under a fixed concentration HIV-1 RT (1 nM) and the substrate concentration was varied from 10 to 900 nM. The IC50 of DHBNH, a sample of which was generously provided by Dr M.A. Parniak, was determined on two separate days and was run in triplicate using 200 nM substrate and 1.5 nM HIV-1 RT in 1% DMSO. Eight concentrations of DHBNH were dispensed by two-fold serial dilutions starting from 50 µM of the inhibitor.
RESULTS AND DISCUSSION
Physical properties of RNA containing PhpC
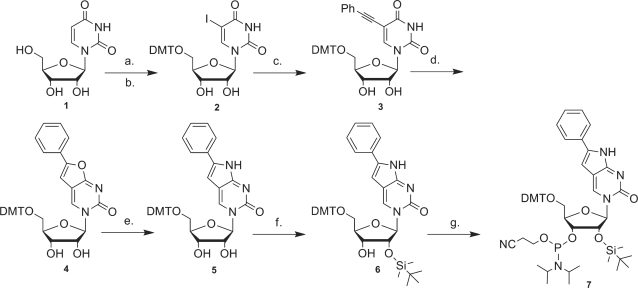
PhpC-containing oligoribonucleotides were obtained by conventional phosphoramidite chemistry in a similar manner as deoxy-PhpC (22–24) with the 2′-OH group protected as the tert-butyldimethylsilyl ether (27,28) (Scheme 3). Thermal denaturation studies of these sequences hybridized to complementary DNA (DNA-1) and RNA (RNA-2) were carried out (Table 1). PhpC-1 showed comparable binding affinity to unmodified controls, whereas the PhpC at a central position, PhpC-2, displayed a 2°C increase in thermal stability that is ascribed to greater π-stacking, which is consistent with the increased hypochromicity observed in the thermal denaturation profiles (SI), and previous reports of PhpC in PNA and DNA (22,24). The PhpC insert was also shown to be exceptionally well-tolerated in RNA/RNA duplexes by CD spectroscopy (SI), as the CD spectra overlapped nearly identically with the native strands in shape and intensity. It is noteworthy that despite the changes in global conformation that accompany the transition for B-form (dsDNA) through hybrid duplexes (RNA:DNA) to A-form (dsRNA) that the modified C5-face of cytosine is well accommodated based on Tm and CD measurements. Although there is a scarcity of data reporting on conformation for C5-modified pyrimidines in dsRNA or hybrid RNA:DNA duplexes, Tm data indicate that modest modifications such as halogens (29) and alkyl/alkynyl (30) may be modestly stabilizing, as we have observed for PhpC.
Scheme 3.
Synthesis of 6-Phenylpyrrolocytidine phosphoramidite (7). a. I2, AgSO4, MeOH. b. DMT-Cl, py. c. phenylacetylene, DMF, Pd(PPh3)4, CuI, NEt3. d. AgNO3, acetone. e. NH4OH, MeOH, 55°C. f. TBDMS-Cl, imidazole, DMF. g. (iPr)2NP(Cl)O(CH2)2CN, THF, NiPr2Et.
Fluorescence properties of RNA containing PhpC
The fluorescence parameters of PhpC as a free nucleoside, and when incorporated in short oligomers, single-stranded 18-mers and duplexed to DNA or RNA are given in Table 2. The PhpC nucleoside (1) and PhpC nucleoside monophosphate (2) displayed similar fluorescence intensity and quantum yields of 0.31 and 0.29, respectively, a 12-fold increase from a reported 6-methylpyrrolocytidine (31) value (0.023). Thus, it appears that the phosphate group does not play a role on the fluorescence intensity. Comparing the trinucleotides, G-PhpC-U (3), C-PhpC-U (4), U-PhpC-U (5) and (6) A-PhpC-U the fluorescence intensity and quantum yield is dependent on the nature of neighboring bases, with 3 (neighboring guanine) showing a reduced quantum yield (Φ) as was reported earlier for MepC (31,32). Theoretical studies on MepC suggested that base-stacking interactions can cause diminution of Φ by affecting the strength of the oscillator for the fluorescence transition (31), which is consistent with our observations, although quenching of the fluorescence by intrastrand electron transfer cannot be discounted. The U–PhpC–U trinucleotide had the highest observed quantum yield of the series (Φ = 0.41), which is likely due to shielding of the fluorophore from solvent (dynamical quenching) while not introducing other compensating non radiative deactivation pathways. Interestingly, the 18-mer single stranded PhpC-1 had the same fluorescence intensity and quantum yield as the trinucleotide 3, where PhpC had the same nearest neighbors. This trend did not continue with PhpC-2 (Φ = 0.13) and its analogous trinucleotide 4 (Φ = 0.24). In this case, it appears that PhpC is quenched in the longer single-stranded RNA sequence by 50%, which may be due to better stacking interactions in a central position; however, length and base composition may play a greater role for shorter PhpC containing oligonucleotides that are conformationally more flexible.
Table 2.
Fluorescent properties of PhpC and oligonucleotides containing single PhpC inserts
| Entry | Name | Fluorescence intensitya | Quantum yieldb Φ | FPc |
|---|---|---|---|---|
| 1 | PhpC | 51 | 0.31 | <0.02 |
| 2 | 5′-PO4-PhpC | 47 | 0.29 | <0.02 |
| 3 | G-PhpC-U | 36 | 0.11 | 0.03 |
| 4 | C-PhpC-U | 104 | 0.24 | 0.03 |
| 5 | U-PhpC-U | 134 | 0.41 | 0.03 |
| 6 | A-PhpC-U | 106 | n.d. | 0.03 |
| 7 | PhpC-1 | 35 | 0.11 | 0.21 |
| 8 | PhpC-1:DNA-1 | 7.8 | 0.035 | 0.28 |
| 9 | PhpC-1:RNA-2 | 9.5 | n.d. | 0.30 |
| 10 | PhpC-2 | 38 | 0.13 | 0.22 |
| 11 | PhpC-2:DNA-1 | 80 | 0.20 | 0.27 |
| 12 | PhpC-2:RNA-2 | 69 | n.d. | 0.28 |
aOne micro molar samples measured in 10 mM phosphate buffer (pH 7.0) and 50 mM NaCl at 25°C, λex = 360 nm; λemm = 465 nm
bSee Supporting Information for Quantum Yield determination.
cFP measured in identical conditions as footnote a.
n.d. signifies not determined.
Oligonucleotides containing pC analogs have consistently shown fluorescence quenching upon duplex formation (22–24,33–35). The fluorescence of PhpC-1 hybridized to DNA (8) and RNA (9) also showed ∼75% quenching. An increase in fluorescence as the duplexes denature could be monitored by thermal denaturation profiles, and the mid-point of the transition corresponds to the Tm determined by UV absorbance (SI). The fluorescence emission when PhpC-2 bound to DNA (11) and RNA (12) complementary strands surprisingly increased. This is unexpected based on our previous work with modified oligonucleotides, and to our knowledge is the first example of a pC analog showing increased fluorescence emission with hybridization. These observations are consistent with Thompson’s recent study that proposed that the changes in quantum yield of pC are not only dominated by base stacking, which is responsible for much of the changes in fluorescence for 2-AP (31), but possibly by collisional deactivation from solvent. We observed a transition in the fluorescent thermal denaturation curve of single-stranded PhpC-1 (SI) and PhpC-2 (not shown) indicating that some quenching in single strands is contributed by base stacking. The dramatic quenching of fluorescence normally observed during duplex formation and specific hydrogen bonding to guanine may be due to base pairing mediated electron transfer, which leads to non-radiative relaxation of the electronically excited state. Currently, this explanation awaits experimental support. It is clear from these results and the limited studies on MepC, that there are a variety of factors that affect the fluorescence of PhpC including the sequence context (neighboring bases) and hybridization state along with microsequence effects.
RNase H assays
The PhpC-containing RNA were determined to be excellent substrates for HIV-1 RT RNase H by standard 5′-32P-label/PAGE assay (Figure 2), unlike the FQ system, which compromises enzymatic activity (SI). Furthermore, no aberrant cleavage products were generated as was previously observed with deoxy-MepC inserts on the polypurine tract of HIV-1 RT (36).
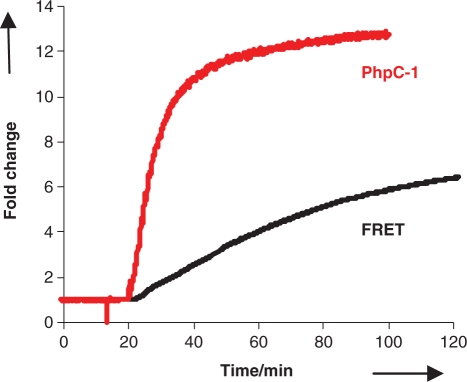
Subsequently, the RNase H activity was monitored fluorimetrically. Duplex 8 proved to be remarkably responsive and gave the largest fluorescence change (14-fold) between the hybrid duplex and the cleavage product (Figure 3). This was easily followed in kinetic or end-point mode. The increase in fluorescence was so dramatic that it is visible to the naked eye (Figure 3, inset). We compared the RNase H cleavage of PhpC-1, to the classic RNA-3′-fluorescein/DNA-5′-dabcyl (FQ) assay (6) (Figure 4) by running them simultaneously at 1 µM substrate concentrations. We found that the FQ assay has higher fluorescence signal due to the greater brightness of fluorescein, but the PhpC-1 assay generated cleavage products with greater relative fluorescence compared with the intact substrates (14- versus 8-fold). As observed in gel-based assays, the PhpC insert did not slow cleavage as did the FQ substrate resulting in a substantial gain in analysis time. The end point of cleavage (90% completion) was achieved in ∼20 min for 8 versus 130 min for FQ.
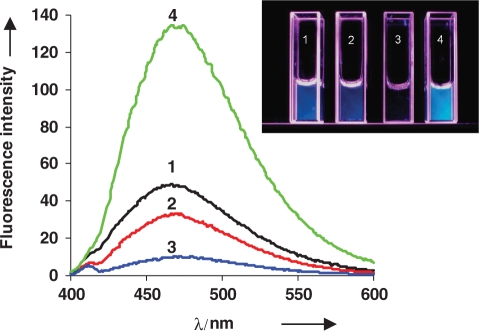
Figure 3.
Fluorescence emission spectra of PhpC nucleotide (1, black), PhpC-1 single stranded (2, red), PhpC-1:DNA-1 duplex (3, blue) and PhpC-1:DNA-1 after RNase H cleavage by HIV-1 RT (4, green). Visual changes in fluorescence of the same solutions under UV365 light, at 1 µM concentration (inset).
Figure 4.
Comparison the change in fluorescence emission intensity for the PhpC-1 RNase H assay (red curve) compared to the classic Fluorescein/Dabcyl assay (black curve). Both substrates were treated with HIV-1 RT RNase H activity simultaneously in identical conditions at a concentration of 1 µM scanning at the λmax excitation and emission for both fluorescein (485/520 nm) and PhpC (360/465 nm).
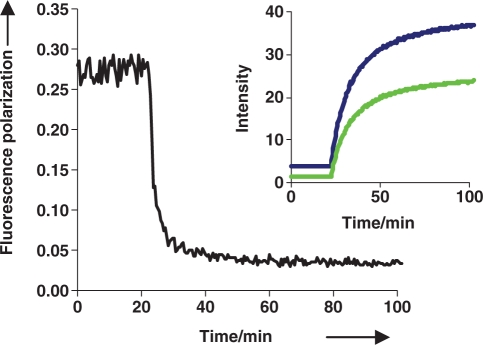
While a labeled DNA strand was found to be fluorimetrically responsive to the cleavage of its unlabeled RNA complement (data not shown), an advantage of incorporation of the fluorescent nucleoside into the RNA strand is that it gets processed by RNase H into fragments that are much smaller than the hybrid duplex. The substantial change in mass/size of the fluorescent moiety may be detected by changes in FP (33). As seen in Table 1, FP values increase with increasing molecular weight as expected for molecules undergoing less Brownian motion. RNase H mediated cleavage of PhpC-1:DNA was readily monitored in real-time by FP (Figure 5) in a similar fashion to the RNase H assay developed by Pfizer (7).
Figure 5.
Monitoring the RNase H activity of HIV-1 RT on PhpC-1:DNA-1 by fluorescence polarization (black curve). The inset shows the fluorescence intensity with the excitation and emission filters parallel (blue curve) and perpendicular (green curve).
For application of this assay to screen for inhibitors of RNase H activity, it is advantageous to monitor polarization as well as fluorescence intensity. FP can discern molecules that quench the fluorophore producing false hits (7). With the versatility of PhpC we have demonstrated that both fluorescence intensity (which reports on hybridization/base stacking) and FP (which reports on the size of the molecule containing the fluorophore) can be monitored simultaneously using only one probe.
RNase H assays on 96-well plates
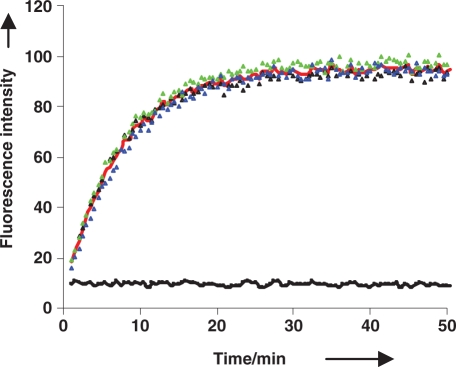
Although fluorescent spectrophotometers are far more sensitive, it is desirable to adapt assays to fluorescent microplate readers that are amenable to HTS. Since duplex 8 was the most fluorescent and responsive substrate for RNase H activity, it was tested in 96-well plate spectrofluorometers by fluorescence intensity (Figure 6) and FP (SI). Using identical reaction conditions used in the cuvettes (1 µM substrate and 25 nM HIV-1 RT), Figure 6 shows very good signal to noise and response to RNase H cleaveage (10-fold increase). We later tested the limits of sensitivity of the assay and found we could still monitor RNase H activity down to 10 nM substrate concentration (60 nM in FP mode), which is comparable to 32P PAGE assays
Figure 6.
Fluorescence HIV-1 RT RNase H assay of 1 µM PhpC-1:DNA-1 monitored by a 96-well plate spectrofluorimeter. Colored triangles represent reactions in three individual wells, with the red curve representing their average. The average of three controls without enzyme is represented by the black curve.
We determined the kinetic parameters Km (54 ±3.3 nM), Vmax (0.31 nmols/min) and a kcat of 6/min of duplex 8 for HIV-1 RT RNase H (SI). Since this assay will ultimately be used to screen potential inhibitors of RNase H, a known inhibitor of HIV-1 RT RNase H, namely DHBNH, was tested in this assay. Our single-label assay demonstrated that DHBNH was able to inhibit RNase H activity in a dose-dependent manner with an IC50 of 5 µM, which is consistent with previous results (37).
CONCLUSIONS
This work demonstrates the significant advantages of base-modified nucleosides, such as PhpC, compared with traditional fluorophores: one single PhpC insert can act as a sensitive reporter group that is non-disruptive to the structure and enzymatic activity. Although fluorescein is a substantially brighter luminophore, the PhpC-based assay for RNase H offers several advantages. The responsiveness, rapidity and ease (single label versus dual) of the RNase H assay has been improved. The fluorescence provided by PhpC is sufficient to compete with gel-based techniques on the basis of sensitivity and the assay can be adapted to multiwell plate format for HTS. This was also possible because the HIV-1 RT RNase H cleavage product, the tetranucleotide, showed remarkably greater fluorescence than even the free nucleoside. Also uncovered in this study, was the surprising and unprecedented observation that oligomer PhpC-2 showed an increase in fluorescence intensity upon duplex formation. Together, these observations indicate that PhpC is significantly different from MepC and warrants further investigation on the affects of sequence length and composition on changes in fluorescence. Overall, this report lays the framework for a sensitive and rapid assay for RNase H activity and inhibitors thereof. We feel that PhpC will contribute to the growing repertoire of useful fluorescent nucleobase analogs such as tC°, which also shows red-shifted fluorescence and a high quantum yield (Φ = 0.3) but whose fluorescence is insensitive to duplex formation (38).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Natural Sciences and Engineering Research Council of Canada is acknowledged for funding for this work through the discovery grants program (RHEH, MJD).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the Natural Sciences and Engineering Research Council of Canada for financial support in the form of grants (RHEH and MJD). We thank Dr Matthias Götte for providing the HIV-1 RT and Dr Michael A. Parniak for the RNase H inhibitor.
REFERENCES
- 1.Coffin JM. HIV Population-dynamics in-vivo - implications for genetic-variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Schatz O, Mous J, Le Grice SFJ. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′→5′ exonuclease activity. EMBO J. 1990;9:1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tisdale M, Schulze T, Larder BA, Moelling K. Mutations within the RNase-H Domain of Human-Immunodeficiency-Virus Type-1 Reverse-Transcriptase Abolish Virus Infectivity. J. Gen. Virol. 1991;72:59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 4.Rizzo J, Gifford LK, Zhang X, Gewirtz AM, Lu P. Chimeric RNA–DNA molecular beacon assay for ribonuclease H activity. Mol. Cell. Probes. 2002;16:277–283. doi: 10.1006/mcpr.2002.0423. [DOI] [PubMed] [Google Scholar]
- 5.McLellan N, Wei X, Marchand B, Wainberg MA, Gotte M. Nonradioactive detection of retroviral-associated RNase H activity in a microplate-based, high-throughput format. Biotechniques. 2002;33:424–429. doi: 10.2144/02332ht03. [DOI] [PubMed] [Google Scholar]
- 6.Parniak MA, Min KL, Budihas SR, Le Grice SFJ, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama GR, Bingham P, Tan D, Maegley KA. A fluorescence polarization assay for screening inhibitors against the ribonuclease H activity of HIV-1 reverse transcriptase. Anal. Biochem. 2006;351:260–265. doi: 10.1016/j.ab.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Yang CJ, Wu Y, Conlon P, Kim Y, Lin H, Tan W. Light-switching excimer beacon assays for ribonuclease H kinetic study. ChemBioChem. 2008;9:355–359. doi: 10.1002/cbic.200700542. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 10.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 11.Rist MJ, Marino JP. Fluorescent nucleotide base analogs as probes of nucleic acid structure, dynamics and interactions. Curr. Org. Chem. 2002;6:775–793. [Google Scholar]
- 12.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides.I. formycin 2-aminopurine riboside 2,6-diaminopurine riboside and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 13.Hawkins ME, Balis FM. Use of pteridine nucleoside analogs as hybridization probes. Nucleic Acids Res. 2004;32:e62. doi: 10.1093/nar/gnh060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tor Y. Exploring RNA-ligand interactions. Pure Appl. Chem. 2009;81:263–272. [Google Scholar]
- 15.Sandin P, Stengel G, Ljungdahl T, Borjesson K, Macao B, Wilhelmsson LM. Highly efficient incorporation of the fluorescent nucleotide analogs tC and tCO by Klenow fragment. Nucleic Acids Res. 2009;37:3924–3933. doi: 10.1093/nar/gkp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson JN, Kool ET. Fluorescent DNA base replacements: reporters and sensors for biological systems. Org. Biomol. Chem. 2006;4:4265–4274. doi: 10.1039/b612284c. [DOI] [PubMed] [Google Scholar]
- 17.Socher E, Bethge L, Knoll A, Jungnick N, Herrmann A, Seitz O. Low-noise stemless PNA beacons for sensitive DNA and RNA detection. Angew. Chem. Int. Ed. 2008;47:9555–9559. doi: 10.1002/anie.200803549. [DOI] [PubMed] [Google Scholar]
- 18.Dodd DW, Hudson RHE. Intrinsically fluorescent base-discriminating nucleoside analogs. Mini-Rev. Org. Chem. 2009;6:378–391. [Google Scholar]
- 19.Inoue H, Imura A, Ohtsuka E. Synthesis of dodecadeoxyribonucleotides containing a pyrrolo[2,3-D]pyrimidine nucleoside and their base-pairing ability. Nippon Kagaku Kaishi. 1987:1214–1220. [Google Scholar]
- 20.Woo J, Meyer RB, Jr, Gamper HB. G/C-modified oligodeoxynucleotides with selective complementarity: synthesis and hybridization properties. Nucleic Acids Res. 1996;24:2470–2475. doi: 10.1093/nar/24.13.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry DA, Jung KY, Wise DS, Sercel AD, Pearson WH, Mackie H, Randolph JB, Somers RL. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides. Tetrahedron Lett. 2004;45:2457–2461. [Google Scholar]
- 22.Hudson RHE, Ghorbani-Choghamarani A. Selective fluorometric detection of guanosine-containing sequences by 6-phenylpyrrolocytidine in DNA. Synlett. 2007:870–873. [Google Scholar]
- 23.Hudson RHE, Dambenieks AK, Viirre RD. Fluorescent 7-deazapurine derivatives from 5-iodocytosine via a tandem cross-coupling-annulation reaction with terminal alkynes. Synlett. 2004:2400–2402. [Google Scholar]
- 24.Wojciechowski F, Hudson RHE. Fluorescence and hybridization properties of peptide nucleic acid containing a substituted phenylpyrrolocytosine designed to engage guanine with an additional H-bond. J. Am. Chem. Soc. 2008;130:12574–12575. doi: 10.1021/ja804233g. [DOI] [PubMed] [Google Scholar]
- 25.King KC, Budihas SR, Le Grice SFJ, Parniak MA, Crouch RJ, Gaidamakov SA, Isaaq HJ, Wamiru A, McMahon JB, Beutler JA. A capillary electrophoretic assay for ribonuclease H activity. Anal. Biochem. 2004;331:296–302. doi: 10.1016/j.ab.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Bellon L. Oligoribonucleotides with 2′-O-(tert-butyldimethylsilyl) groups. In: Beaucage SL, Herdewijn P, Matsuda A, editors. Curr. Protoc. Nucleic Acid Chem. 2001. John Wiley & Sons, Vol. 3.6, pp. 1–8. [DOI] [PubMed] [Google Scholar]
- 27.Ogilvie KK, Schifman AL, Penney CL. Synthesis of Oligoribonucleotides. 3. Use of Silyl Protecting Groups in Nucleoside and Nucleotide Chemistry.8. Can. J. Chem. 1979;57:2230–2238. [Google Scholar]
- 28.Damha MJ, Ogilvie KK. Oligoribonucleotide synthesis. The silyl-phosphoramidite method. Methods Mol. Biol. 1993;20:81–114. doi: 10.1385/0-89603-281-7:81. [DOI] [PubMed] [Google Scholar]
- 29.Ziomek K, Kierzek E, Biala E, Kierzek R. The thermal stability of RNA duplexes containing modified base pairs at internal and terminal positions of the oligorionucleotides. Biophys. Chem. 2002;97:233–241. doi: 10.1016/s0301-4622(02)00074-1. [DOI] [PubMed] [Google Scholar]
- 30.Terrazas M, Kool ET. RNA major groove modifications improve siRNA stability and biological activity. Nucleic Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardman SJO, Botchway SW, Thompson KC. Evidence for a nonbase stacking effect for the environment-sensitive fluorescent base pyrrolocytosine-comparison with 2-aminopurine. Photochem. Photobiol. 2008;84:1473–1479. doi: 10.1111/j.1751-1097.2008.00368.x. [DOI] [PubMed] [Google Scholar]
- 32.Hardman SJO, Thompson KC. Influence of base stacking and hydrogen bonding on the fluorescence of 2-aminopurine and pyrrolocytosine in nucleic acids. Biochemistry. 2006;45:9145–9155. doi: 10.1021/bi060479t. [DOI] [PubMed] [Google Scholar]
- 33.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu CH, Martin CT. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 35.Saito Y, Shinohara Y, Bag SS, Takeuchi Y, Matsumoto K, Saito I. Ends free and self-quenched molecular beacon with pyrene labeled pyrrolocytidine in the middle of the stem. Tetrahedron. 2009;65:934–939. [Google Scholar]
- 36.Dash C, Rausch JW, Le Grice SFJ. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 2004;32:1539–1547. doi: 10.1093/nar/gkh307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark AD, Knight JL, Julias JG, Clark PK, et al. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem. Biol. 2006;1:702–712. doi: 10.1021/cb600303y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandin P, Borjesson K, Li H, Martensson J, Brown T, Wilhelmsson LM, Albinsson B. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.