Abstract
Human telomeric DNA G-quadruplex has been considered as an attractive target for cancer therapeutic intervention. The telomeric sequence shows intrinsic structure polymorphism. Here we report a novel intramolecular G-quadruplex structure formed by a variant human telomeric sequence in K+ solution. This sequence forms a basket-type intramolecular G-quadruplex with only two G-tetrads but multiple-layer capping structures formed by loop residues. While it is shown that this structure can only be detected in the specifically truncated telomeric sequences without any 5′-flanking residues, our results suggest that this two-G-tetrad conformation is likely to be an intermediate form of the interconversion of different telomeric G-quadruplex conformations.
INTRODUCTION
Telomeres are specialized DNA sequences that cap the ends of linear chromosomes (1–3). The structure and stability of telomeres are closely related with cancer (4–7), aging (8,9) and genetic stability (3,10,11). Human and vertebrate telomeres consist of highly conserved tandem repeats of the hexanucleotide d(TTAGGG)n 5–10 kb in length 5′ to 3′ toward the chromosome end, terminating in a single-stranded 3′-overhang of 100–200 bases (12–15). In normal cells, each cell replication results in a 50- to 200-base loss of the telomere. After a critical shortening of the telomeric DNA is reached, the cell undergoes apoptosis (16). However, telomeres of cancer cells do not shorten on replication, due to the activation of a reverse transcriptase, telomerase, that extends the telomeric sequence at the chromosome ends (17). Telomerase is activated in 80–85% of human cancer cells and has been suggested to play a key role in maintaining the malignant phenotype by stabilizing telomere length and integrity (18,19). The G-rich telomeric sequence can form G-quadruplex DNA secondary structures that consist of stacked G-tetrad planes connected by a network of Hoogsteen hydrogen bonds and stabilized by monovalent cations, such as Na+ and K+. The G-quadruplex formation by the single-stranded human telomeric DNA can inhibit the activity of telomerase (20). Compounds that stabilize the intramolecular DNA G-quadruplexes formed in the human telomeric sequence have been shown to inhibit the activity of telomerase and disrupt telomere capping and maintenance, making the intramolecular human telomeric DNA G-quadruplex an attractive target for cancer therapeutic intervention (4,6,7,21–23).
The human telomeric sequence was first shown more than a decade ago to form a basket-type intramolecular structure in Na+ solution by NMR, using the 22-nt telomeric sequence d[AGGG(TTAGGG)3] (24). The same 22-nt telomeric sequence was reported to form a parallel-stranded intramolecular G-quadruplex in crystalline state in the presence of K+ (25). Recent results show that the hybrid-type intramolecular G-quadruplex structures appear to be the major conformations formed in human telomeric sequences in K+ solution, with a dynamic equilibrium between hybrid-1 and hybrid-2 conformations (26–33). The hybrid forms appear to be the major form for human telomeric sequences in K+ solution even in the co-presence of high concentration of Na+ (26). The two hybrid-type telomeric G-quadruplexes are distinct and yet closely related, and appear to be largely determined by the 3′-flanking sequence and thus the different capping structures (27,28). The highly conserved human telomeric sequence shows intrinsic structure polymorphism [see recent review (34)]; however, little is known about the interconversion of different telomeric G-quadruplex conformations. Interestingly, while the energy difference between the two hybrid-type forms appears to be low as both conformations coexist in K+ solution, the kinetics of the interconversion between the two hybrid forms is rather slow and very few exchange peaks were observed on the NMR time scale (27), indicating a high energy barrier of the intermediate state(s). In addition, recent reports have shown a multi-step melting transition of telomeric G-quadruplexes in K+ or Na+ solution (35,36).
In this study, we have determined an interesting intramolecular G-quadruplex structure formed by a variant human telomeric sequence with a G-to-I substitution in K+ solution. This sequence forms a basket-type intramolecular G-quadruplex with only two G-tetrads but multiple-layer capping structures formed by loop residues. The stable formation of this two-G-tetrad basket-type structure is driven by the extensive capping structures and loop interactions. We have also shown that this structure can be clearly detected in a specifically truncated telomeric sequence, GGG(TTAGGG)3T; however, the addition of any 5′-flanking residue(s) abolishes this conformation. A very similar NMR structure has very recently been reported using an 8-Br-incorporated telomeric sequence (37). The structure was suggested to be a new form of the natural human telomeric sequence in K+ solution and was reported to be more stable than the hybrid-type structures. Our results differ from the previous report in several aspects. First, our results show substantial differences in the capping structures and loop interactions. Most importantly, our results suggest that this two-G-tetrad conformation is likely to be an intermediate form of the interconversion of different telomeric G-quadruplex conformations, rather than a stable new form of the extended human telomeric sequence, as this structure can hardly be detected in any telomeric sequence with a 5′-flanking segment. We have previously proposed a model for the interconversion between the hybrid-type K+ form and the basket-type Na+ form telomeric G-quadruplex structures through a strand-reorientation mechanism (26). This two-G-tetrad basket-type structure appears to fit the intermediate state in our proposed transition model, and may also be a transition state of the interconversion of the two hybrid-type conformations. This will be the first report of a NMR-determined structure that represents an intermediate state of G-quadruplex structure interconversion.
MATERIALS AND METHODS
Sample preparation
The DNA oligonucleotides were synthesized using β-cyanoethylphosphoramidite solid-phase chemistry on an ExpediteTM 8909 Nucleic Acid Synthesis System (Applied Biosystem, Inc) in DMT-on mode, and were purified using C18 reverse-phase HPLC chromatography, as described previously (26–28,38–41). Samples in D2O were prepared by repeated lyophilization and final dissolution in 99.96% D2O. Samples in water were prepared in 10%/90% D2O/H2O solution. The final NMR samples contained 0.1–2.5 mM DNA in 25 mM K-phosphate buffer (pH 7.0) and 70 mM KCl.
NMR experiments
NMR experiments were performed on a Bruker DRX-600 spectrometer as described previously (26–28,38–41). Standard 2D NMR experiments, including NOESY, TOCSY and DQF-COSY, were collected at 1, 7, 15, 20, 25 and 30°C to obtain the complete proton resonance assignment. Non-exchangeable protons were estimated based on the NOE cross-peak volumes at 50–300 ms mixing times, with the upper and lower boundaries assigned to ±30% of the estimated distances. The methyl base proton Me pseudoatom–H6 distance (2.99 Å) was used as a reference. The distances involving the unresolved protons, e.g. methyl protons, were assigned using pseudo-atom notation in X-PLOR.
Distance geometry and simulated annealing (DGSA) calculations
Metric matrix distance geometry (MMDG) and simulated annealing calculations were carried out in X-PLOR (42) to embed and optimize 100 initial structures based on an arbitrary extended conformation for the single-stranded I14Tel23 sequence, as described previously (27,28,38,40). The experimentally obtained distance restraints and G-tetrad hydrogen-bonding distance restraints were included during the calculation.
NOE-distance restrained molecular dynamics calculations
All of the 100 molecules obtained from the DGSA calculations were subjected to NOE-restrained molecular dynamics refinement in XPLOR (42) with a distance-dependent dielectric constant, as described previously (27,28,38,40). The force constants were scaled at 10–30 and 80–100 kcal/mol Å2 for NOE and hydrogen bond distance restraints, respectively. A total of 508 NOE distance restraints, of which 276 are from inter-residue NOEs, were incorporated into the NOE-restrained structure calculation. Dihedral angle restraints were used to restrict the glycosidic torsion angle (χ) for the experimentally assigned syn and anti conformations. A dihedral angle restraint of 60(±40)° was applied to the syn G-tetrad guanines, and a dihedral angle restraint of 240(±40)° was applied to the anti G-tetrad guanines. The force constants of dihedral angle restraints were 10 kcal/mol rad2. The eight best molecules were selected based both on their minimal energy terms and number of NOE violations and have been deposited in the Protein Data Bank (PDB ID 2kka).
CD spectroscopy
CD spectra of the oligonucleotides were recorded on a Jasco J-810 spectropolarimeter (Jasco, Easton, MD). A quartz cell of 1 mm optical path length and an instrument scanning speed of 100 nm/min with 1 s response time was used for measurements. The measurements are the averages of three repetitions between 200 and 320 nm at room temperature. Spectra were baseline-corrected and the signal contributions of the buffer were subtracted. DNA solutions for CD measurements were made in 25 mM K(/Na)-PO4, 70 mM K(/Na)Cl, pH 7.0 buffer with 10 µM strand concentrations.
RESULTS
A variant human telomeric sequence forms a stable intramolecular G-quadruplex structure in K+ solution
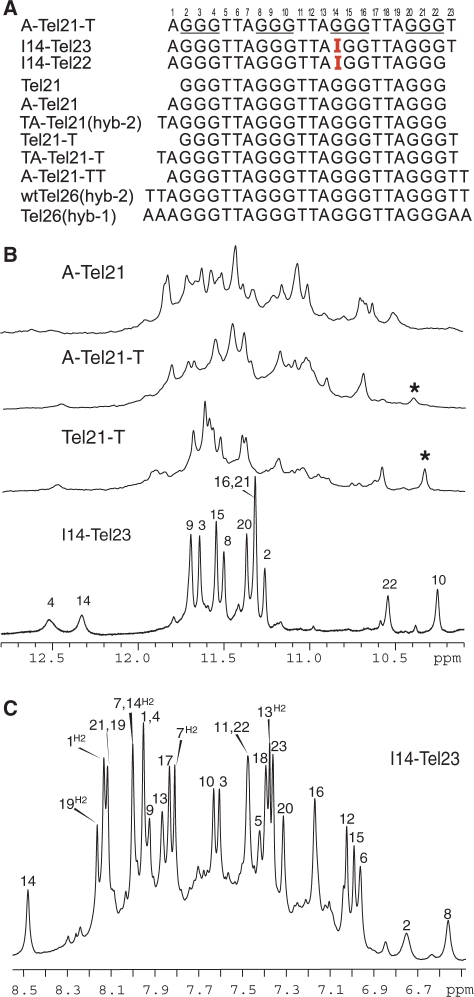
The 22-nt human telomeric sequence A–Tel21, d[AGGG(TTAGGG)3] (Figure 1A), has been shown to form a single stable unimolecular G-quadruplex in Na+ solution (24) and was used for the crystallography study in the presence of K+ (25). However, we found this sequence forms a mixture of multiple G-quadruplex conformations in K+ solution as indicated by 1D 1H NMR (Figure 1B) (26). We have carried out single G-to-I substitution at various guanine positions of the A–Tel21 sequence. Inosine differs from guanine by lacking the NH2 group at the C2 position. We found that the G-to-I substitution at position 14 results in a sequence (I14–Tel22, Figure 1A) that gives rise to a very different but well-resolved NMR spectrum (Supplementary Figure S1), and the presence of well-resolved peaks between 11 and 12 ppm indicates the formation of a G-quadruplex structure. Minor conformations are present in I14–Tel22 as indicated by the presence of weak resonances, which are much more evident in 2D NOESY spectra (data not shown). The addition of a thymine residue at the 3′-end (I14–Tel23, Figure 1A) stabilizes the major structure and improves the NMR spectral quality (Figure 1B). The 1H NMR spectrum of I14–Tel23 is much cleaner as the population of minor conformations is clearly reduced as compared to I14–Tel22. The 1D 1H NMR spectrum of I14–Tel23 in K+ solution shows eight well-resolved imino proton resonances at 11–12 ppm with sharp line widths (∼6 Hz at 20°C) (Figure 2B), indicating the formation of a stable G-quadruplex structure. The melting temperature of this G-quadruplex is concentration-independent as shown by both NMR and CD, indicating that the I14–Tel23 G-quadruplex structure is unimolecular.
Figure 1.
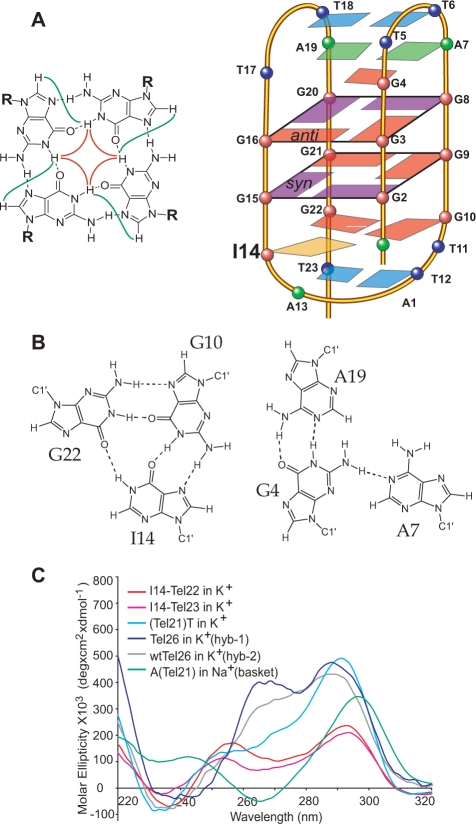
(A) Native and variant four-G-tract human telomeric DNA sequences. Tel21 is used as the base sequence to show various flanking segments. The numbering system is shown above A–Tel21–T. The sequences used for structure determination are labeled, with the structure determined in parentheses. (B) Imino regions of 1D 1H NMR spectra of several telomeric sequences shown in (A). The assignment for I14–Tel23 is shown. The G10NH1 proton of Tel21–T and A–Tel21–T are labeled with asterisks. (C) The aromatic region of 1D 1H NMR spectra of I14–Tel23 in K+ solutions at 20°C, with assignments. Conditions: 25 mM KPO4, 70 mM KCl, pH 7.0, 20°C.
Figure 2.
The imino proton region with assignment of the 1D 1H NMR spectrum of I14–Tel23 in K+ solution (top), and imino proton assignments of I14–Tel23 using 1D 15N-filtered experiments on site-specific labeled oligonucleotides. Conditions: 25 mM KPO4, 70 mM KCl, pH 7.0, 20°C, 0.50–0.6 mM DNA. G4NH1 was assigned using the I14–Tel22 sequence.
Assignment of proton resonances for the I14–Tel23 G-quadruplex in K+ solution
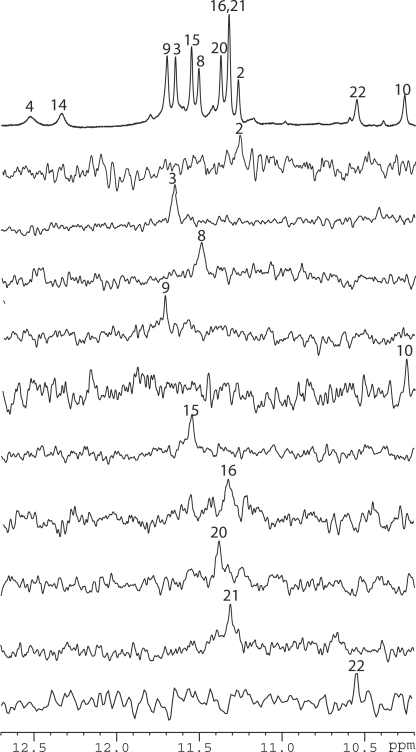
Interestingly, I14–Tel23 shows only eight strong imino peaks in the 1D proton spectrum between 10.6 and 12 ppm in K+ solution (Figure 2B bottom). This result indicates that, instead of three G-tetrads, only two G-tetrads appear to form in the I14–Tel23 G-quadruplex. We have site-specifically labeled each guanine of I14–Tel23, one at a time, with low-enrichment (6%) incorporation of 15N-labeled guanine nucleoside. Using 15N-filtered experiments as previously reported (26–28,39,40), the imino (one-bond connection to N1) and base aromatic H8 (two-bond connection to N7) protons of guanines were unambiguously assigned. The assignment of imino protons of guanines of I14–Tel23 is shown in Figure 2. In addition, I14–Tel23 also shows four weaker imino peaks, with two more upfield (10.1–10.5 ppm) and two more downfield (12.1–12.6 ppm) relative to the eight strong imino protons of the two G-tetrads (Figure 1B bottom). We were also able to assign the three weaker imino guanine protons (G10, G22 and G4) using 15N-filtered experiments. The base H6 and methyl proton resonances of thymines have been unambiguously assigned by substituting deoxyuridine (dU) for each thymine one at a time. Complete assignment of the proton resonances of I14–Tel23 was accomplished by using 2D-NOESY, TOCSY and COSY data at various temperatures, and the standard DNA sequential assignment procedure (Figure 3 and Table 1). We were also able to assign the weak imino proton at ∼12.3 ppm at 20°C to be I14NH1 (Figure 1B bottom) based on the strong NOE with its H2 proton (Figure 3A). The VT studies of I14–Tel23 by NMR shows most imino protons of the two G-tetrads can be detected at 60°C (Supplementary Figure S2). Notably, the imino proton of G10 can still be detected at 50°C. The melting temperature of this G-quadruplex is 54°C as measured by CD (Supplementary Figure S3).
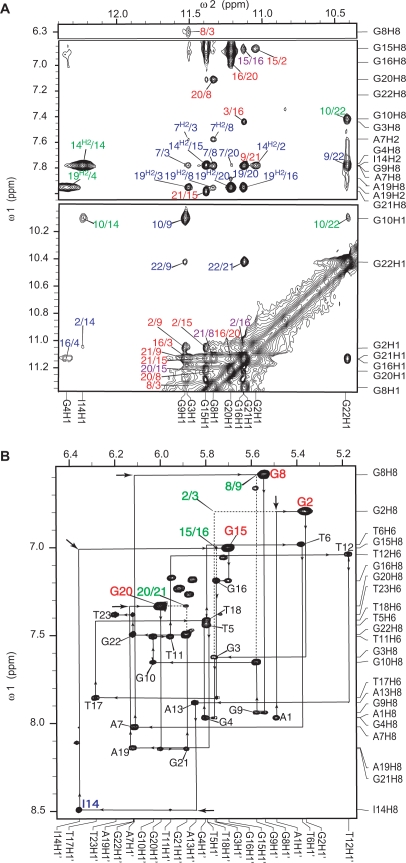
Figure 3.
(A) The expanded H1–H8/H2/H6 (top) and H1–H1 (bottom) regions, with labeling, of the exchangeable proton 2D JR-NOESY spectrum of I14–Tel23 in K+ solution at 1°C. Only the aromatic H2 protons are specified in the labels. NOEs are labeled as following: intra-tetrad NOEs are labeled in red, inter-tetrad NOEs are labeled in purple, intra-triple NOEs are labeled in green, NOEs between the G-tetrads and the stacking triple structures are labeled in blue. (B) The expanded H8/H6–H1′ region of the non-exchangeable 2D-NOESY spectrum of I14–Tel23 in K+ solution at 20°C. The sequential assignment pathway is shown. The H8–H1′ NOEs are labeled with residue names. The residue names of the guanines with syn conformation are in red. A3 and A15 show much broader peaks, likely related with the dynamic motion. Missing connectivities are labeled with arrows. The characteristic G(i)H8/G(i + 1)H1′ NOEs for the syn G(i)s are labeled in green. Conditions: 25 mM pH 7.0 K-phosphate, 70 mM KCl, 2.0 mM DNA.
Table 1.
Proton chemical shifts for the I14–Tel23 at 20°Ca
| H6/H8 | H2/Me | H1′ | H2′/H2″ | H3′ | H4′ | H5′/H5″ | NH1b | NH2a/NH2bb | |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 7.97 | 8.16 | 5.49 | 1.90/2.70 | 4.53 | 3.97 | 3.78,3.93 | ||
| G2d | 6.80 | 5.36 | 2.93/2.71 | 4.80 | 4.36 | 4.29 | 11.05 | 6.52/9.16 | |
| G3 | 7.63 | 5.76 | 2.11/2.34 | 4.91 | 4.38 | 3.94,4.16 | 11.51 | 5.95/9.31 | |
| G4 | 7.97 | 5.80 | 2.76/2.42 | 4.94 | 4.61 | 4.16,4.23 | 12.35 | 6.92/8.40 | |
| T5 | 7.44 | 1.77 | 5.79 | 1.67/1.41 | 4.37 | 3.78 | 3.54,3.83 | ||
| T6 | 6.99 | 1.19 | 5.38 | 1.64/1.96 | 4.36 | 3.72 | 2.97,3.30 | ||
| A7 | 8.02 | 7.83 | 6.11 | 2.11/3.06 | 4.64 | 4.18 | 3.65,3.80 | ||
| G8 | 6.59 | 5.54 | 2.99/2.71 | 4.80 | 4.26 | 4.15,4.21 | 11.34 | 6.01/9.43 | |
| G9 | 7.94 | 5.58 | 2.49/2.45 | 4.94 | 4.30 | 4.10,4.14 | 11.53 | 5.93/9.47 | |
| G10 | 7.66 | 6.03 | 2.52/2.52 | 4.96 | 4.45 | 4.17 | 10.10 | 5.91c/– | |
| T11 | 7.51 | 1.83 | 5.96 | 2.09/2.45 | 4.60 | 4.18 | 3.94,4.01 | ||
| T12 | 7.04 | 1.27 | 5.17 | 0.91/1.50 | 4.33 | 3.74 | 3.84,3.88 | ||
| A13 | 7.88 | 7.40 | 5.85 | 2.43/2.11 | 4.51 | 3.67 | 3.15 | ||
| I14 | 8.49 | 8.02 | 6.36 | 2.81/2.77 | 4.73 | 4.23 | 3.89,4.36 | 12.24 | |
| G15 | 7.00 | 5.70 | 3.02/2.66 | 4.73 | 4.49 | 4.13,4.37 | 11.39 | 6.78/9.25 | |
| G16 | 7.19 | 5.75 | 2.48/2.48 | 5.08 | 4.24 | 4.10,4.15 | 11.13 | 6.57/8.96 | |
| T17 | 7.86 | 1.95 | 6.28 | 2.27/2.37 | 4.89 | 4.43 | 4.10,4.27 | ||
| T18 | 7.42 | 1.61 | 5.80 | 1.44/1.98 | 4.74 | 4.17 | 3.79,3.94 | ||
| A19 | 8.14 | 8.19 | 6.12 | 2.77/2.68 | 4.95 | 4.30 | 3.79,3.96 | ||
| G20 | 7.33 | 6.00 | 3.36/2.87 | 4.89 | 4.38 | 4.16,4.32 | 11.22 | 6.89/8.624 | |
| G21 | 8.15 | 5.88 | 2.64/2.64 | 4.98 | 4.39 | 4.15,4.23 | 11.13 | 6.01/9.43 | |
| G22 | 7.50 | 6.12 | 2.34/2.55 | 4.91 | 4.38 | 4.21,4.23 | 10.42 | 5.98/– | |
| T23 | 7.38 | 1.30 | 6.20 | 2.18/2.25 | 4.45 | 4.01 | 3.99,4.10 |
aThe chemical shifts are measured in 25 mM K-phosphate, 70 mM KCl, pH 7.0 referenced to DSS.
bThe chemical shifts of NH1, NH21 and NH22 are measured at 1°C.
cThis chemical shift is measured at 20°C.
dGuanines in syn glycosidic conformation are in bold.
I14–Tel23 adopts a basket-type G-quadruplex structure with only two G-tetrads and extensive loop interactions in K+ solution
Many inter-residue NOE interactions are observed in 2D-NOESY of I14–Tel23 in K+. Critical inter-residue NOE interactions of I14–Tel23 are schematically summarized in Figure 5, which clearly define the overall structure of the unique G-quadruplex structure formed in I14–Tel23. I14–Tel23 forms a basket-type intramolecular G-quadruplex with two G-tetrads and multiple layers of capping structures in K+ solution (Figure 4A). The two G-tetrads are G2–G15–G21–G9 and G3–G16–G20–G8, with reversed arrangements of sugar glycosidic conformations, i.e. syn-syn-anti-anti and anti-anti-syn-syn, respectively. The assignment of the imino and base H8 protons of guanines leads to the direct determination of the folding topology of the G-quadruplex structure formed in I14–Tel23 in K+ solution (Figure 4A). In a G-tetrad plane with a Hoogsteen-type H-bond network, the imino proton NH1 of a guanine is in close spatial vicinity to the NH1s of the two adjacent guanines, and to the base H8 of one of the adjacent guanines (Figure 4A, left). Two G-tetrad planes were determined based on the NOE connectivities of exchangeable protons (Figure 3A). For example, the GNH1/GNH1 NOE interactions, including G2NH1/G15NH1, G15NH1/G21NH1, G21NH1/G9NH1, and G9NH1/G2NH1 (Figure 3A, bottom); and the GH1/GH8 NOE interactions, including G2NH1/G15H8, G15NH1/G21H8 and G21NH1/G9H8 (Figure 3A, top), define the G-tetrad plane of G2–G15–G21–G9 (Figure 4A). Similarly, G3NH1/G8NH1, G8NH1/G20NH1, G20NH1/G16NH1, G16NH1/G3NH1 and G3NH1/G8H8, G8NH1/G20H8, G20NH1/G16H8, G16NH1/G3H8 (Figure 3A) define the G-tetrad plane of G3–G16–G20–G8 (Figure 4A). The strong NOE interactions of G2NH1/G16NH1, G15NH1/G20NH1, and G21NH1/G8NH1 (Figure 3A) indicate that the two G-tetrads have reversed G-arrangements (Figure 4A). An expanded region for base and sugar H1′ protons of non-exchangeable proton NOESY with sequential assignment is shown in Figure 3B. Four guanine residues are in syn-conformation, including G2, G8, G15 and G20, as indicated by the very strong H8–H1′ NOE intensities (Figure 3B). The regular sequential NOE connectivities are either missing or very weak at the N(i)- synG(i + 1) steps, i.e., A1–G2, A7–G8, I14–G15 and A19–G20. The characteristic synG(i)H8/G(i + 1)H1′ NOEs are observed, including those of G2–G3, G8–G9, G15–G16 and G20–G21 (Figure 3B). A characteristic downfield shift is observed for the H2′/H2″ sugar protons of the syn-guanines (Figure 3B and Table 1). For the N(i)- antiG(i + 1) steps, the sequential NOE connectivities of the base H8 protons to the 5′-flanking residue sugar H1′/H2′/H2″ protons, typical for right-handed DNA twist, are clearly observed (Figure 3B).
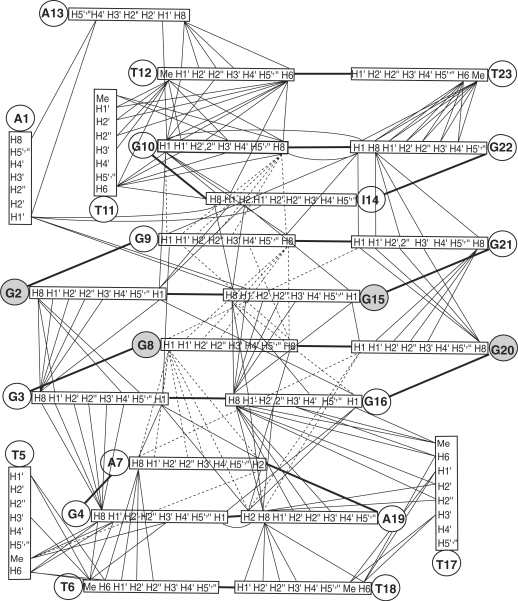
Figure 5.
Schematic diagram of inter-residue NOE connectivities of I14–Tel23 G-quadruplex formed in K+ solution. The guanines in syn conformation are represented using gray circles. The NOE connectivities clearly define the G-quadruplex conformation and provide distance restraints for structure calculation.
Figure 4.
(A) Schematic drawing of the folding topologies of the intramolecular G-quadruplex formed in I14–Tel23 in K+ solution (right). I14 is in orange. Red box = (anti) guanine, purple box = (syn) guanine, green box = adenine, blue box = thymine; red ball = guanine, green ball = adenine, blue ball = thymine. A G-tetrad with H1–H1 and H1–H8 connectivity patterns detectable in NOESY experiments is also shown in the inset. (B) Schematic drawing of the G10–I14–G22 and G4–A7–A19 triple structures with potential H-bonds. (C) CD spectra of several telomeric sequences shown in Figure 1A in pH7 K+ or Na+ solution at 25°C.
Remarkably, the two G-tetrads in the I14–Tel23 G-quadruplex show extensive interactions with the loop residues. Multi-layer capping structures appear to form involving the loop residues, covering the centrally located two G-tetrads. Specifically, the G2–G15–G21–G9 G-tetrad is covered by a G10–I14–G22 triple which is further capped by a T12–T23 base pair, while the G3–G16–G20–G8 G-tetrad is covered by a G4–A7–A19 triple which is further capped by a T6–T18 base pair (Figure 4A). Each loop residue exhibits extensive inter-residue NOE interactions as summarized in Figure 5 and Supplementary Table S1. The four layers of the capping structures are defined by extensive NOE interactions.
We have also site-specifically labeled each guanine of I14–Tel22 and have collected a full set of 2D NMR data on I14–Tel22 to have a complete assignment of I14–Tel22 (data not shown). Our NMR data indicate that the major G-quadruplex formed in I14–Tel22 is the same as that formed in I14–Tel23. However, minor conformations are much more evident in I14–Tel22, as the G10–I14–G22 triple structure was not stably formed in I14–Tel22 due to the absence of T23.
NOE-derived molecular structure of I14–Tel23 in K+ shows well-defined, multi-layer capping structures formed by loop residues
Solution structures of the I14–Tel23 G-quadruplex were calculated using a NOE-restrained distance geometry (DGSA) and molecular dynamics (RMD) approach (Figure 6, PDB ID 2kka), starting from an arbitrary extended single-stranded DNA. A total of 508 NOE distance restraints, of which 276 are from inter-residue NOE interactions, were incorporated into the NOE-restrained structure calculation (Table 2). Dihedral angle restraints were used for the glycosidic torsion angle (χ) based on the experimentally determined syn and anti conformations. The structure statistics are listed in Table 2.
Figure 6.
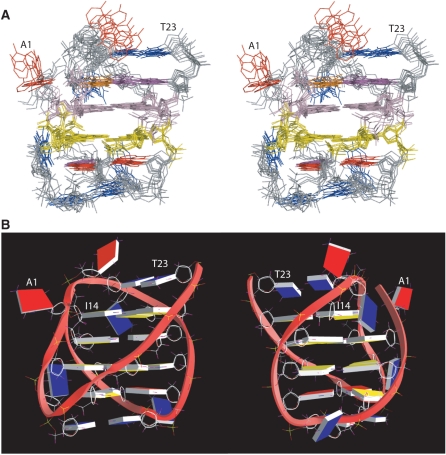
(A) Stereo view of the superimposed 8 NMR-refined structures of the two-G-tetrad G-quadruplex formed in I14–Tel23 in K+ solution. The G2–G15–G21–G9 tetrad is in light pink, and the G3–G16–G20–G8 is in yellow. The guanines in the G10–I14–G22 and G4–A7–A19 triples are in magenta, I14 in orange, adenines are in red, and thymines in blue. (B) A representative model of the NMR-refined I14–Tel23 G-quadruplex structure from two different views, prepared using GRASP (43) (guanine = yellow, adenine = red, thymine = blue).
Table 2.
Structural statistics for the I14–Tel23 G-quadruplex
| NMR distance and dihedral constraints | |
|---|---|
| Distance restraints | |
| Total NOE | 508 |
| Intraresidue | 232 |
| Interresidue | 276 |
| Sequential (|i-j| = 1) | 184 |
| Non-sequential (|i-j| > 1) | 92 |
| Hydrogen bonds | 21 |
| Total dihedral angle restraints | 16 |
| Structure statistics | |
| NOE violations | |
| numbers (>0.2Å) | 2.6 ± 0.9 |
| r.ms.d. of violation (Å) | 0.040 ± 0.014 |
| Deviations from idealized geometry | |
| Bond length (Å) | 0.007 ± 0.000 |
| Bond angle (°) | 1.345 ± 0.016 |
| Impropers (°) | 0.983 ± 0.005 |
| pairwise r.m.s.d. of heavy atoms (Å) | |
| Two G-tetrads | 1.10 ± 0.16 |
| With two triads | 1.21 ± 0.10 |
| With two T–T base pairs | 1.26 ± 0.10 |
| All residues | 1.59 ± 0.17 |
The ensemble of eight structures is selected based both on the minimal energy terms and number of NOE violations.
The I14–Tel23 in K+ forms a well-defined G-quadruplex with multi-layer capping structures formed by loop residues (Figure 6A). The two central G-tetrad layers are each covered by a middle triple structure, namely the G10–I14–G22 triple and the G4–A7–A19 triple, which are each further capped by an outer T–T base pair, i.e., T12–T23 and T6–T18. A representative model of the I14–Tel23 G-quadruplex structure in K+ is shown in two different views in Figure 6B. For the 23–nt I14–Tel23 sequence, except for the 8 guanines involved in the G-tetrad formation, there are 15 loop and flanking residues. Remarkably, 10 of the 15 loop residues are involved in the well-defined capping structures. For the remaining 5 loop residues, three thymines, T5, T11, and T17, are located in the grooves; while A1 and A13 have more flexible conformations.
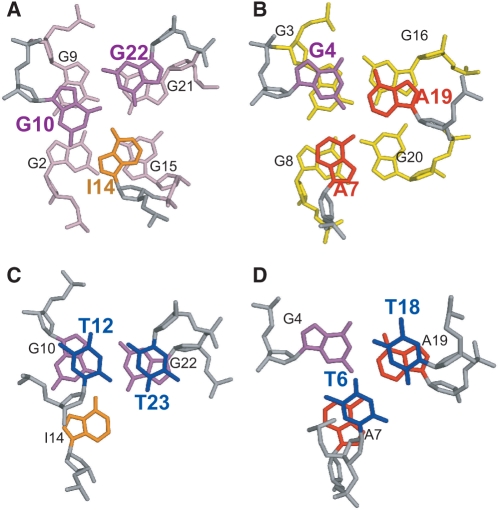
A well-defined G10–I14–G22 triple structure is found to cover the G2–G15–G21–G9 tetrad, with G10 stacking over G9 and G2, I14 stacking over G2 and G15, and G22 stacking over G21 and G9 (Figure 7A). Potential H-bonds are formed within the triple structure. G10NH1 is H-bonded with I14O6, I14NH1 is H-bonded with G22O6, and G22NH1 is H-bonded with G10O6, while G10NH2b is H-bonded with I14N7, and G22NH2b is H-bonded with G10N7 (Figure 4B, left, and Figure 7A). The imino protons of all three residues, i.e. G10NH1, I14NH1, and G22NH1, can be clearly detected in 1D 1H NMR up to 30°C between 10–12.5 ppm (Figure 1B and Supplementary Figure S2). The G10–I14–G22 triple structure is supported by experimental NMR data. The imino proton of G10 can be detected even at 50°C (Supplementary Figure S2). It is thus indicated that the imino protons of all three residues are involved in H-bonding, with the G10NH1 being the most stable. The chemical shifts of the imino protons of G10 and G22 are consistent with the NH-O hydrogen bonding, while the downfield shifting of I14 is typical for an inosine base. In addition, the NOEs between the imino protons and amino protons NH2b of G10 and G22 are both observed by NMR, indicating that the amino proton of the two guanines are also involved in H-bonding (data not shown). Moreover, NOE interactions between G10, I14 and G22 are observed, such as G22NH1/G10H8, G10NH1/I14H8, and G10NH1/I14NH1, and G10NH1/G22NH1 (Figures 3A and 5). The three residues of the G10–I14–G22 triple are well-stacked with the G2–G15–G21–G9 G-tetrad, as defined by extensive NOEs (Figure 5 and Supplementary Table S1). For example, G10 is above G9 along the G9–G2 side of the G2–G15–G21–G9 G-tetrad, as G10NH1 shows a strong NOE with G9NH1 and a weaker NOE with G2NH1 (Figures 3A and 5). I14 appears to stack above G15 along the G15–G2 side, with a strong NOE observed between I14H2/G15NH1 (Figure 3A). G22 appears to stack above G21–G9, with a strong NOE observed between G22NH1/G21NH1 and a clear NOE between G22NH1/G9NH1 (Figure 3A).
Figure 7.
Loop conformations in the two-G-tetrad G-quadruplex formed by I14–Tel23 in K+ solution. (A) The top view (as in Figure 6) of the G10–I14–G22 triple capping the G2–G15–G21–G9 G-tetrad. (B) The bottom view of the G4–A7–A19 triple capping the G3–G16–G20–G8 G-tetrad. (C) The top view of the T12–T23 base pair capping the G10–I14–G22 triple. (D) The bottom view of the T6–T18 base pair capping the G4–A7–A19 triple. The color code is the same as that in Figure 6A.
A second well-defined G4–A7–A19 triple structure is found to cover the G3–G16–G20–G8 tetrad, with G4 stacking over G3 and G16, A7 stacking over G8, and A19 stacking over G20 and G16 (Figure 7B). Potential H-bonds are formed within the G4–A7–A19 triple structure (Figure 4B, right). The imino proton of G4 is observed in 1H NMR (Figure 1B). The G4–A7–A19 triple structure is also supported by experimental NMR data (Figure 5 and Supplementary Table S1). The downfield-shifted chemical shift of G4NH1 indicates a N–H—N hydrogen bonding. In addition, the amino protons of G4 show clear NOEs with G4NH1 and appear to be H-bonded. Within the G4–A7–A19 triple, a very strong NOE is detected between G4NH1 and A19H2, and a clear NOE is detected between A7H2 and A19H2. Three residues of this triple structure, A19, A7, and G4, are all shown to stack well with the G3–G16–G20–G8 tetrad by NOE interactions (Figure 5 and Supplementary Table S1). A19H2 shows strong NOEs with G20NH1 and G16NH1, medium NOE with G3NH1 and weak NOE with G8NH1 (Figure 3A, top), indicating A19 is stacked right above the G20–G16 edge of the G3–G16–G20–G8 tetrad. A7H8 and H2 show clear NOEs with G8 and G3 (weaker) (Figure 3A, top), indicating A7 is stacked also above the G3–G16–G20–G8 tetrad but towards the G8–G3 edge. G4 shows nice stacking with G3, as indicated by inter-residue NOEs. In addition, G4NH1 shows a clear NOE with G16NH1.
In addition to the two triple structures immediately covering the two G-tetrads, each triple structure is further covered and stabilized by a T–T capping structure at each end of the I14–Tel23 G-quadruplex. Specifically, the G10–I14–G22 triple is capped by the T12–T23 base pair (Figure 7C), while the G4–A7–A19 triple is covered by the T6–T18 base pair (Figure 7D). The two T–T base pairs are also well supported by experimental NMR data. For the T12–T23 base pair, while T23 is well stacked with G22, T12 shows very good stacking with G10 (Figure 5 and Supplementary Table S1). Indeed, the presence of T23, and thus the formation of T23–T12 base pair, appears to be important in stabilizing the unique two-G-tetrad G-quadruplex. While the stabilization effect of the T23–T12 base pair to the G10–I14–G22 triple appears to improve the stability of the overall I14–Tel23 G-quadruplex, the T23–T12 base pair is critical in stabilizing the two-G-tetrad G-quadruplex in the Tel21–T sequence (37). Similarly, T18 and T6 also appear to stack well with A19 and G4, respectively. A notable upfield-shifting is observed for the sugar and base protons of T12 and T6 (Table 1), indicating that the two residues are positioned right above the neighboring triple structure and experiencing strong ring-current effect.
The I14–Tel23 G-quadruplex can only be clearly detected in the specifically truncated telomeric sequences with no 5′-flanking
We have previously shown that Hybrid-1 and Hybrid-2 types of intramolecular G-quadruplexes co-exist in extended four-G-tract human telomeric sequences with various lengths of 5′- and 3′- flanking segments (27). It is important to find out whether this new two-G-tetrad G-quadruplex can also form in the human telomeric sequences. We have tested all four-G-tract human telomeric sequences with various lengths of 5′- and 3′-flanking TTA segments. The formation of the new two-G-tetrad G-quadruplex can be monitored by the characteristic imino peak of G10 (Figure 1B). We found that the new two-G-tetrad G-quadruplex can only be clearly detected in the truncated telomeric sequences with no 5′-flanking. A 3′-flanking thymine (T23) is critical in stabilizing the two-G-tetrads G-quadruplex. The two-G-tetrad G-quadruplex is mostly clearly detected in Tel21–T and accounts for ∼60–65% of the total population (Figure 1B). This can be explained by the structural data, in that T23–T12 base pair stabilizes the G10–I14–G22 triple conformation and thus the overall two-G-tetrad G-quadruplex. However, as soon as a 5′-flanking segment is added, even a single adenine residue, the population of this two-G-tetrad form is prohibited. A–Tel21–T is the only telomeric sequence with a 5′-flanking residue (Figure 1A) in which the two-G-tetrad form can be detected; however, the population of the two-G-tetrad form is reduced to less than 10% of the total population in A–Tel21–T (Figure 1B), and is undetectable in any other four-G-tract human telomeric sequences with a 5′-flanking segment (Figure 1B and Supplementary Figure S1).
CD spectroscopic study
We have measured CD spectra for I14–Tel23, I14–Tel22, and wild-type four-G-tract telomeric sequences with various flanking segments in 100 mM K+ solution, pH 7. CD spectra of several sequences are shown in Figure 4C. I14–Tel23 and I14–Tel22 show very similar CD profiles in K+ solution. It is noteworthy that the CD profile of A–Tel21 (Figure 1A) in Na+ solution, which has also been shown to form a basket-type G-quadruplex (24), is very different from that of I14–Tel23. In addition, the Tel21–T sequence (Figure 1A), the only wild-type telomeric sequence in which the reported structure can be clearly detected (Figure 1B), also gives rise to a CD profile that is quite different from that of I14–Tel23. The CD spectra in K+ solution of wtTel26 and Tel26, which mainly form the Hybrid-2 and Hybrid-1 G-quadruplex, respectively, are also shown for reference. A peak at 255 nm appears to be associated with the reported G-quadruplex structure. This result indicates that the CD profile is mainly determined by the base-stacking rather than the strand-orientation of a G-quadruplex structure. The folding structure of a G-quadruplex cannot be determined purely on the CD profiles.
The CD melting profile of I14–Tel23 in K+ is shown in Supplementary Figure S3. The melting temperature of the I14–Tel23 quadruplex is about 54°C. We found that truncated telomeric sequences with multi-G endings give higher melting temperatures (Supplementary Figure S3), likely due to the presence of multiple structures including some higher-order structures. For example, both the A–Tel21 and Tel21–T sequences show markedly higher melting temperatures, while the two-G-tetrad form can only be detected in the Tel21–T sequence.
DISCUSSION
We report here a unique G-quadruplex structure formed in a variant human telomeric sequence, I14–Tel23 (Figures 6 and 7). This G-quadruplex is a basket-type structure with only two G-tetrads but multiple layers of capping structures. The extensive capping structures and loop interactions appear to stabilize the formation of this two-G-tetrad G-quadruplex. The two G-tetrads are sandwiched between the two triple capping structures, which are each further capped and stabilized by a T–T base pair (Figure 4). Specifically, the first G-tetrad G2–G15–G21–G9 (from the 5′-end of the sequence) is covered by the G10–I14–G22 triple, which is further capped by the T12–T23 base pair. The substitution of inosine for G14 of the human telomeric sequence (Figure 1A) appears to drive the formation of this two-G-tetrad G-quadruplex form, and the 3′-flanking T (T23) further stabilizes this structure, indicating that the formation of the G10–I14–G22 triple capping structure is critical for the stable formation of this two-G-tetrad quadruplex structure. The second G-tetrad G3–G16–G20–G8 is covered by the G4–A7–A19 triple, which is further capped by the T6–T18 base pair.
The four layers of the capping structures are very well defined by extensive NOE interactions (Figure 5 and Supplementary Table S1). For example, the G10–I14–G22 triple structure (Figure 7) is very well defined by the NMR experimental data. The imino protons of G10, I14 and G22 are clearly observed in 1D 1H NMR at 20°C (Figure 1B), indicating that all three imino protons are involved in H-bonding. This observation is consistent with the G10–I14–G22 triple conformation, in which G10NH1 is H-bonded with I14O6, I14NH1 is H-bonded with G22O6, and G22NH1 is H-bonded with G10O6 (Figure 4B). The G10–I14–G22 triple structure appears to be quite stable with the imino proton of G10 detectable even at 50°C (Supplementary Figure S2). A similar structure was very recently reported using a 8–Br-G-substituted Tel21–T sequence (BrG7–Tel22) in K+ (37). The structure formed by BrG7–Tel22 is also a basket-type two-G-tetrad G-quadruplex with two triple capping structures; however, our structure differs from that structure in the capping structures and loop interactions in several aspects. While both structures showed two capping triples, the conformations of both triples in our structure differ from that reported for BrG7–Tel22. Specifically, for the G10–I14–G22 triple, the conformation of I14 in our structure is different from G13 of the equivalent G9–G13–G21 triple in the structure reported for BrG7–Tel22 (37). Note that our numbering system is based on Tel 23 (A–Tel21–T, see Figure 1A) and thus is one number larger. G13NH1 is not shown to form any H-bond in the BrG7–Tel22 structure. However, this imino proton is clearly detected at ∼11.2 ppm at 20°C for the native Tel22 sequence (Supplementary Figure S1, G14 of Tel21–T). In addition, the remarkable stability of G10NH1 [also observed for the equivalent G9NH1 of BrG7–Tel22 (37)] may be better explained by the G10–I14–G22 triple conformation in our structure (Figure 4B), in which the additional hydrogen bonds formed by G10NH2b:I14N7 and I14NH1:G22O6 can stabilize the G10NH1:I14O6 hydrogen bond. For the G4–A7–A19 triple, the conformation of A7 in our structure is different from the equivalent A6 in the structure of BrG7–Tel22 (37). The G4–A7–A19 triple conformation in our structure (Figure 4B) is supported by the extensive NOE interactions (Figure 5 and Supplementary Table S1). For example, a clear NOE interaction is observed between A7H2 and A19H2, which are rather far in distance in the structure of BrG7–Tel22 (37). In addition, the T6–T18 capping structure was not shown to form in the molecular structure of BrG7–Tel22, but the possibility has been mentioned in the text (37).
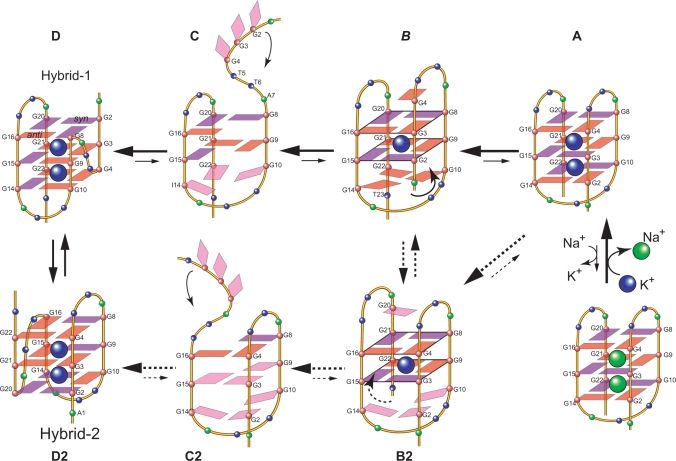
Most importantly, our results show that this structure can only be detected in the specifically truncated telomeric sequences without any 5′-flanking residues, and thus do not support the conclusion by Lim et al. that the two-G-tetrad structure is a stable new form for the extended human telomeric sequences (37). As soon as a 5′-flanking segment is present, even with a single adenine residue, the formation of this two-G-tetrad form is abolished (Supplementary Figure S1). A–Tel21–T is the only telomeric sequence with a 5′-flanking segment (Figure 1A) in which the two-G-tetrad form can be weakly detected; however, the population of the two-G-tetrad form is less than 10% in A–Tel21–T. The two-G-tetrad form cannot be detected in any other four-G-tract human telomeric sequences with a 5′-flanking segment (Figure 1B and Supplementary Figure S1). Therefore, this two-G-tetrad form is unlikely to be a general form of the G-quadruplex formed in the extended human telomeric sequences. Instead, our results suggest that this two-G-tetrad conformation is likely to be an intermediate transition form of the interconversion between different telomeric G-quadruplex conformations (Figure 8). We have previously proposed a model for the interconversion between the hybrid-type K+ form and the basket-type Na+ form telomeric G-quadruplex structures through a strand-reorientation mechanism (26). The 5′-G-strand of the basket-type G-quadruplex (Figure 8A) may dissociate from the core G-tetrads (Figure 8C) and swing back to the other side of the second G-stand to form a parallel-stranded structural motif with a double-chain-reversal loop (Figure 8D). Intriguingly, the two-G-tetrad form appears to fit the intermediate state between the transition from Figure 8A to C: when the 5′-strand is sliding up to dissociate from the core G-tetrads, a two-G-tetrad conformation can be formed transiently, which may then be stabilized by specific loop interactions of a specific sequence (Figure 8B). For example, in the case of I14–Tel23, the substitution of G14 by inosine appears to first destabilize the 5′-G-tetrad of the three G-tetrad form and subsequently capture the formation of the two-G-tetrad form. Specifically, as inosine lacks the amino group at C2, the G-to-I substitution at G14 loosens the H-bonding connection between G14 and G2 in the 5′-G-tetrad of the three G-tetrad basket form (Figure 8A). The 5′-end of the basket-type form becomes more flexible and can start to slide up, which is subsequently captured at the two-G-tetrad stage (Figure 8B) due to the stable formation of the G10–I14–G22 capping structure (which is further stabilized by the presence of T23 due to the T23–T12 capping structure). Interestingly, the glycosidic conformations of the tetrad guanines in the two-G-tetrad form are the same as those in the hybrid-1 form (Figure 8D); thus the presence of I14 appears to loosen up the 5′-end and favor the 5′-end dissociation (Figure 8B) for the conversion to the hybrid-1 form by the 5′-end swing-back rearrangement. A similar sliding (Figure 8B2) and dissociation (Figure 8C2) process could also happen at the 3′-end of the basket-form structure for the conversion to the hybrid-2 form G-quadruplex by the 3′-end swing-back rearrangement mechanism (Figure 8D2). In addition, the two-G-tetrad form may also be an intermediate state of the interconversion between the two hybrid-type forms (Figure 8). As previously shown, while the energy difference of the two hybrid-forms is quite small so that the two forms can co-exist in the extended telomeric sequences, the kinetics of the interconversion between the two forms appears to be rather slow, which cannot be detected on the NMR time scale (ms) (27), indicating the energy barrier of the intermediate state(s) is likely to be high. In addition, it has been shown that the unfolding processes of the telomeric G-quadruplexes have multiple intermediate states (35,36). Taken together, a possible model may be proposed for the interconversion of the two hybrid forms with multiple intermediate states, including the two-G-tetrad forms (Figure 8, D↔C↔B↔B2↔C2↔D2).
Figure 8.
Schematic diagram of a proposed model for interconversions between the Na+ basket form (A) and the K+ hybrid form of telomeric G-quadruplexes (D and D2), and between the two hybrid forms telomeric G-quadruplexes (D and D2). Guanines with known anti conformation are in red, and guanines with syn conformation are in purple. The A–Tel21 forms a single stable basket-type intramolecular G-quadruplex in Na+ solution. In the presence of K+ the basket form (A) is not favored and A–Tel21 can convert to the hybrid forms (D and D2) through a strand-reorientation mechanism (C and C2). The two-G-tetrad conformation is likely to be the intermediate state of this strand reorientation and may be stabilized by specific loop interactions of a specific sequence. For example, the presence of I14 loosens the H-bonding connection between G14 and G2 in the basket-form (A) and thus frees the 5′-end for its sliding-up dissociation from the core G-tetrads. However, this strand sliding-up reorientation is captured at the two-G-tetrad stage (B) due to the stable formation of the G10–I14–G22 capping structure. A similar sliding (B2) and dissociation (C2) could also potentially happen at the 3′-end of the basket-form structure to convert to the hybrid-2 form G-quadruplex by the 3′-end swing-back (D2). The two-G-tetrad form may also be an intermediate state of the interconversion between the two hybrid-type forms (D↔C↔B↔B2↔C2↔D2 circle).
It is noteworthy that the CD profile of this basket-type two-G-tetrad G-quadruplex is very different from the CD profile of the basket-type telomeric G-quadruplex formed by A–Tel21 in Na+ solution (Figure 4C). In addition, the CD melting profile shows that the melting temperature of the I14–Tel23 quadruplex is about 54°C (Supplementary Figure S3). The two-G-tetrad form does not appear to be more stable than the hybrid forms, as suggested in the previous report (37). Although the Tel21–T sequence shows a higher melting temperature, the increased Tm is likely caused by the presence of multiple structures, as evident by its 1D 1H NMR spectrum (Figure 1B). Furthermore, the A–Tel21 sequence, which does not appear to form the two-G-tetrad structure (Figure 1B), shows a similarly increased melting temperature (Supplementary Figure S3).
ACCESSION NUMBER
PDB ID 2kka.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (1S10RR16659 and CA122952). The China Scholarship Council for Chinese Ph.D. candidates to study abroad (to Z.Z.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr Emmanuel Hatzakis for his assistance with the CD experiments. They also thank Megan Carver for proofreading the paper.
REFERENCES
- 1.Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 2.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 3.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 4.Mergny JL, Helene C. G-quadruplex DNA: A target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 5.Sun DY, Hurley LH. Targeting telomeres and telomerase. In: Chaires JB, Waring MJ, editors. Methods in Enzymology, Drug-Nucleic Acid Interactions. Vol. 340. San Diego: Academic Press Inc; 2001. pp. 573–592. [DOI] [PubMed] [Google Scholar]
- 6.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 7.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 8.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 9.Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutation Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's; Syndrome Helicase Unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 11.Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 13.Allshire RC, Dempster M, Hastie ND. Human Telomeres Contain at Least 3 Types of G-Rich Repeat Distributed Non-Randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol. Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 18.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 21.Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem. Soc. Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 22.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han HY, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 23.Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Patel DJ. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 26.Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai JX, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang DZ. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)(3)] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 30.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 33.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai JX, Carver M, Yang DZ. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)(4) repeat in a potassium solution. Biochemistry. 2007;46:4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- 36.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim KW, Amrane S, Bouaziz S, Xu WX, Mu YG, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrus A, Chen D, Dai JX, Jones RA, Yang DZ. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter implications for g-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 39.Dai JX, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang DZ. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai JX, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai JX, Punchihewa C, Mistry P, Ooi AT, Yang DZ. Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug. J. Biol. Chem. 2004;279:46096–46103. doi: 10.1074/jbc.M404053200. [DOI] [PubMed] [Google Scholar]
- 42.Brünger AT. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. New Haven, CT: Yale University Press; 1993. [Google Scholar]
- 43.Nicholls A, Sharp KA, Honig B. Protein folding and association – insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.