Abstract
The activity of the telomerase ribonucleoprotein enzyme is essential for the maintenance of genome stability and normal cell development. Despite the biomedical importance of telomerase activity, detailed structural models for the enzyme remain to be established. Here we report a single-molecule assay for direct structural analysis of catalytically active telomerase enzymes. In this assay, oligonucleotide hybridization was used to probe the primer-extension activity of individual telomerase enzymes with single nucleotide sensitivity, allowing precise discrimination between inactive, active and processive enzyme binding events. FRET signals from enzyme molecules during the active and processive binding events were then used to determine the global organization of telomerase RNA within catalytically active holoenzymes. Using this assay, we have identified an active conformation of telomerase among a heterogeneous population of enzymes with distinct structures.
INTRODUCTION
Telomerase is a specialized reverse transcriptase that solves the end replication problem by adding short G-rich telomere DNA repeats to the ends of eukaryotic chromosomes (1,2). The critical importance of telomerase regulation in promoting proper cell development and the maintenance of genome stability is evidenced by the hyper-activation of telomerase in greater than 85% of human tumors (3). Moreover, mutations that result in impaired telomerase function have been linked to hereditary premature aging diseases such as dyskeratosis congenita (4). Although these findings suggest telomerase detection and inhibition may prove useful in the diagnosis and treatment of these diseases, efforts to develop telomerase-based therapies are limited by an incomplete understanding of telomerase structure and catalytic mechanism.
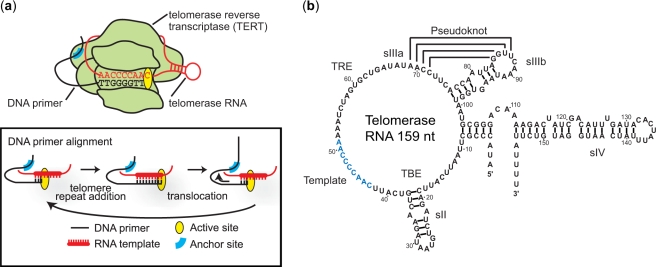
The telomerase holoenzyme is a ribonucleoprotein (RNP) comprised of several components, including (i) the telomerase RNA, which provides the template for telomere synthesis, (ii) the telomerase reverse transcriptase (TERT), which serves as the catalytic subunit, and (iii) several accessory protein cofactors involved in the regulation of enzyme assembly and activity (1,5). The telomerase catalytic cycle has three stages: DNA primer alignment, telomere repeat addition and translocation (Figure 1a) (6). During DNA primer alignment, telomerase binds a single-strand DNA substrate via Watson–Crick base-pairing with the RNA template and through TERT-DNA interactions at the ‘anchor site’ (7). During the subsequent telomere repeat addition phase, the TERT subunit catalyzes the addition of one telomere DNA repeat according to the sequence specified by the RNA template. Upon completion of one full repeat sequence, a series of molecular rearrangements take place to reposition TERT, telomerase RNA and the DNA substrate during a translocation process, such that the enzyme is positioned to catalyze the addition of the next telomeric repeat.
Figure 1.
Telomerase catalytic cycle and RNA secondary structure. (a) Cartoon showing a telomerase RNP bound to a DNA primer. Inset: the three general stages of the telomerase catalytic cycle, primer alignment, telomere repeat addition and translocation, are depicted. (b) The 159 nucleotide Tetrahymena thermophila telomerase RNA consists of several conserved structural elements important for enzyme assembly and catalysis. The TBE at the base of stemloop II (sII) serves as a high affinity binding site for TERT and helps to define the end of the RNA template (blue) used in the synthesis of telomeric DNA (32). The TRE functions to position the adjacent template sequence in the active site and is important for repeat addition processivity (33). Stemloop IIIa (sIIIa) and IIIb (sIIIb) together form a putative pseudoknot structure required for telomerase activity (14,22,34). The stemloop IV (sIV) is proposed to potentiate folding of the pseudoknot and plays critical role in enzyme assembly and activity (14,16).
Secondary structure models of the telomerase RNA derived from ciliates (8), yeast (9,10) and vertebrates (11) reveal several evolutionarily conserved RNA structural motifs, suggesting that the RNA may serve more than a template function during reverse transcription. Indeed, extensive biochemical studies have identified specific regions of the RNA that are required for enzyme assembly, catalysis and processivity (Figure 1b). In particular, conformational dynamics within the RNA pseudoknot of human telomerase RNA have been proposed to function as a molecular switch that drives enzyme translocation (12,13). Moreover, mutation studies have implicated stemloop IV of Tetrahymena telomerase RNA in the regulation of telomerase activity and repeat addition processivity (14,15). Chemical footprinting experiments suggested that the folding of a predicted pseudoknot motif within telomerase RNA may be potentiated by stemloop IV (16), providing a possible structural basis for the reported catalytic defects observed in the absence of this conserved structural element. Although these traditional biochemical and structural studies have provided considerable insight into aspects of telomerase function, the lack of high-resolution structure of active telomerase RNP and appropriate assays for studying functionally relevant conformational dynamics of telomerase has rendered structure-function models for the enzyme speculative. It is therefore of critical importance to determine the functional structure of the telomerase RNA.
Ensemble methods for determining enzyme structure typically require large quantities of input material, and often suffer from an implicit assumption of structural and functional homogeneity of the enzyme. Satisfying the latter requirement for telomerase is particularly challenging due to the difficulty in preparing a homogeneous population of functionally active telomerase enzymes. The widely employed reconstitution strategy utilizing partially purified cell lysates is believed to produce a heterogeneous mixture of both active and inactive telomerase, which likely assume different structures (17). To circumvent this difficulty, we developed a novel telomerase structure-function assay based on single-molecule fluorescence resonance energy transfer (FRET) (18–20). Single-molecule approaches have been used to study the folding, assembly and composition of telomerase (21–23). Here we exploit single-molecule FRET to detect the conformation of individual enzymes and to directly correlate the enzyme conformation with catalytic outcome. Using this assay, we have characterized the relative orientation of stemloop IV and the RNA pseudoknot region as well as the conformational state of the template region of Tetrahymena telomerase RNA within active holoenzyme.
MATERIALS AND METHODS
Preparation of FRET-labeled telomerase RNA
Preparation of labeled telomerase RNA was described previously (21). Briefly, wild-type telomerase RNA were transcribed in vitro using T7 RNA polymerase from PCR templates containing the full length telomerase RNA sequence downstream of a T7 promoter. Synthetic RNA precursors containing site-specific amine modified uracil (5NU, Dharmacon) were labeled with either Cy3 or Cy5 monoreactive dye (GE Healthcare) according to manufacturer’s protocol. The RNA precursors were then gel purified with 8% 7M Urea denaturing Poly-Acrylamide Gel Electrophoresis (PAGE) and HPLC purified by reverse phase chromatography on a C-8 column (GE Healthcare). All ligation precursors, including both synthetic and transcribed RNA, were treated with RNAse H (NEB) in the presence of chimeric 2′-O-methyl RNA/DNA oligonucleotides (Dharmacon) to generate the desired ligation ends, followed by PAGE purification. RNA precursors were then annealed in water to DNA splints at 95°C for 4 min, then 37°C for 10 min. Ligation reactions were incubated at 37°C for 1–2 h in the presence of T4 RNA Ligase 2 (NEB) and ligation buffer (NEB). The full-length ligation products of labeled telomerase RNA were subsequently PAGE purified from unligated precursors and partial products. (See Supplementary Table S1 online for the oligonucleotides used in the present study.)
Reconstitution and purification of FRET-labeled telomerase
FRET-labeled telomerase RNAs were reconstituted into functional telomerase RNPs using TnT Coupled Rabbit Reticulocyte Lysate System (Promega). Specifically, reconstitution reactions containing 1.25 ng/µl of labeled RNA, 41.7 ng/µl pFLAG-TERT (gift from Dr Tracy Bryan), and 8.3 ng/µl pCITE-p65 (gift from Dr Kathy Collins) were set up according to manufacturer’s protocol. After 90 min incubation at 30°C, a small aliquot of each reaction was flash frozen in liquid nitrogen and saved for FRET experiments on surface-anchored enzymes. In these single-molecule experiments, the enzymes were anchored to a slide surface coated with anti-FLAG antibodies through the FLAG-anti-FLAG linkage. Such surface-attachment also serves the purification purpose, and thus an additional purification step was not needed. The remaining reconstitution reaction was immediately purified with anti-FLAG M2 affinity agarose beads (Sigma-Aldrich) as previously described (17). Elution of the purified telomerase RNPs from the beads was accomplished by incubation with a wash buffer (20 mM Tris acetate pH 7.5, 10% v/v glycerol, 1 mM EDTA, 5 mM MgCl2) supplemented with 0.75 mg/ml 3× FLAG peptide at 4°C for 1 h with agitation followed by centrifugation at 1500×g for 2 min. Supernatant containing the eluted enzyme was divided into aliquots and immediately flash frozen in liquid nitrogen and stored at –80°C. These purified enzymes were subsequently used in the ensemble primer extension assay and single-molecule structure-function assay. Concentrations of the eluted enzyme were determined using a dot blot assay with a 5′ radiolabeled probe (5′ TAT CAG CAC TAG ATT TTT GGG GTT GAA TG 3′) against telomerase RNA.
Ensemble primer extension assay of telomerase activity
Equal amounts of dye-labeled or unlabeled purified telomerase RNPs were diluted in 1× telomerase buffer (50 mM Tris–Cl pH 8.0, 1.25 mM MgCl2, 10% v/v glycerol), plus 1 µM biotinylated-(TG)8T2G4T2 primer, 100 µM dTTP, 10 µM [α-32P] dGTP at 80 Ci/mmol (PerkinElmer Life Sciences) and incubated at room temperature for 1 h. Reaction products were analyzed by denaturing PAGE and autoradiography. To quantify activity, the total intensity of all major bands, normalized by the intensity of a loading control, was measured for each reaction (ImageQuant). To quantify processivity, the intensities of individual repeat bands, after adjustment of dG content, were normalized against the loading control and plotted against the repeat number in a semi-log plot. A linear fit to each sample was determined and the negative inverse of the slope was defined as the processivity. The relative activity and processivity values were expressed as a percentage of those determined for the unlabeled enzyme.
Single-molecule FRET
Quartz slides were coated with polyethylene glycol (PEG), biotinylated PEG and streptavidin as described (24). A blocking step with 10 mg/ml BSA (NEB) was added prior to deposition of streptavidin to reduce non-specific adsorption of molecules onto the slide surface. Unless otherwise specified, all FRET measurements were performed at room temperature (23°C) in imaging buffer (1 × telomerase buffer plus 0.5 mg/ml BSA, 2 mM Trolox to suppress Cy5 blinking (25), and an oxygen scavenger system consisting of 10% w/v glucose, 300 µg/ml glucose oxidase, and 40 µg/ml catalase) using a prism-type total internal reflection fluorescence (TIRF) microscope with an back-illuminated electron multiplying CCD camera (Andor Ixon 887) controlled by a custom-written LabView software. The FRET donor Cy3 was excited by a 532 nm Nd:YAG laser (Crystal Laser). Emissions from donor and acceptor were separated from the excitation light using a 550LP filter and split using a 630 nm dichroic mirror (Chroma Technology) before imaging onto the two halves of the CCD chip. Emission filters were used in the Cy3 and Cy5 emission paths (545/135 for Cy3, 665LP for Cy5). In some experiments, we used an alternating laser excitation scheme (ALEX) (26) to unambiguously identify active Cy5 dyes (not photobleached or blinking). In this scheme, the molecules were illuminated with alternating four frames of 532 nm excitation and one frame of 635 nm excitation with a diode CUBE laser (Coherent) at a frame rate of 5 Hz. Data analysis software written in IDL was used to determine the time-dependent donor (ID) and acceptor (IA) intensities for each molecule. FRET is defined as IA/(ID+ IA) after subtracting the leakage of donor signal into the acceptor channel. With the 630 nm dichroic mirror and emission filters used, we characterized an 8% of leakage of donor signal into the acceptor channel.
Single-molecule FRET measurements of surface-anchored enzyme
Biotinylated protein-G (Sigma-Aldrich) and M2 Anti-FLAG antibody (Sigma-Aldrich) were sequentially deposited onto the streptavidin-coated quartz slides at 0.2 mg/ml in blocking buffer (20 mM Tris acetate pH 7.5, 10% v/v glycerol, 1 mM EDTA, 5 mM MgCl2, 100 mM potassium glutamate, 0.5 mg/ml BSA, 0.05 mg/ml glycogen, 0.1 mg/ml yeast tRNA). Reconstituted Cy3-Cy5-labeled telomerase RNPs (5–15 µl of unpurified lysate reaction plus blocking buffer to a total volume of 100 µl) were then attached to the surface through FLAG-anti-FLAG linkage. After exchanging with imaging buffer, FRET measurements were taken at 10 Hz. FRET histograms were constructed from the first 20 frames of each molecule excluding data derived from molecules containing bleached or blinked Cy5 (as verified by direct excitation). We have constructed the FRET distribution by using both a single excitation scheme (exciting FRET donor only) and an ALEX scheme as described in the ‘single-molecule FRET’ section. Both schemes yielded identical FRET distributions (data not shown).
Single-molecule FRET assay for probing telomerase activity
For proof of principle studies, Alexa Fluor-488 labeled standard primers corresponding to the three types of expected products from telomerase reaction (i.e. no extension product: (TG)8T2G4T2, single base extension product: (TG)8T2G4T2G, processive extension product: (TG)8T2G4T2G4T), each containing a 5′ biotin, were anchored to the streptavidin-coated quartz slides as described above. The Alexa Fluor-488 fluorophore allowed initial identification of DNA primer density and locations without interfering with subsequent Cy3-Cy5-based FRET measurements. Note that our telomerase reaction buffer used for the single-molecule structure-function assay as described later contained dGTP and the chain-terminating ddTTP. Thus we expect a processive primer extension to end after addition of a ddTTP nucleotide. Hence we used (TG)8T2G4T2G4T to represent the processive extension product here. After surface anchoring of the standard primers, a Cy3 containing labeling oligonucleotide (LO: Cy3-(CA)8) was then diluted to 20 nM in imaging buffer and flowed onto the slide to introduce a Cy3 dye to each primer through stable annealing to the non-telomeric region (the (TG)8 portion) of the primer. Subsequently, Cy5-labeled detection oligonucleotide (DO: Cy5-C2A2C3), complementary to the telomeric sequence, was diluted to 300 nM in imaging buffer and flowed onto the slide to probe the length of the surface-anchored primers through DNA hybridization. FRET between Cy3 and Cy5 was recorded at 30 Hz at room temperature with continuous excitation with 532 nm laser to monitor the kinetics and stability of hybrid formation between the primers and the DO.
To quantify the difference in hybrid stability among the three standard primer types, the hybridization free energy between a surface-anchored primer and the DO, ΔGhyb, was derived from their hybridization equilibrium: ΔGhyb = –kBT. ln (total duration of high FRET/total duration of low FRET). For each trajectory, 900 frames of data were used to calculate a free energy of hybridization without requiring a minimum number of binding events. Using the ΔGhyb distributions obtained from the three standard primer types, we determined two ΔGhyb threshold values that were used to distinguish the telomerase reaction products. First, the mean ΔGhyb values for the standard no extension primers and the standard single base extension primers were both incremented by the same multiple of their respective standard deviations until the ΔGhyb values of the two cases equalized; the resulting ΔGhyb was then taken to be the first threshold value ΔGhyb_1. Likewise, the mean ΔGhyb values for the standard single base extension primers and the standard processive extension primers were both incremented by the same multiple of their respective standard deviations until the ΔGhyb values of the two cases equalized; the resulting ΔGhyb was then taken to be the second threshold value ΔGhyb_2. For a given reaction product, if its ΔGhyb value satisfies ΔGhyb ≥ ΔGhyb_1, we assign it as a no extension product; if its ΔGhyb value satisfies ΔGhyb_1 > ΔGhyb ≥ ΔGhyb_2, we assign it as a single base extension product; if its ΔGhyb value satisfies ΔGhyb < ΔGhyb_2, we assign it as a processive extension product.
Single-molecule structure-function assay for telomerase
Alexa Fluor-488 labeled and 5′ biotinylated (TG)8T2G4T2 primers were anchored to the streptavidin-coated quartz slides as described above. Fluorescence emission was recorded at 5 Hz during the first 60 frames under 460 nm excitation, which allowed visualization of the Alexa Fluor-488 labeled primers. Subsequently, the purified Cy3-Cy5-labeled telomerase RNPs were diluted in imaging buffer containing 10 µM dGTP and 100 µM chain-terminating ddTTP and flowed onto the slide to initiate the enzyme binding phase of the experiment. Data were collected with alternating four frames of 532 nm excitation and one frame of 635 nm excitation at 0.2 s/frame for 20 min. At the end of the binding phase, the enzyme solution was exchanged with the labeling solution (imaging buffer containing 20 nM Cy3-labeled LO) that simultaneously washed out the enzymes and labeled the primers with a Cy3 donor dye. Subsequently, a detection solution (imaging buffer containing 300 nM Cy5-labeled DO) was immediately flowed onto the slide to wash out the excess LO and to initiate the activity detection phase of the experiment. Data were acquired at 30 Hz during the detection phase with continuous 532 nm excitation. The enzyme binding and activity detection phases were then combined and analyzed. Enzyme binding events were identified and classified according to their catalytic outcomes. Histograms of FRET from individual telomerase binding events were constructed by taking the first 10 frames of each binding event, excluding data derived from bleached or blinked Cy5 (as verified by direct excitation).
RESULTS
Generation of FRET-labeled telomerase RNP
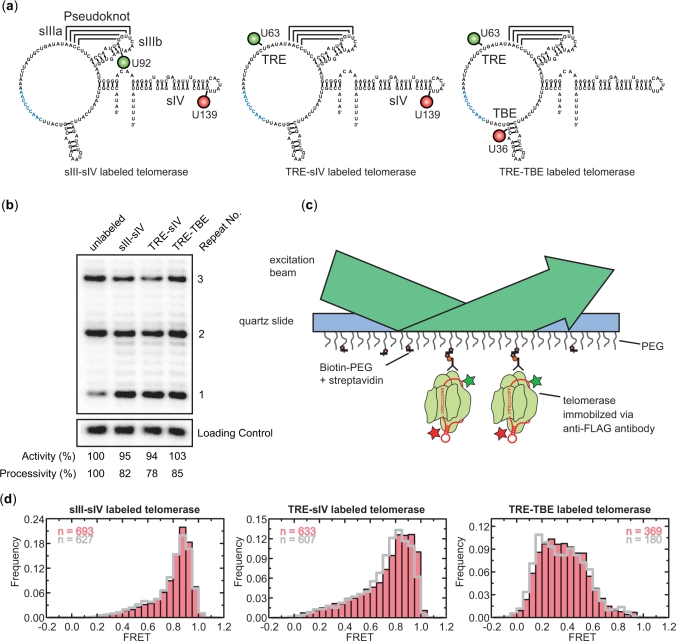
FRET-labeled telomerase RNAs were generated to monitor the conformation of functionally important regions of the RNA. Specifically, Cy3-Cy5 FRET pairs introduced into RNA positions U92-U139 (Figure 2a, left), U63-U139 (Figure 2a, middle) and U63-U36 (Figure 2a, right) were used to monitor the relative distances between stemloop III (sIII) and stemloop IV (sIV), Template Recognition Element (TRE) and sIV, and TRE and Template Boundary Element (TBE), respectively. The FRET-labeled RNAs were co-assembled with FLAG-tagged TERT and a protein cofactor p65 using standard telomerase rabbit reticulocyte lysate reconstitution protocols. Assembled telomerase holoenzymes were affinity purified from the lysate via the FLAG tag. The catalytic activity of the labeled enzymes was then verified to be similar to that of unlabeled enzyme in an ensemble primer extension assay utilizing the primer (TG)8T2G4T2 (Figure 2b).
Figure 2.
The FRET-labeled telomerase RNPs display structural heterogeneity. (a) The three FRET-labeled telomerase RNA constructs used in this study are shown. Cy3/Cy5 FRET pairs introduced into the positions U92/U139, U63/U139 and U63/U36 were used to monitor the relative distance between sIII and sIV, between TRE and sIV and between TRE and TBE of telomerase RNA, respectively. Cy3 and Cy5 are shown as green and red circles, respectively. (b) Ensemble primer extension assay performed with telomerase RNPs reconstituted from both labeled and unlabeled telomerase RNAs demonstrates the dye labeling did not significantly affect enzyme function. Activity and processivity were quantified and normalized against the values derived from unlabeled enzyme. A 16 bases oligonucleotide radio-labeled with 32P was used as a loading and recovery control in the activity assay. (c) Schematics showing the surface anchoring scheme. Antibodies against the FLAG tag at the N-terminus of TERT were used to anchor telomerase RNPs directly onto the quartz slide. Data were collected using a prism type total internal reflection fluorescence (TIRF) microscope. (d) FRET histograms derived for the sIII-sIV labeled, TRE-sIV labeled and TRE-TBE labeled enzymes in the absence (pink bars) and presence (gray lines) of 1 µM (TG)8T2G4T2 primer, 10 µM dGTP and 100 µM ddTTP.
FRET measurement reveals enzyme structural heterogeneity
To assess the conformation of telomerase RNA within reconstituted enzymes, the labeled telomerase RNPs were anchored on quartz slides through binding of the FLAG epitope on TERT to anti-FLAG antibodies on the slide surface. FRET values from individual telomerase RNPs were recorded using a prism-type total internal reflection microscope and a CCD camera (Figure 2c). FRET histograms were constructed from the measured FRET values of many molecules, each molecule contributing an equal number of points. Specifically, the histogram derived from the sIII-sIV labeled telomerase displayed a primary peak centered on 0.85 with a minority population sampling lower FRET values (Figure 2d, left). Similarly, the FRET distribution of TRE-sIV labeled telomerase also displayed a primary peak centered around 0.85; however, a significant fraction of these molecules (∼30%) sampled FRET values below 0.7, thus contributing to the broad histogram seen for this construct (Figure 2d, middle). Finally, the TRE-TBE labeled enzyme, with the FRET dye pair flanking the template region, exhibited even more heterogeneous behavior in which molecules with FRET values ranging from 0.2 to 0.6 were observed without a clear dominant species (Figure 2d, right). The addition of DNA primer and dNTPs at saturating concentrations failed to suppress the observed heterogeneity in FRET for all three telomerase constructs (gray outline in Figure 2d). Moreover, the FRET time traces of individual molecules do not exhibit significant fluctuations (Supplementary Figure S1 online), indicating that the observed broad distributions of FRET primarily arose from relatively static heterogeneity among the telomerase RNPs rather than conformational fluctuations within individual enzyme.
The heterogeneous FRET behaviors seen in these constructs could be due to either the presence of a misfolded and thus inactive population of the enzyme or to the existence of alternate active conformations of telomerase RNPs. In order to distinguish these possibilities and to gain insights into the catalytically active conformation of the telomerase RNP complex, we developed a single molecule structure-function assay to directly correlate an observed FRET behavior in the enzyme and its ability to catalyze DNA primer extension.
Probing the catalytic reaction product of individual telomerase RNPs
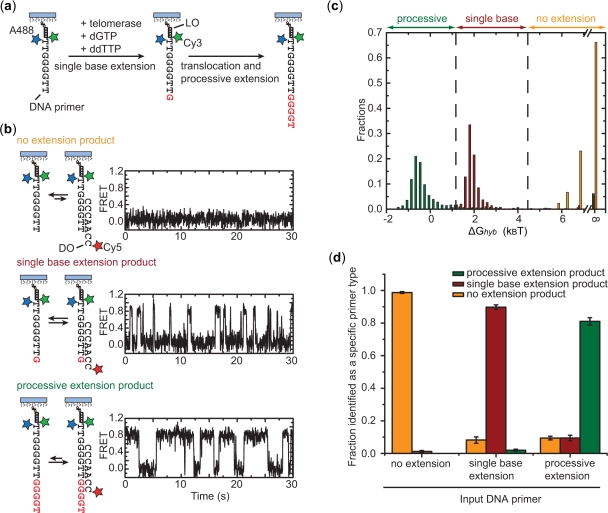
To probe telomerase catalysis, we first developed a method to identify reaction products resulting from a single enzyme–DNA primer encounter (Figure 3). In this assay, rather than anchoring FRET-labeled telomerase on the surface, we allowed soluble telomerase enzymes to bind to surface-anchored DNA primers terminating with the sequence T2G4T2 (Figure 3a). The sequence of the DNA primer was specifically designed to be one nucleotide shorter than the complete telomere DNA repeat. In the presence of dGTP and chain-terminating ddTTP, three classes of reaction products may be expected for this DNA substrate (Figure 3a): (i) the enzyme fails to extend the primer prior to dissociation, resulting in no extension; (ii) the enzyme generates a single base extension product by catalyzing the addition of a single dGTP to the primer, now ending with the sequence T2G4T2G; and (iii) upon reaching the template boundary after one nucleotide addition, the enzyme translocates and adds additional nucleotides until incorporating the chain-terminating ddTTP, characteristic of processive extension. As shown in Figure 2b, which is consistent with previous biochemical data (27), the telomerase-catalyzed primer extension primarily pauses or stops at the end of each repeat sequence with little accumulation of products corresponding to incomplete repeat synthesis. Therefore, the predominant processive extension product here in the presence of ddTTP will end with the sequence T2G4T2G4T, terminated by the incorporation of ddTTP. To differentiate these three possible reaction products, we designed a single-molecule hybridization assay in which a detection oligonucleotide (DO) was used to distinguish primers of different lengths. The sequence of the DO was designed to form a different number of base pairs with each class of reaction products. Consequently, the change in observed hybrid stability provided a direct measure of the primer length. We probed DNA hybridization in real time by recording the FRET signal between a donor dye (Cy3) on the primer and an acceptor dye (Cy5) on the DO.
Figure 3.
Single-molecule telomerase activity assay. (a) Schematics illustrating the three types of expected products from a telomerase-catalyzed primer extension reaction in the presence of dGTP and chain-terminating ddTTP. The Alexa Fluor-488 (A488, blue star) dye on the primer serves the purpose of primer density determination. Twenty nanomolar Cy3-labeled LO with a sequence complementary to the non-telomeric region of the primer is added to introduce the FRET donor Cy3 (green star) to the primer. (b) The three classes of primer-extension products show distinct hybridization kinetics with the Cy5-labeled DO, which is present at 300 nM in the imaging buffer. The hybridization kinetics were probed by FRET between the Cy3 (green star) on the primer and Cy5 (red star) on the DO. (c) The distribution of the hybridization free energy ΔGhyb derived from many primer molecules shows little overlap for standard primers representing the no extension (orange), single base extension (red) and processive extension (green) products of a telomerase reaction. The distribution for the no extension primers appears asymmetric because hybridizations between primer and DO that lasted shorter than one camera frame were indistinguishable and therefore were all binned as having a ΔGhyb equal to infinity. The vertical dashed lines represent the threshold values used to determine the length of an unknown primer. (d) Characterization of the error rates of the detection scheme using standard primers of known lengths: the orange, red and green bars indicate the fractions of primers identified as a no extension, single base extension and processive extension products. Error bars represent SD of the mean obtained from three independent experiments.
As a proof of principle, standard DNA primers corresponding to each of the expected products from the telomerase reaction (i.e. no extension (TG)8T2G4T2, single base extension (TG)8T2G4T2G, processive extension (TG)8T2G4T2G4T) were anchored to streptavidin-coated quartz slides via a terminal 5′ biotin modification. An Alexa Fluor-488 fluorophore was introduced into the primer, which allowed identification of DNA primer locations without interfering with subsequent Cy3-Cy5 based FRET measurements. Next, a Cy3-labeled oligonucleotide with the sequence (CA)8, referred to as the labeling oligonucleotide (LO), was stably annealed with the non-telomeric (TG)8 portion of each primer to introduce the FRET donor Cy3 to the primer. A Cy5-labeled detection oligonucleotide (DO) with complementary sequence (C2A2C3) to the telomeric region of the primer was then added to the slides to probe the length of the surface-anchored primers. FRET traces from individual primers exhibited characteristic fluctuation of FRET between two well defined levels (FRET = 0 and FRET = 0.8), indicating repetitive binding and dissociation of the Cy5-labeled DO (Figure 3b). A binding equilibrium, and consequently the hybridization free energy (ΔGhyb), can be derived from these traces directly. We note that the zero FRET events were not due to blinking of the Cy5 acceptor, which was rarely observed for Cy5 attached to DNA duplexes under the experimental condition used [in the presence of a blinking-suppression reagent, Trolox (25)]. Thus the FRET = 0 events observed here are primarily due to dissociation of the DO from the primer. This notion was further supported by the observation that the frequency and lifetime of the FRET = 0 events depended on the primer sequence and DO concentration (data not shown).
Analysis of the FRET time traces revealed substantial differences in the hybrid stability among the three primer types (Figure 3b). To quantify this effect, we derived the hybridization free energy (ΔGhyb) of the DO with each primer molecule and constructed a histogram of ΔGhyb for each primer type from hundreds of molecules (Figure 3c). The ΔGhyb distributions for the three primer types are well separated without significant overlap. We then defined the following quantitative criteria for product determination: no extension product (ΔGhyb ≥ 4.5 kBT), single base extension product (4.5 kBT > ΔGhyb ≥ 1.2 kBT), and processive extension product (ΔGhyb < 1.2 kBT). The false positive and false negative rates of this detection scheme were characterized by scoring primers of known sequences according to the above criteria and evaluating the probability of erroneous assignment (Figure 3d). Both false positive (i.e. scoring a primer as longer than its actual length) and false negative (i.e. scoring a primer as shorter than its actual length) detection rates were found to be low. Importantly, the false positive rate of under 2% ensures the accurate determination of the active structure of telomerase, since an inactive binding event of the enzyme would rarely be mistaken as an active one. Moreover, this activity detection scheme has single nucleotide sensitivity and thus would allow further differentiation between non-processive and processive telomere synthesis activities.
Single-molecule structure-function assay for telomerase
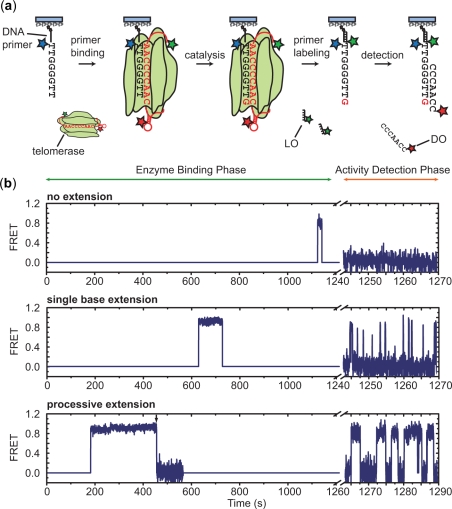
Next, we integrated the activity detection scheme with an enzyme binding phase to directly correlate structural measurements on individual primer-bound enzymes with the catalytic outcome of each binding event (Figure 4a). First, Alexa Fluor-488 labeled and biotinylated (TG)8T2G4T2 primers, which are one nucleotide shorter than a complete telomere DNA repeat and correspond to the standard no extension primers, were surface-anchored and imaged to identify the positions of each primer in the experiment. Subsequently, purified Cy3-Cy5-labeled telomerase was diluted in imaging buffer containing 10 µM dGTP and 100 µM ddTTP and flowed onto the slide to initiate the enzyme binding phase. During this period, binding of telomerase RNP enzymes to the surface-anchored DNA primers allowed FRET measurements to be made between the Cy3 and Cy5 dyes engineered onto the enzyme. Thus, this assay provides a direct measure of the enzyme conformation when bound to a DNA substrate. When Cy3 and Cy5 were attached to the telomerase enzyme, the observed dye blinking and bleaching frequency was higher than that observed in the DO-primer hybridization assay (i.e. activity detection assay), presumably because of the proximity of dyes to proteins in the telomerase enzyme. We therefore used alternating 532 nm excitation (for exciting Cy3) and 635 nm excitation (for direct excitation of Cy5) scheme (26) to identify primer-bound enzymes possessing only a single dye (Cy3 or Cy5) and to discriminate transitions to FRET = 0 caused by Cy5 bleaching and blinking from genuine structural transitions (Supplementary Figure S2 online). Upon completion of the binding phase, the enzyme solution was displaced with imaging buffer containing LO, which introduced a Cy3 dye onto the surface-anchored DNA primers. The activity detection phase was subsequently initiated with a solution containing Cy5-DO, and FRET between the DO and the primer was detected with continuous 532 nm excitation. The enzyme binding and activity detection phases were combined to generate FRET time traces which clearly revealed binding events that resulted in no extension, single base extension and processive extension (Figure 4b).
Figure 4.
Single-molecule structure-function assay. (a) Schematics showing the experimental flow. The Alexa Fluor-488 labeled primers are anchored on the surface and the Alexa Fluor-488 images are first used to identify primers. During the enzyme binding phase, FRET is recorded from a Cy3- and Cy5-labeled telomerase RNP that binds to a surface-anchored primer in the presence of 10 µM dGTP and 100 µM ddTTP. The enzyme is then exchanged with a solution containing a 20 nM Cy3-labeled LO to label the primer with a Cy3 dye. In the subsequent activity detection phase, 300 nM of Cy5-labeled DO is added and FRET between Cy3 and Cy5 is used to determine the catalytic outcome of the primer-enzyme interaction. (b) The FRET time traces (blue) during both the enzyme binding phase and the activity detection phase illustrate three binding events that led to no extension, single base extension and processive extension of the DNA primer by TRE-sIV labeled enzyme, respectively. During the enzyme binding phase, the background signal during the period when enzyme is not bound to the primer is set to a FRET = 0. An enzyme binding event is detected by the appearance of either Cy3 or Cy5 signal using alternating 532 nm and 635 nm excitation. A FRET signal is recorded for the structural characterization of the bound enzyme only when both dyes are present. The alternate excitation scheme also allows identification of the apparent transition to FRET = 0 in the processive extension trace (indicated by the black arrow) as caused by photobleaching of Cy5 (Supplementary Figure S2). The axis break corresponds to the period during which the buffer exchange took place for removing telomerase and introducing the LO and DO into the solution.
Telomerase RNA adopts a single active conformation
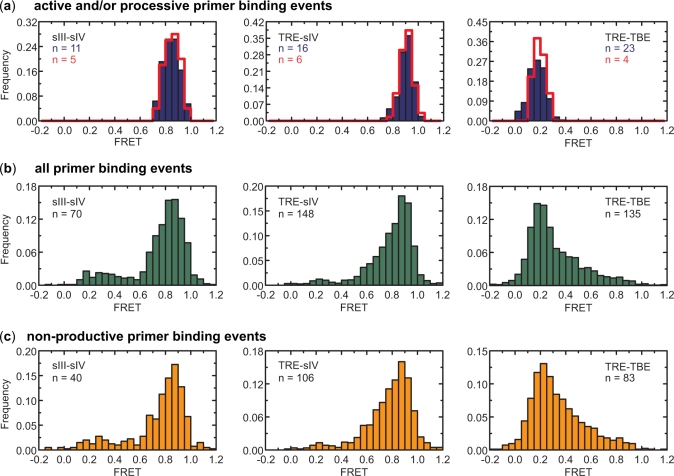
To investigate the functionally relevant conformation of telomerase RNA, we applied the structure-function assay described above to all three labeled enzymes (i.e. sIII-sIV, TRE-sIV and TRE-TBE) and analyzed the FRET values from primer-bound enzymes during each binding event. The free energy of hybridization between a primer and detection oligonucleotide was calculated from the activity detection phase of the experiment for all primers that had encountered a telomerase during the enzyme binding phase (Supplementary Figure S3 online). This distribution clearly separates into three peaks mirroring those seen in Figure 3c. From the hybridization free energies and the threshold values established earlier to separate the three types of primer extension products, the catalytic outcome of each binding event was determined and allowed classification of enzyme binding events into those that resulted in no extension, single base extension or processive extension of the primer respectively. During a typical enzyme binding phase (∼20 min), only about 10% of the primers in a field of view give rise to binding events of doubly labeled enzyme due to the relatively low enzyme concentration. Of all binding events, about 15% led to single base extension and 7% led to processive extension of the primer (of 379 binding events scored, 55 resulted in single base extension and 29 resulted in processive extension of the primer). In all three labeling schemes, the events that led to single base or processive extension of the primer yielded FRET distributions that were substantially narrower than the FRET distributions measured from all binding events (compare Figure 5a to b), indicating that a large fraction of the enzymes that were capable of binding primers assumed inactive, misfolded structures. Consistent with the observation that the majority of the binding events did not lead to any primer extension, the non-productive enzyme-binding events exhibited similar FRET distributions to those observed for all binding events (Figure 5c). Furthermore, FRET distributions derived from telomerase RNP complexes directly anchored on the surface, reflecting the conformations of all enzymes regardless of whether they were capable of primer binding (pink bars in Figure 2d), were even broader than those derived from all primer binding events (Figure 5b). Upon addition of primers and dNTPs to these surface-anchored enzymes, the FRET distributions did not change substantially (Figure 2d), suggesting that a substantial fraction of the enzyme preparation misfolded into conformations that were not able to bind a primer. Within the resolution of the assay, our results suggest the existence of a single active conformation for each of the labeling schemes (FRETTRE-sIV = 0.91, FRETsIII-sIV = 0.85 and FRETTRE-TBE = 0.16). Moreover, these experiments demonstrate substantial structural and functional heterogeneity within enzyme preparations produced using widely employed telomerase reconstitution protocols, substantiating our efforts to characterize telomerase structural properties using novel single molecule approaches. Finally, from the FRET time traces, the dwell time of each binding event can be measured (Supplementary Figure S4 online). We found a weak correlation between binding time and primer extension activity, which is consistent with a previous observation (28).
Figure 5.
Structural characterization of conserved telomerase RNA motifs. (a) FRET histogram of active binding events (blue, consisting of both single base extension and processive extension events), and processive binding events (red outline, consisting of only processive extension events) in the presence of 10 µM dGTP and 100 µM ddTTP. (b) FRET histogram of all binding events irrespective of catalytic outcome in the presence of 10 µM dGTP and 100 µM ddTTP. (c) FRET histogram of non-productive binding events. We note that the binding events that led to single base or processive extension allow us to unambiguously determine the active conformation of the enzyme, whereas the FRET values derived from the non-productive binding events do not necessarily correspond to inactive enzyme conformations, as some binding events may simply be too short to extend the primer. FRET traces with multiple binding events were excluded from the active and processive binding events analysis because it was impossible to determine which binding event resulted in the observed activity. Binding events followed by fast photobleaching in the detection phase were also excluded from the nonproductive, active and processive binding events analysis as it was difficult to unambiguously determine the catalytic outcome in this case. However, these traces were included in the analysis of all binding events, and thus the number of all binding events is larger than the sum of the numbers of the nonproductive, active and processive binding events.
DISCUSSION
In summary, we report a single-molecule telomerase activity assay with single nucleotide resolution and structural sensitivity. In contrast to traditional biochemical assays, this method allows the conformation of individual primer-bound telomerase enzymes to be captured and correlated to enzyme activity and processivity. Application of the assay to sIII-sIV, TRE-sIV and TRE-TBE labeled telomerase allowed the identification of an active enzyme conformation within a heterogeneous preparation of enzyme. Notably, the FRET values for the active conformations of TRE-sIV and sIII-sIV labeled telomerase were both high, suggesting proximity of the stemloop IV to the pseudoknot structure formed between stemloops IIIa and IIIb. These observations are consistent with the proposed role of stemloop IV in promoting catalysis and processivity (16,29). For TRE-TBE labeled telomerase, the low FRET value of the active enzyme signifies that the template region of the telomerase RNA exists in an extended conformation within an active enzyme, consistent with a recently published crystal structure of the TERT protein whose local geometry near the active site can theoretically accommodate an extended single stranded RNA template (30).
The inactive enzyme fraction observed in our assay most likely represents misfolded or misassembled telomerase RNPs that are impaired in their ability to bind primer and/or support catalytic function. While the heterologous lysate reconstitution system is likely the source of the observed structural and functional heterogeneities, we cannot exclude other factors that might have given rise to the misfolded and inactive fraction of the enzyme. For example, the telomerase assembly process itself is perhaps inefficient and thus prone to produce off-pathway products that are inactive even in the native environment of the cell. Interestingly, a recent study reports that enzymatically inactive TERT protein can function in transcription regulation (31). It is possible that the misassembled telomerase RNPs have a function independent of telomere maintenance in vivo. Such a possibility may be tested by applying the single-molecule structure-function assay to endogenously assembled telomerase to determine whether these enzymes assume different conformations and to determine the functional significance of such structural heterogeneity.
By labeling other sites of the telomerase RNA or modifying the labeling scheme to include site-specific protein labeling, the structure-function assay reported here can be applied to investigate other structural domains of the telomerase and test a wide range of existing structural models of telomerase function. The method can also be adapted to telomerase RNPs from other organisms, including human. We anticipate this new single-molecule structure-function assay to complement other biochemical and structural analyses, and to help further elucidate how the static and dynamic structure of telomerase enables its function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work is in part supported by Howard Hughes Medical Institute. X.Z. is a Howard Hughes Medical Institute investigator. J.Y.W is a National Science Foundation Graduate Research fellow. M.D.S. is a NIH Ruth L. Kirschstein NSRA Fellow. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Dr Kathy Collins and Dr Tracy Bryan for materials and protocols for telomerase reconstitution, and Mariana Mihalusova and Elio Abbondanzieri for discussions.
REFERENCES
- 1.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. Tracking telomerase. Cell. 2004;116:S83–S86. doi: 10.1016/s0092-8674(04)00053-4. 81 p. following S86. [DOI] [PubMed] [Google Scholar]
- 3.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 5.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins K. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 7.Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 9.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 11.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 12.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. Proc. Natl Acad. Sci. USA. 2002;99:16998–17003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 17.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol. Biol. Cell. 2003;14:4794–4804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 21.Stone MD, Mihalusova M, O'C;onnor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Wen JD, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alves D, Li H, Codrington R, Orte A, Ren X, Klenerman D, Balasubramanian S. Single-molecule analysis of human telomerase monomer. Nat. Chem. Biol. 2008;4:287–289. doi: 10.1038/nchembio.82. [DOI] [PubMed] [Google Scholar]
- 24.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 26.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl Acad. Sci. USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greider CW. Telomerase is processive. Mol. Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards RJ, Wu H, Trantirek L, O'C;onnor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 31.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl Acad. Sci. USA. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilley D, Blackburn EH. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl Acad. Sci. USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.