Abstract
Apurinic/apyrimidinic endonuclease-1 (APE1) is an essential enzyme in the base excision repair (BER) pathway. Here, we show that APE1 is a target of the SIRTUIN1 (SIRT1) protein deacetylase. SIRT1 associates with APE1, and this association is increased with genotoxic stress. SIRT1 deacetylates APE1 in vitro and in vivo targeting lysines 6 and 7. Genotoxic insults stimulate lysine acetylation of APE1 which is antagonized by transcriptional upregulation of SIRT1. Knockdown of SIRT1 increases cellular abasic DNA content, sensitizing cells to death induced by genotoxic stress, and this vulnerability is rescued by overexpression of APE1. Activation of SIRT1 with resveratrol promotes binding of APE1 to the BER protein X-ray cross-complementing-1 (XRCC1), while inhibition of SIRT1 with nicotinamide (NAM) decreases this interaction. Genotoxic insult also increases binding of APE1 to XRCC1, and this increase is suppressed by NAM or knockdown of SIRT1. Finally, resveratrol increases APE activity in XRCC1-associated protein complexes, while NAM or knockdown of SIRT1 suppresses this DNA repair activity. These findings identify APE1 as a novel protein target of SIRT1, and suggest that SIRT1 plays a vital role in maintaining genomic integrity through regulation of the BER pathway.
INTRODUCTION
The major mammalian Apurinic/apyrimidinic endonuclease APE1, also known as Redox Factor-1 (Ref-1), is a multifunctional protein. It plays a vital role in the repair of single-strand DNA breaks induced by oxidative and alkylating agents, abasic sites generated during the repair of DNA bases chemically modified by such genotoxic agents, and spontaneously generated abasic DNA sites (1–4). In this essential role in the base excision repair (BER) pathway, APE1 interacts with other DNA repair proteins including DNA polymerase β (Polβ) (5), flap endonuclease-1 (FEN1) (6), and X-ray cross-complementing-1 (XRCC1) (6). In addition to this DNA repair function, APE1, through its properties as a reducing protein, has been identified as an activator of several redox (reduction-oxidation)-sensitive transcription factors including p53 (7), AP-1 (8), and NF-κB (9). The final known function of APE1 relates to its identification as one of the proteins that binds to negative calcium-response elements (nCaREs) in the parathyroid hormone (PTH) and renin genes, down-regulating their transcription in response to increase in extracellular calcium (10,11).
Independent of transcriptional control, APE1 is regulated by many post-translational modifications with some of these modifications having an impact on one or more of its functions. Casein kinase II (CKII)-mediated phosphorylation impairs the DNA repair activity of the protein (12), while stimulating its redox function toward the AP-1 transcription complex (13). Protein kinase C (PKC) phosphorylates APE1 as well, also stimulating its ability to promote DNA binding of AP-1 (14). The nuclear export of APE1 is regulated by S-nitrosylation of cysteines 93 and 310 (15). In addition, APE1 is targeted by the cellular acetylation-deacetylation machinery. APE1 is acetylated by the p300 acetyltransferase on lysines 6 and 7, and this acetylation enhances its binding to the nCaRE in the PTH promoter, stimulating PTH promoter activity (16). Moreover, acetylated APE1 has been implicated in oxidative stress-induced upregulation of phosphinositol phosphatase and tensin homologue (PTEN) (17), a negative regulator of the phosphoinositide 3-kinase/Akt signaling pathway. Although studies have demonstrated that APE1 binds to class I histone deacetylases (HDAC), the role of these HDAC in deacetylating APE1 has not been examined. Furthermore, although APE1 does not appear to bind to class II HDAC, its association with class III HDACs, as well as the role of class III HDAC in regulating its acetylation has not been reported.
SIRTUIN 1 (SIRT1) is a class III HDAC. It is the closest mammalian ortholog of yeast silent information regulator (sir2) protein which senses changes in energy availability and responds by regulating metabolic pathways (18). SIRT1 expression and function are regulated by external stressors including a decrease in available nutrition and genotoxic agents (19,20). In response to such stressors, SIRT1 not only orchestrates transcription through its function as a histone deacetylase, but also deacetylates many non-histone proteins that are important in regulating energy metabolism and stress response such as p53 (21,22), nuclear factor-κB (23), peroxisome proliferator-activated receptor-γ (PPAR-γ) (24), and PPAR-γ coactivator-1α (PGC-1α) (25). Although SIRT1 affects the cell cycle and promotes stress resistance in response to cytotoxic stimuli (21,26–28), few substrates of SIRT1 have been identified that are directly involved in the maintenance of genomic integrity. A recent report shows that SIRT1, by directly targeting Nijmegen Breakage syndrome (NBS1) protein for deacetylation and thereby facilitating its phosphorylation by the ataxia telangiectasia mutated (ATM) protein kinase, plays a central role in the cellular response to agents that cause double-stranded DNA breaks (29). To identify other DNA repair proteins that are novel targets for deacetylation by SIRT1 we investigated the possibility that SIRT1 deacetylates APE1, and thereby has an important part in the cellular BER pathway.
MATERIALS AND METHODS
Materials
Antibodies against SIRT1, APE1, Myc-tag and p300 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against acetyl-lysine, Ac-K 382-p53 and XRCC1 were purchased from Cell Signaling Technology (Beverly, MA). Monoclonal antibody against FLAG-tag was from Sigma-Aldrich (St Louis, MO). All reagents were obtained from Sigma-Aldrich unless otherwise stated.
Cell culture
HEK 293 cells and HeLa cells were obtained from ATCC (Manassas, VA). Cells were maintained at 37°C in 5% CO2 in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. Transfection of plasmid and/or RNAi was accomplished using LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s instructions. Approximately 2µg of plasmid DNA was transfected per 5 × 106 cells (+) or multiples thereof. Validated Stealth RNAi for APE1 and SIRT1 as well as the appropriate control scrambled oligonucleotides were purchased from Invitrogen.
Plasmids
The cDNA of human APE1 was cloned into p3xFLAG-CMV-7.1 mammalian expression plasmid (Sigma-Aldrich). The same cDNA was cloned into pET-41b (Novagen, Madison, WI) for bacterial expression of APE1 and into pDsRed-N1 (Clonetech, Mountain View, CA) for cell fluorescence studies. Myc-tagged mammalian expression vectors carrying human SIRT1 (wild type or H363Y mutant) were kindly provided by T Kouzarides. The same human SIRT1 cDNA was cloned into pEGFP-C2 (Clonetech) for co-localization experiments. The cDNA of human p300 was a generous gift from Dr. Wangsen Cao and was cloned into pcDNA 3.1(−).
Microscopy
APE1 cloned into pDsRed-N1 and SIRT1 cloned into pEGFP-C2 were co-transfected into HEK 293 cells. Red and green fluorescent images were taken using an Axiovert 200 Fluorescence microscope (Carl Zeiss, Jena, Germany). Merged images were created using Adobe Photoshop software.
Co-immunoprecipitation
Cells were collected by brief centrifugation. The pellet was suspended in IP lysis buffer (50 mM Tris–HCl (pH 7.5), 1% (v/v) Triton X-100, 5% (v/v) glycerol, 5 mM EDTA, 150 mM NaCl, 10 mM NaF, 1 mM PMSF, 1× protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), 10 mM nicotinamide (NAM), 5 µM trichostatin A (TSA)) and incubated for 20 min on ice. After centrifugation (14 000×g, 15 min, 4°C), cell lysates were collected and protein concentration was determined by Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA). Cell lysates were incubated with 2 µg of antibody for 16–20 h at 4°C followed by the addition of 20 µl of Protein-A or -G Sepharose beads (50% slurry). After further 2 h incubation at 4°C, beads were washed four times with IP lysis buffer and then washed once with PBS. Samples were mixed with 2× SDS sample buffer and subjected to SDS–PAGE, followed by immunoblotting using the specified primary antibody and the appropriate secondary antibody.
In vitro APE1 acetylation/deacetylation
Recombinant human APE1 protein was expressed in BL21 (DE3) Escherichia coli (Stratagene, La Jolla, CA) as GST fusion protein. GST-APE1 protein was purified using Glutathione Sepharose 4B beads (GE Healthcare Bio-Sciences, Piscataway, NJ). GST portion was cleaved off by Thrombin Clean Cleave Kit (Sigma) and removed using Glutathione Sepharose 4B beads. After exchanging the buffer with 50 mM Tris–HCl (pH 8.0), protein concentration was determined.
Recombinant APE1 protein (4.2 µg) was incubated with 200 ng of p300 acetyltransferase (Active Motif, Carlsbad, CA) in acetylation buffer (50 mM Tris–HCl (pH 8.0), 0.1 mM EDTA, 10% (v/v) glycerol, 1 mM DTT, 50 mM AcCoA) for 1 h at 30°C with shaking. After the incubation, the mixture was supplemented with 10 units of SIRT1 (Biomol, Plymouth Meeting, PA), 1 mM NAD+, 150 mM NaCl, 3 mM KCl, 4 mM MgCl2 to initiate the deacetylation reaction. The deacetylation reaction was performed with or without 1 µM TSA or 10 mM NAM. After the incubation for 90 min at 37°C, the reaction was terminated by adding 3× SDS sample buffer and samples were subjected to SDS–PAGE and immunoblotting to evaluate APE1 acetylation.
In vivo APE1 acetylation/deacetylation
Cells were collected by brief centrifugation. The pellet was suspended in pre-boiled (10 min) denaturing cell lysis buffer [50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 1% (w/v) SDS, 70 mM β-ME] and was boiled for another 10 min. After sonication (15 s × 3) and following centrifugation (14 000×g, 15 min, 4°C), supernatants were transferred to new tubes. The samples were diluted by the addition of 9 vol of IP lysis buffer and protein concentration was determined. Three µg of APE1 antibody was added to the samples containing equal amount of protein, and rotated overnight at 4°C. After adding 30 µl of Protein G-Sepharose beads (50% slurry) samples were rotated for 3–6 h at 4°C. Beads were washed four times with IP lysis buffer and then washed once with PBS. Thirty microliters of 2× SDS sample buffer was added to the beads and boiled for 5 min. Samples were subjected to SDS–PAGE and immunoblotting using acetyl-lysine antibody to evaluate APE1 acetylation.
Real-time PCR
Total RNA from cells was isolated by the acid guanidinium thiocyanate/phenol/chloroform method. Real time PCR was performed using the Prism 7000 Sequence Detection System (Applied Biosystems) with the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen). The primer sequences for human SIRT1 are: forward 5′-TCGCAACTATACCCAGAACATAGACA-3′, reverse 5′-CTGTTGCAAAGGAACCATGACA-3′. Human GAPDH was used as an internal control. The primer sequences for human GAPDH are: forward 5′-ATG ACA TCA AGA AGG TGG TG-3′, reverse 5′-CAT ACC AGG AAA ATG AGC TTG-3′. Dissociation curves were monitored to check the aberrant formation of primer-dimers.
Promoter-reporter assay
A 1403 base pair fragment of the human SIRT1 promoter (–1266 to +137 relative to transcription start site) was cloned into the pGL4.1 firefly luciferase reporter vector (Promega). The SIRT1 promoter-reporter plasmid was co-transfected with a constitutive renilla reporter plasmid using Lipofectamine2000 (Invitrogen) as per manufacturer’s recommendations. Firefly and renilla luciferase activity were measured using the Dual Luciferase reporter kit (Promega) as per manufacturer’s recommendations, and firefly activity was normalized to renilla activity to correct for differences in transfection efficiency. Results presented are from a representative experiments performed in triplicate.
Apoptosis
Apoptotic cell death was quantified using Cell Death Detection ELISA kit (Roche) according to the manufacturer’s instructions.
AP sites
Apurinic/apyrimidinic (AP) sites were quantified using the DNA Damage Quantification kit (Oxford Biomedical Research, Oxford MI), as per manufacturer’s recommendations. This colorimetric kit quantifies apurinic/apyrimidinic sites by using an aldehyde reactive probe which tags them with biotin, followed by detection with HRP-streptavidin.
AP endonuclease activity assay
A 25-mer oligonucleotide containing tetrahydrofuran (THF; Midland Certified Reagents, Midland, TX) at position 15 in the sequence 5′-AATTCACCGGTACCXCCTAGAATTCG-3′ (X: THF) was 5′-terminally labeled with [γ-32P]ATP using DNA 5′-end-labeling System (Promega, Madison, WI). The labeled oligonucleotide was annealed to the complementary strand with T opposite THF and purified by PAGE.
HeLa cells transfected with Flag-APE1 plasmid and subsequently treated with NAM (5 mM) or resveratrol (50 µM) for 6 h were harvested and washed with PBS. Cells were resuspended in PBS containing 2 mM DTT and pulse-sonicated three times for 15 s each. After centrifugation at 14 000×g for 10 min at 4°C, protein concentration of cell extract was determined by Bio-Rad Protein Assay reagent. XRCC1 immunoprecipitated from 1500 µg was incubated in a total volume of 20 µl of assay buffer [50 mM HEPES–NaOH (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1% BSA, 0.05% Triton X-100] containing the labeled duplex oligonucleotide (400 fmol) for 10, 20 and 30 min at 37°C. Reactions were terminated by adding 10 µl of formamide loading buffer (96% formamide, 10 mM EDTA, 0.1% bromophenol blue). The samples were then heated at 95°C for 5 min to denature the oligonucleotides, and were analyzed on a 20% polyacrylamide, 7 M urea gel. After electrophoresis, the gel was autoradiographed to locate the labeled substrate and product oligonucleotides. AP endonuclease activity was similarly performed in vitro using purified recombinant GST-tagged APE1 that was acetylated with recombinant active p300 and deacetylated with recombinant SIRT1.
RESULTS
APE1 and SIRT1 co-precipitate and co-localize in cells
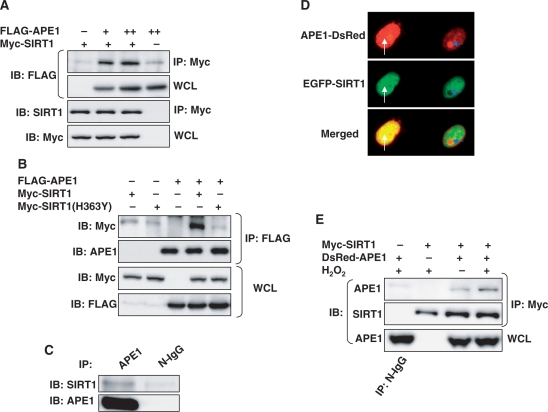
To examine the relationship between APE1 and SIRT1 we first determined whether there exists a physical association between the two proteins. Tagged APE1 co-precipitated with SIRT1 in HEK293 but not with a catalytically inactive, dominant negative, SIRT1(H363Y) mutant (Figure 1A and B). Endogenous SIRT1 also co-precipitated with endogenous APE1 (Figure 1C). Moreover, APE1 and SIRT1 co-localized to the nuclei of HEK 293 cells (Figure 1D). Postulating that this association may have functional relevance, we examined the effect of genotoxic stress on APE1-SIRT1 binding. Because APE1 and SIRT1 protect cells against hydrogen peroxide (H2O2) (30–35), an oxidative stress that leads to DNA damage resulting in abasic DNA sites (36), we asked whether H2O2 affects the association between the two proteins. In HEK 293 cells H2O2 promoted the binding of APE1 to SIRT1 (Figure 1D), suggesting that this association may have importance in the context of genotoxic stress. Thus, APE1 and SIRT1 bind to and localize with each other and genotoxic stress, against which APE1 and SIRT1 offer protection, increases this association.
Figure 1.
APE1 and SIRT1 associate with each other. (A) APE1 and SIRT1 bind to each other in HEK 293 cells. Immunoprecipitation of epitope-tagged SIRT1 co-precipitates epitope-tagged APE1 expressed in HEK 293 cells. (B) APE1 does not bind to catalytically inactive dominant negative SIRT1. Immunoprecipitation of epitope-tagged APE1 co-precipitates epitope-tagged wild-type SIRT1 but not dominant negative SIRT1 (H363Y) expressed in HEK 293 cells. (C) Endogenous SIRT1 binds to endogenous APE1. Co-immunoprecipitation of endogenous SIRT1 and APE1 in HEK 293 cells. (D) Co-localization of fluorescent epitope-tagged SIRT1 and APE1 expressed in HEK 293 cells. Co-localization of extra-nucleolar (white arrow) APE1 but not nucleolar (blue arrow) APE1 with SIRT1 is shown. (E) Hydrogen peroxide (H2O2) promotes binding of APE1 to SIRT1. H2O2 (500 µM, 30 min) increases co-precipitation of epitope-tagged SIRT1 in immunoprecipitates of epitope-tagged APE1 expressed in HEK 293 cells. WCL: whole cell lysate. N-IgG: non-immune immunoglobulin. FLAG-APE1 and Myc-SIRT1 were expressed in A and B, DsRed-APE1 and EGFP-SIRT1 in C, and DsRed-APE1 and Myc-SIRT1 in D.
SIRT1 deacetylates APE1 in vitro and in vivo
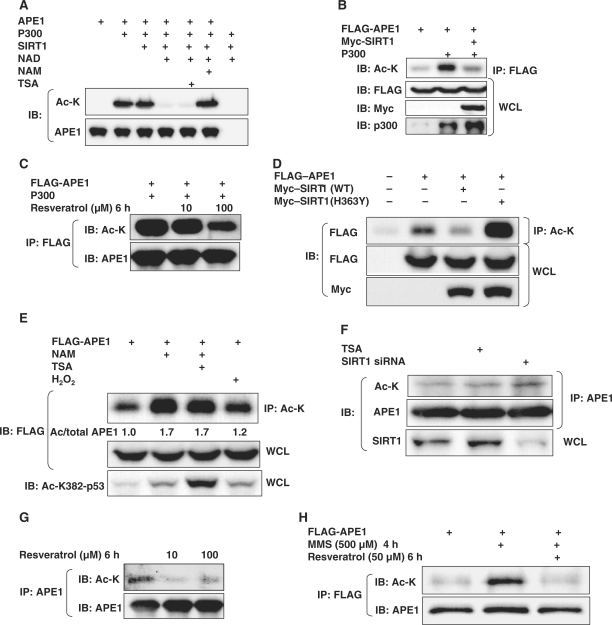
We then asked if SIRT1 targets APE1 for deacetylation. First, we examined if APE1 is a target of SIRT1 deacetylase activity in vitro. Recombinant APE1 was enzymatically acetylated with the p300 acetyltransferase in vitro, followed by incubation with active SIRT1. SIRT1 in the presence, but not absence of its co-factor nicotinamide adenine dinucleotide (NAD), deacetylated acetylated APE1 (Figure 2A). Addition of Trichostatin A (TSA), a class I and II HDAC inhibitor, to the reaction mixture did not affect SIRT1-induced deacetylation of APE1, whereas the SIRT1 inhibitor nicotinamide (NAM) negated deacetylation of APE1, indicating that acetylated APE1 is a direct target of SIRT1. In HEK 293 cells as well, APE1 was acetylated by overexpression of the p300 acetyltransferase, and this increase in lysine acetylation was abrogated by overexpression of SIRT1 (Figure 2B). Similarly, stimulation of endogenous SIRT1 with resveratrol decreased p300-induced lysine acetylation of APE1 (Figure 2C). Notwithstanding the limitation that our data do not allow us to draw conclusions about site-specific acetylation and deacetylation, they show that APE1 is acetylated by the p300 acetyltransferase, and deacetylated by SIRT1, in vitro and in vivo.
Figure 2.
SIRT1 deacetylates APE1. (A) In vitro deacetylation of recombinant APE1 by SIRT1. Recombinant APE1 enzymatically acetylated by the p300 acetyltransferase was deacetylated by recombinant active SIRT1 in presence of the SIRT1 co-factor NAD+ (1 mM). Deacetylation was inhibited by the SIRT1 inhibitor nicotinamide (NAM, 10 mM), but not the class I and II HDAC inhibitor Trichostatin A (TSA, 1 µM). (B) APE1 acetylated by the p300 acetyltransferase is deacetylated by SIRT1 in vivo. (C) Resveratrol inhibits p300-induced lysine acetylation of exogenous APE1. (D) Expression of wild-type SIRT1 (WT) decreases and dominant negative SIRT1 (H363Y) increases, acetylation of exogenous APE1 in HEK293 cells. (E) The SIRT1 inhibitor NAM and H2O2 increases acetylation of exogenous APE1 in HEK 293 cells. Cells were treated with H2O2 (500 µM), NAM (5 mM) and the class I and II HDAC inhibitor Trichostatin A (TSA: 5 µM) for 30 min. Values of lysine acetylated/total APE1 from a representative experiment, normalized to untreated cells, are shown. Acetylation of endogenous p53 is shown at bottom for comparison. WCL: whole cell lysate. (F) TSA (5 µM) and siRNA-mediated knockdown of endogenous SIRT1 increase lysine acetylation of endogenous APE1 in HEK 293 cells. (G) Resveratrol suppresses basal lysine acetylation of APE1 in HEK 293 cells. (H) Activation of endogenous SIRT1 with resveratrol antagonizes MMS-induced lysine acetylation of exogenous APE1.
Next, we determined the role of SIRT1 in regulating basal acetylation of APE1. Tagged APE1 was expressed in HEK 293 cells and its acetylation on lysine residues was determined in the presence and absence of SIRT1 overexpression. SIRT1 overexpression led to a decrease in basal lysine acetylation of APE1 (Figure 2D). In contrast to wild-type SIRT1, expression of the dominant negative SIRT1(H363Y) increased acetylation of APE1. In addition, treatment of cells with the SIRT1 inhibitor NAM increased basal lysine acetylation of APE1 (Figure 2E). However, addition of TSA to NAM-treated cells did not further increase acetylation of APE1 (Figure 2E). This is in contrast to the synergistic effect of TSA and NAM on lysine acetylation of p53 (Figure 2E). In line with the effect of SIRT1 manipulation on lysine acetylation of exogenous APE1, knockdown of endogenous SIRT1 expression with siRNA promoted acetylation of endogenous APE1 (Figure 2F). In comparison with SIRT1 knockdown, addition of the class I and II HDAC inhibitor TSA resulted in a smaller increase in lysine acetylation of APE1 (Figure 2F). In addition, stimulation of endogenous SIRT1 with resveratrol decreased lysine acetylation of endogenous APE1 (Figure 2G). These findings show that in addition to deacetylating APE1 that is hyperacetylated by the p300 acetyltransferase, SIRT1 also regulates basal acetylation of APE1. In addition, they suggest that class I and II HDAC inhibitors also play a role, albeit lesser than SIRT1, in regulating basal acetylation of APE1.
APE1 acetylation by genotoxic stress is antagonized by SIRT1
Because we had noted that H2O2 increases the binding of APE1 to SIRT1 (Figure 1E), we also asked whether this is accompanied by a change in lysine acetylation of APE1. In HEK 293 cells, H2O2 led to an increase in acetylation of APE1 (Figure 2E). To determine whether the increase in lysine acetylation of APE1 observed with H2O2 is a phenomenon common to other genotoxic agents that induce abasic DNA damage, we next examined the effect of methyl methanesulfonate (MMS), another well-known genotoxic stress that results in abasic DNA sites. Similar to the effect of H2O2, MMS treatment for a short time (4 h) resulted in an increase in lysine acetylation of endogenous APE1 (Figure 2H). Importantly, stimulation of endogenous SIRT1 with the SIRT1 activator resveratrol abrogated the increase in APE1 lysine acetylation induced by MMS.
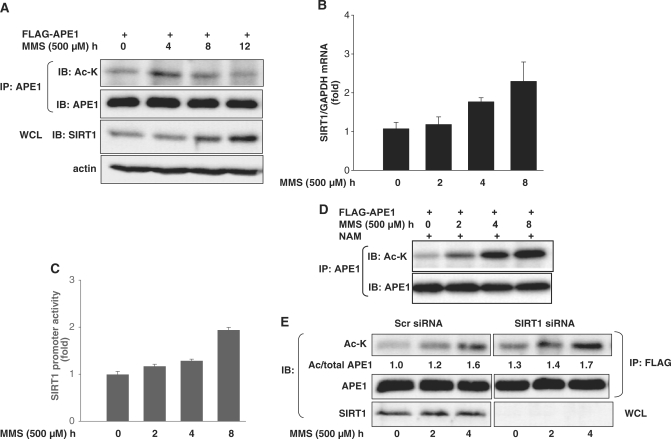
We next investigated the kinetics of APE1 acetylation with genotoxic stress. A time course of treatment with MMS revealed that APE1 acetylation was increased at an early time point, followed by a decline to near basal levels at later time points (Figure 3A). Interestingly, the decline in APE1 acetylation at later times was accompanied by up-regulation of SIRT1 protein (Figure 3A). This time-dependent progressive upregulation of SIRT1 with MMS was corroborated with SIRT1 mRNA expression and SIRT1 promoter-reporter assays (Figure 3B and C). The reciprocal relationship between APE1 acetylation and SIRT1 expression suggested to us that upregulation of endogenous SIRT1 may be a feedback mechanism for regulation of APE1 acetylation induced by MMS. Indeed, inhibition of endogenous SIRT1 activity resulted in more robust lysine acetylation of APE1, even at later time points (Figure 3D). Moreover, knockdown of endogenous SIRT1 also promoted lysine acetylation of APE1 induced by MMS (Figure 3E). These findings demonstrate an important role for endogenous SIRT1 in antagonizing the lysine acetylation of APE1 induced by the genotoxic agent MMS, and implicate transcriptional upregulation of SIRT1 by MMS as an endogenous feedback mechanism that modulates genotoxic stress-induced APE1 acetylation.
Figure 3.
SIRT1 is upregulated by MMS and antagonizes MMS-induced acetylation of APE1. (A) Reciprocal temporal relationship between MMS-induced APE1 acetylation and SIRT1 expression. Time course of APE1 lysine acetylation and SIRT1 protein expression in HEK 293 cells challenged with MMS. (B) MMS increases SIRT1 mRNA. Time course of SIRT1 mRNA upregulation in HeLa cells challenged with MMS. (C) MMS increases SIRT1 promoter activity. Time course of human SIRT1 promoter induction in HeLa cells challenged with MMS. (D) SIRT1 inhibition augments MMS-induced acetylation of APE1. Time course of APE1 lysine acetylation in HEK 293 cells pre-treated with the SIRT1 inhibitor NAM (5 mM) and challenged with MMS. (E) SiRNA-mediated suppression of SIRT1 in HEK 293 cells promotes MMS-induced APE1 acetylation. Lysine acetylated/total APE1 ratio from a representative experiment is shown and is normalized to untreated control siRNA cells. WCL: whole cell lysate.
Lysines 6 and 7 in APE1 are deacetylated by SIRT1
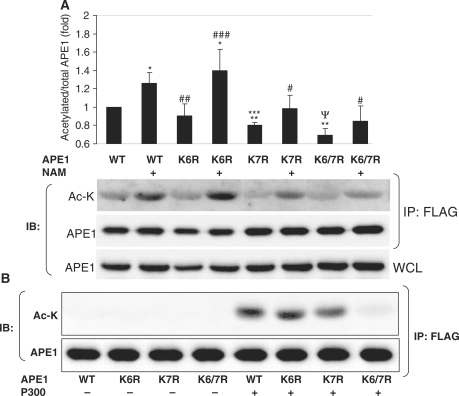
We then investigated the specific lysine residues in APE1 that SIRT1 targets for deacetylation. APE1 is acetylated on lysine residues 6 and 7 at its N-terminus in response to changes in extracellular calcium (16). We therefore looked at these specific residues as targets of SIRT1. Lysines 6 and 7 in APE1 were mutated to non-acetylatable arginines, individually or in concert, and these non-acetylatable forms of APE1 were expressed in HEK 293 cells. Basal acetylation and change in acetylation induced by NAM-mediated inhibition of endogenous SIRT1 activity was then examined and quantified on wild-type as well as non-acetylatable APE1 lysine mutants. Compared with wild-type protein, mutation of lysine 6 did not result in a significant decrease in either basal or NAM-induced acetylation of APE1 (Figure 4A). However, APE1 that was mutated on lysine 7 did show a significant decrease in both basal and NAM-stimulated acetylation, when compared with wild-type protein. Moreover, APE1 that was non-acetylatable at both lysines 6 and 7 showed an even greater decrease in basal and NAM-induced acetylation. These findings show that both lysines 6 and 7 are targeted for deacetylation by SIRT1, and also suggest synergism between deacetylation by SIRT1 of the two lysine residues. Consistent with this notion, p300-stimulated lysine acetylation of APE1 was essentially unchanged when lysine 6 was non-acetylatable, diminished when lysine 7 was non-acetylatable, and almost completely abolished when both lysines 6 and 7 were non-acetylatable (Figure 4B). However, because replacement of both lysines 6 and 7 with non-acetylatable residues did not completely abolish the increase in acetylation in response to NAM, deacetylation of lysines other than 6 and 7 by SIRT1 remains a possibility.
Figure 4.
SIRT1 targets lysines 6 and 7 in APE1 for deacetylation. (A) Basal lysine acetylation, and increase in lysine acetylation by treatment with the SIRT1 inhibitor nicotinamide (NAM: 5 mM, 16 h), of epitope-tagged wild-type and non-acetylatable mutants of APE1 expressed in HEK 293 cells. Lysine acetylated/total APE1 was quantified and is expressed relative to wild-type APE1 in untreated cells. *P < 0.05 and #P > 0.05 compared with untreated cells transfected with the same construct. ##P > 0.05, and **P < 0.005 compared with untreated cells transfected with WT APE1. ###P > 0.05, ***P < 0.05, and ΨP < 0.005 compared with NAM-treated cells transfected with WT APE1. N = 4 in all conditions. (B) Targeting of the N-terminal lysines in APE1 for acetylation by the p300 acetyltransferase. Epitope-tagged wild-type (WT) APE1, or the non-acetylatable lysine mutants of APE1, were expressed in HEK 293 cells, with and without overexpression of the p300 acetyltransferase. WCL: whole cell lysate.
APE1 mediates SIRT1-dependent protection against genotoxic stress
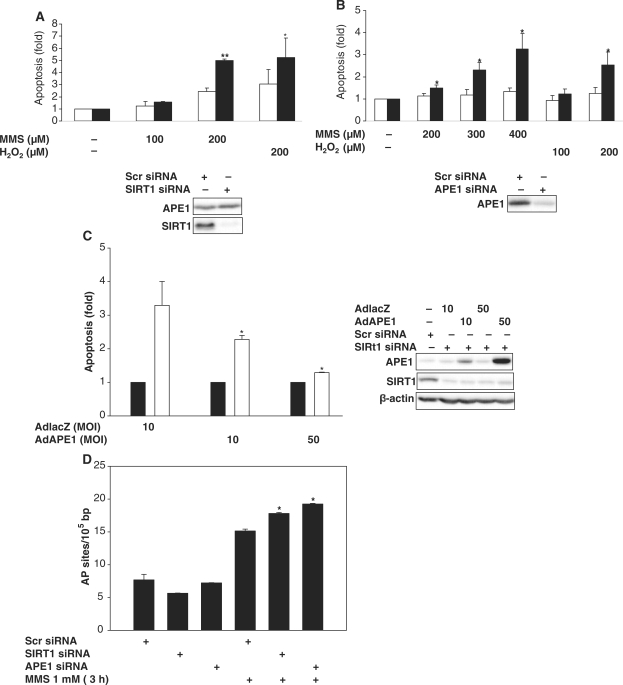
Next, we determined the relationship between APE1 and SIRT1 in cytoprotection against genotoxic stress. First, we determined if APE1 and SIRT1, independently, protect cells from such stress. Knockdown of endogenous SIRT1 increased susceptibility of HeLa cells to apoptosis induced by MMS and H2O2 (Figure 5A). Similarly, knockdown of endogenous APE1 also made HeLa cells more vulnerable to death by H2O2 and MMS (Figure 5B). Then, to determine if APE1 mediates SIRT1-dependent cytoprotection, the effect of increasing APE1 expression on the vulnerability to cell death associated with knockdown of SIRT1 was examined. In HeLa cells in which SIRT1 expression was knocked down, susceptibility to MMS-induced apoptosis was rescued by adenoviral overexpression of APE1 (Figure 5C). These findings demonstrate that both endogenous SIRT1 and APE1 protect from cell death induced by genotoxic stimuli, and suggest that stimulation of APE1 activity is responsible for the cytoprotection conferred by SIRT1. However, we cannot definitively conclude the role of SIRT1-mediated APE1 deacetylation in protection against genotoxic stress.
Figure 5.
SIRT1 protects from genotoxic stress-induced cell death through APE1. Apoptotic death with oxidative stress (H2O2 for 24 h) and abasic DNA damage (MMS for 24 h) in HeLa cells in which SIRT1 expression (A) or APE1 expression (B) is knocked down with SIRT1 or APE1 siRNA (black bars). Scrambled (scr) siRNA (white bars) was used as control. Apoptosis is expressed as fold change compared with untreated cells. *P < 0.05 and **P < 0.01 compared with control siRNA. Knockdown of SIRT1 and APE1 is shown at bottom. (C) APE1 overexpression rescues cells with SIRT1 down-regulation from MMS-induced apoptosis. Apoptotic cell death induced by MMS (200 µM, 24 h) in HeLa cells treated with SIRT1 siRNA (white bars) or scrambled (scr) siRNA (black bars) were infected with a control virus (AdLacZ) that expresses the inert E. coli LacZ gene, or and adenovirus that expresses APE1 (AdAPE1). Apoptosis is expressed as fold change compared with control siRNA. *P < 0.05 compared with cells infected with AdLacZ. Knockdown of SIRT1 and adenoviral overexpression of APE1 is shown at right. (D) SIRT1 plays a role in abasic DNA repair. SIRT1 or APE1 was knocked down in HeLa cells with siRNA. Apurinic/apyrimidinic DNA sites were quantified in untreated cells and cells treated with MMS for 3 h. *P < 0.05 compared with control siRNA.
APE1 is essential for repair of abasic DNA. Therefore, to further establish a functional link between cellular APE1 and SIRT1, we examined the role of SIRT1 in the cellular DNA repair machinery. The effect of SIRT1 knockdown on the formation of apurinic/apyrimidinic DNA sites generated by genotoxic stress was determined. Knockdown of SIRT1 increased cellular apurinic/apyrimidinic sites in response to MMS (Figure 5D). This effect was similar to that observed with knockdown of APE1. These findings show that endogenous SIRT1, like endogenous APE1, plays an important role in the repair of abasic DNA sites.
SIRT1 promotes the association of APE1 and XRCC1
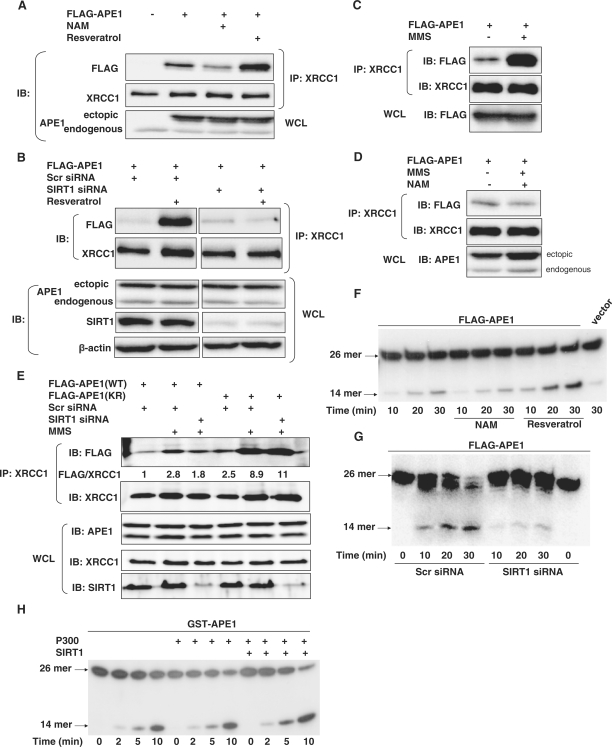
We then sought to further elucidate the APE1-mediated mechanism responsible for cytoprotection conferred by SIRT1. Hypothesizing that SIRT1, by deacetylating APE1, may modulate its binding to other proteins that play a part in the BER pathway, we sought out known binding partners of APE1. X-ray cross-complementing-1 (XRCC1) is a protein that binds with enzymes involved at each step in the BER pathway (37), and functions to organize proteins at the damaged DNA site. XRCC1 binds to APE1 and the N-terminal region of APE1 has been shown to mediate this interaction (38). Knowing that the N-terminal lysine residues of APE1 are targeted for deacetylation by SIRT1 prompted us to investigate whether SIRT1 affects binding of APE1 to XRCC1. In HeLa cells, APE1 co-precipitated with XRCC1 (Figure 6A). Inhibition of endogenous SIRT1 with NAM suppressed binding of APE1 to XRCC1 (Figure 6A). Conversely, stimulation of endogenous SIRT1 activity with resveratrol promoted the association of APE1 and XRCC1. Moreover, resveratrol-stimulated association of APE1 and XRCC1 was abrogated by knockdown of endogenous SIRT1 (Figure 6B). These findings indicate that SIRT1 promotes the association of APE1 and XRCC1.
Figure 6.
SIRT1 promotes cellular endonuclease activity toward abasic DNA sites by stimulating the binding of APE1 to XRCC1. (A) Endogenous SIRT1 promotes binding of APE1 to XRCC1. Co-immunoprecipitation of endogenous XRCC1 with epitope-tagged APE1 expressed in HeLa cells treated with the SIRT1 inhibitor NAM (5 mM), or the SIRT1 activator resveratrol (50 µM) for 6 h. (B) Resveratrol-stimulated association between APE1 and XRCC1 is mediated by SIRT1. Co-immunoprecipitation of epitope-tagged APE1 and endogenous XRCC1 in HeLa cells, with and without resveratrol (50 µM, 6 h), that are treated with scrambled siRNA (Scr siRNA) or SIRT1 siRNA. Separated panels are from the same immunoblot, with irrelevant lanes deleted. (C) Genotoxic stress promotes binding of APE1 to XRCC1. Co-immunoprecipitation of epitope-tagged APE1 and endogenous XRCC1 in HeLa cells treated with MMS (500 µM, 6 h) (D) SIRT1 mediates genotoxic stress-induced binding of APE1 to XRCC1. Co-immunoprecipitation of epitope-tagged APE1 and endogenous XRCC1 in HeLa cells treated with MMS (500 µM, 6 h) with and without treatment with SIRT1 inhibitor NAM (5 mM). (E) SIRT1 mediates genotoxic stress-stimulated binding of wild-type, but not acetylation-deficient APE1. Co-immunoprecipitation of endogenous XRCC1 and epitope-tagged wild-type APE1 (WT) or mutated APE1 (KR) that is non-acetylatable on lysines 6 and 7, in HeLa cells treated with MMS (500 µM, 6 h), with or without knockdown of SIRT1. Cells were co-transfected with scr siRNA or SIRT1 siRNA. WCL: whole cell lysate. (F and G) SIRT1 promotes AP endonuclease activity in XRCC1-bound protein complex. AP endonuclease activity toward abasic DNA in XRCC1 immunoprecipitates from (F) HeLa cells expressing FLAG-tagged APE1 and treated with the SIRT1 activator resveratrol (50 µM), or the SIRT1 inhibitor NAM (5 mM) for 6 h and (G) HeLa cells in which SIRT1 expression is knocked down with siRNA. (H) P300-induced acetylation, and SIRT1-induced deacetylation, of recombinant APE1 does not affect its AP endonuclease activity. In vitro AP endonuclease activity toward abasic DNA of recombinant GST-APE1 that is acetylated by the p300 acetyltransferase, followed by deacetylation by SIRT1.
Armed with the knowledge that SIRT1 facilitates the interaction of APE1 and XRCC1, and SIRT1 antagonizes MMS-induced acetylation of APE1, we then asked whether MMS affects the binding of APE1 to XRCC1, and whether SIRT1 modulates this effect. Challenge with MMS increased the association of APE1 and XRCC1 in HeLa cells (Figure 6C). However, inhibition of endogenous SIRT1 activity with NAM negated MMS-induced increase in APE1-XRCC1 binding (Figure 6D). Because NAM may have off-target effects, we also examined the effect of SIRT1 knockdown on MMS-stimulated binding of APE1 to XRCC1. Knockdown of SIRT1 with siRNA suppressed MMS-stimulated binding of APE1-XRCC1 (Figure 6E). Collectively, these findings suggest that genotoxic stress upregulates SIRT1 and acetylates APE1, and the increase in SIRT1 expression acts to deacetylate APE1, thereby allowing formation of the APE1:XRCC1 complex in the BER pathway.
Because SIRT1 targets the N-terminal lysine residues in APE1 for deacetylation, we next asked whether these lysine residues mediate the stimulatory effect of SIRT1 on binding of APE1 to XRCC1. When compared with wild-type protein, APE1 that was non-acetylatable at lysines 6 and 7 bound with greater affinity to XRCC1 under basal conditions (Figure 6E). In addition, MMS stimulated binding of the non-acetylatable APE1 to XRCC1 to a greater extent than it did of wild-type APE1 (Figure 6E). Importantly, unlike its effect on MMS-stimulated binding of wild-type APE1 to XRCC1, knockdown of SIRT1 did not suppress MMS-stimulated binding of non-acetylatable APE1 to XRCC1 (Figure 6E). These findings show that non-acetylatable APE1 binds preferentially to XRCC1, both under resting conditions and with genotoxic stress. Moreover, they suggest that rendering lysines 6 and 7 non-acetylatable may impact on additional residues within this N-terminal region, thus facilitating genotoxic stress-induced binding of APE1 to XRCC1.
Cellular AP endonuclease activity is regulated by SIRT1
Finally, we asked whether SIRT1-mediated increase in binding of APE1 to XRCC1 is associated with an increase in the abasic endonuclease activity in protein complexes containing XRCC1. AP endonuclease activity was measured in XRCC1 immunoprecipitates from lysates of APE1-expressing HeLa cells that were treated with NAM or resveratrol. Compared with untreated cells, treatment with NAM led to a decrease, whereas treatment with resveratrol increased, the AP endonuclease activity of XRCC1 immunoprecipitates (Figure 6F). In addition, siRNA-mediated knockdown of SIRT1 suppressed AP endonuclease activity of XRCC1 immunoprecipitates (Figure 6G). However, neither p300-induced acetylation, nor SIRT1-induced deacetylation, changed the in vitro AP endonuclease activity of recombinant APE1 (Figure 6H). These findings are consistent with the notion that SIRT1-mediated deacetylation of APE1 does not directly affect the AP endonuclease activity of APE1, but rather, SIRT1 stimulates cellular AP endonuclease activity by promoting the association of APE1 to XRCC1.
DISCUSSION
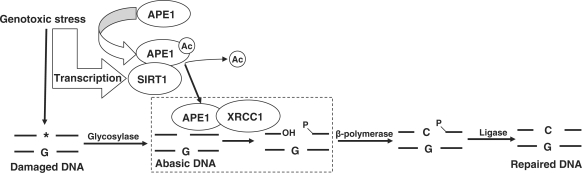
The principal findings in this study are (i) APE1 is a target for deacetylation by SIRT1, (ii) SIRT1 promotes association of APE1 with XRCC1, (iii) SIRT1 stimulates cellular AP endonuclease activity, and (iv) APE1, in part, mediates the cytoprotective effect of SIRT1. These findings add to the growing evidence supporting a role for SIRT1 in maintenance of genomic stability (39,40). SIRT1 serves this role through deacetylation-mediated regulation of multiple target proteins that play a part in orchestrating cell cycle arrest and apoptosis in response to DNA damage (21,26–28,35,41). Despite multiple known targets of SIRT1, few are directly involved in DNA repair. NBS1, a component of the nuclease complex that senses and participates in the response to double-stranded DNA breaks, is one target of SIRT1 that plays a direct role in DNA repair (29). Our findings add APE1 to the target proteins of SIRT1 that are directly involved in repair of damaged DNA. Based on our experimental findings we postulate a model in which SIRT1 plays an important role in the BER pathway, both under basal conditions and with genotoxic stress (Figure 7). This model predicts that SIRT1 tonically binds to and deacetylates APE1, and this physical and functional interaction increases when the demand for BER increases such as with genotoxic insults.
Figure 7.
Scheme depicting the role of SIRT1 in the BER of damaged DNA. Rectangle shows the AP endonuclease step. Ac: acetyl.
Prior work has demonstrated that APE1 is acetylated by the p300 acetyltransferase on lysine 6 and/or 7, and APE1 that is non-acetylatable on these residues is impaired in its capacity to bind to the nCaRE element in the PTH promoter, and repress PTH promoter activity in response to an increase in extracellular Ca2+ concentration (16). This study also showed that inhibition of class I and II HDACs with TSA increased lysine acetylation of APE1 to some degree, suggesting that acetylation of APE1 is regulated by such HDAC. However, despite stimulating lysine acetylation of APE1, TSA did not promote the ability of APE1 to repress PTH promoter activity with increase in extracellular Ca2+. These findings suggest that direct deacetylation of APE1 by class I and II HDACs does not play a major part in the negative calcium response mediated by APE1. Our findings are consistent with this previous report in that they show that class I and II HDACs do play some part in modulating the basal lysine acetylation of APE1. Other studies have shown a more robust increase in lysine acetylation of APE1 with inhibition of class I and II HDAC for a longer period of time (17), and this may reflect difference in deacetylation kinetics between such HDAC and SIRT1. Although we do not underestimate the importance of class I and II HDAC in regulating APE1 acetylation, our data also points toward a novel role for the class III HDAC SIRT1 in inhibiting both basal and genotoxic stress-induced lysine acetylation of APE1, and suggest that, similar to the scenario with p53 acetylation on lysine 382 (22), SIRT1 and certain class I and II HDAC may target some of the same lysine residues in APE1. Moreover, our data show for the first time that acetylation of these lysine residues is reversible, and identify SIRT1 as one of the deacetylases responsible for this.
APE1 forms a multi-molecular DNA repair complex directed toward abasic DNA sites. XRCC1, which may serve as a scaffolding protein in this complex, is known to interact with APE1 through the N-terminal region of APE1, and stimulates the AP endonuclease activity of APE1 (38). The finding that the acetylation status of two lysine residues in the N-terminal region of APE1 is important in determining its binding to XRCC1 is consistent with recent reports showing that this region, which has an intrinsically disordered structure (2,42), is essential for its interaction with other nuclear proteins. A recent study shows that the interaction of APE1 with nucleophosmin-1 (NPM1), a nucleolar protein, is mediated by the 33 amino acid N-terminal region of APE1 (43). Moreover, this interaction stimulates the AP endonuclease activity of APE1 in a cell-free system, a finding that suggests a direct effect on NPM1 on APE1 endonuclease activity. Our experimental data did not show an effect of acetylation or SIRT1-induced deacetylation on the endonuclease activity of recombinant APE1 in vitro, nor did we find that the acetylation of APE1 in vitro changes the polymerase activity of β-polymerase (T. Yamamori and K. Irani, unpublished observations), another enzyme in the BER pathway that physically associated with APE1 (5). These findings support our hypothesis that SIRT1 impacts on cellular AP endonuclease activity indirectly by modulating the interaction of APE1 with other proteins of the BER machinery, but does not directly change the activity of APE1 or APE1-associated DNA repair enzymes. Nevertheless, based on the interesting finding that the interaction of APE1 with NPM1 directly stimulates its endonuclease activity, it would be worthwhile to see if acetylation and SIRT1-induced deacetylation affects APE1 endonuclease activity stimulated by NPM1 through modulation of APE1-NPM1 binding. Such studies may be especially revealing in light of the roles of NPM1 in ribosomal biogenesis (43), and SIRT1 in ribosomal RNA synthesis (44).
APE1 also interacts with the Y-box-binding protein 1 (YB-1) enhancing its binding to the Y-box element, and stimulating multi-drug resistance (MDR1) gene expression (45). This interaction also occurs though the N-terminal region of APE1, reinforcing the importance of this region in binding of APE1 to other proteins. Notably, APE1 that is non-acetylatable on lysines 6 and 7 displays lesser binding to YB-1 than wild-type protein. Our findings suggest that APE1 that is deacetylated by SIRT1 preferentially binds to XRCC1. We did not examine the role of acetylation in mediating the interaction of APE1 with SIRT1. However, we did observe that H2O2 increased acetylation of APE1 and its association with SIRT1, hinting at the possibility that as far as the APE1:SIRT1 interaction is concerned, acetylation of APE1 may facilitate its binding to SIRT1. A similar increase in binding of SIRT1 to FOXO transcription factors, a target of SIRT1, with H2O2 has been reported (46). Increase in this APE1:SIRT1 interaction by genotoxic stress would allow the deacetylation of APE1 thus permitting APE1:XRCC1 binding. It is not clear what purpose acetylation of APE1 may serve in the context of genotoxic insults. However, one can hypothesize that the degree of acetylation of APE1 may serve as a switch that guides the cell down one of two paths: hyperacetylation of APE1 may induce a transcriptional program of apoptosis, perhaps by activating transcription factors as it does YB-1, whereas SIRT1-stimulated hypocetylation may promote DNA repair and cell survival. This scenario, if true, would be reminiscent of how acetylation and SIRT1-mediated deacetylation of FOXO transcription factors guides the cell toward a fate of survival or death (26).
The effect of genotoxic agents on the acetylation of APE1 deserves attention. Although both H2O2 and MMS increased acetylation of APE1, this effect was modest and became much more apparent when endogenous SIRT1 activity was inhibited. Importantly, MMS increased SIRT1 expression and this increase followed a time course that was reciprocal to MMS-stimulated APE1 acetylation. These findings suggest that endogenous SIRT1 functions to negate the acetylation of APE1 by these genotoxic stimuli. Although these studies establish the role of SIRT1 in negatively regulating acetylation of APE1 in response to these genotoxic stimuli, we did not explore the mechanism for acetylation of APE1 by these stimuli. It is noteworthy however, that MMS increases the acetylation of Werner (WRN), another important protein in base excision repair pathways, in a p300-dependent fashion (47), suggesting that H2O2 and MMS-induced acetylation of APE1 may also be mediated by p300. However, we cannot exclude the possibility that other acetyltransferases may also target APE1 for lysine acetylation in response to DNA damaging agents, as has been observed for p53 acetylation (48).
We observed that MMS leads to transcriptional upregulation of SIRT1 expression. SIRT1 is known to be transcriptionally induced by the DNA damaging agent etopside (41). This transcriptional induction is mediated by the transcription factor E2F1, with a conserved E2F1-binding element present in the human and mouse SIRT1 promoters. Therefore, it is likely that induction of SIRT1 by MMS is also dependent on E2F1. In support of this, reporter analysis of truncated human SIRT1 promoters have shown that the sequence 115 bp upstream of the transcription start site, which includes the E2F1 binding site, is sufficient for induction of promoter activity by MMS (T. Yamamori and K. Irani, unpublished observations).
Finally, although our study focused on exploring the consequence of the APE1:SIRT1 interaction on lysine acetylation of APE1, it is tempting to speculate that this association may also facilitate a yet unknown effect of APE1 on SIRT1. One attractive possibility is that this interaction may allow the regulation of SIRT1 by APE1, similar to the role that APE1 plays in the regulation of AP-1 transcriptional activity through its physical association with thioredoxin (49).
FUNDING
Funding for open access charge: National Institutes of Health (R01 HL070929, R01 HL094959, and P01 HL065608) and the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 2.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 3.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 4.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 5.Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 7.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty DM, Monick MM, Hunninghake GW. AP endonucleases and the many functions of Ref-1. Am. J. Respir. Cell Mol. Biol. 2001;25:664–667. doi: 10.1165/ajrcmb.25.6.f220. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J. Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J. Biol. Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 12.Yacoub A, Kelley MR, Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res. 1997;57:5457–5459. [PubMed] [Google Scholar]
- 13.Fritz G, Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene. 1999;18:1033–1040. doi: 10.1038/sj.onc.1202394. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh MM, Hegde V, Kelley MR, Deutsch WA. Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res. 2001;29:3116–3122. doi: 10.1093/nar/29.14.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fantini D, Vascotto C, Deganuto M, Bivi N, Gustincich S, Marcon G, Quadrifoglio F, Damante G, Bhakat KK, Mitra S, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic. Res. 2008;42:20–29. doi: 10.1080/10715760701765616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 20.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol. Metab. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 22.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 23.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat. Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 24.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 26.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 27.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 28.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grosch S, Fritz G, Kaina B. Apurinic endonuclease (Ref-1) is induced in mammalian cells by oxidative stress and involved in clastogenic adaptation. Cancer Res. 1998;58:4410–4416. [PubMed] [Google Scholar]
- 31.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D. Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic. Biol. Med. 2006;40:2175–2182. doi: 10.1016/j.freeradbiomed.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J. Biol. Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 33.Tomicic M, Eschbach E, Kaina B. Expression of yeast but not human apurinic/apyrimidinic endonuclease renders Chinese hamster cells more resistant to DNA damaging agents. Mutat. Res. 1997;383:155–165. doi: 10.1016/s0921-8777(96)00055-9. [DOI] [PubMed] [Google Scholar]
- 34.Walker LJ, Craig RB, Harris AL, Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 37.Fan J, Wilson DM., 3rd Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 42.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 43.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'A;mbrosio C, Scaloni A, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol. Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol. Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J. Biol. Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 47.Muftuoglu M, Kusumoto R, Speina E, Beck G, Cheng WH, Bohr VA. Acetylation regulates WRN catalytic activities and affects base excision DNA repair. PLoS ONE. 2008;3:e1918. doi: 10.1371/journal.pone.0001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl Acad. Sci. USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]