Abstract
Large-scale and genome-wide studies have concluded that ∼80% of the yeast (Saccharomyces cerevisiae) genome is occupied by positioned nucleosomes. In vivo this nucleosome organization can result from a variety of mechanisms, including the intrinsic DNA sequence preferences for wrapping the DNA around the histone core. Recently, a genome-wide study was reported using massively parallel sequencing to directly compare in vivo and in vitro nucleosome positions. It was concluded that intrinsic DNA sequence preferences indeed have a dominant role in determining the in vivo nucleosome organization of the genome, consistent with a genomic code for nucleosome positioning. Some other studies disagree with this view. Using the large amount of data now available from several sources, we have attempted to clarify a fundamental question concerning the packaging of genomic DNA: to what extent are nucleosome positions in vivo determined by histone-DNA sequence preferences? We have analyzed data obtained from different laboratories in the same way, and have directly compared these data. We also identify possible problems with some of the experimental designs used and with the data analysis. Our findings suggest that DNA sequence preferences have only small effects on the positioning of individual nucleosomes throughout the genome in vivo.
INTRODUCTION
Eukaryotic chromosomal DNA is packaged into nucleosomes, each consisting of 147 bp wrapped tightly around a core histone octamer (1,2). A recent genome-wide high-resolution microarray study states that ∼80% of the yeast (Saccharomyces cerevisiae) genome consists of (translationally) positioned nucleosomes (3). This result is consistent with the conclusions of an earlier microarray study (4) and a subsequent parallel sequencing study (5) in which only portions of the yeast genome were examined. There are a variety of possible mechanisms that could lead to nucleosome positioning. For example, the base-pair-specific binding of a non-histone protein to DNA could exclude a nucleosome from that DNA region. Then, a regularly spaced array of adjacent nucleosomes would be positioned in the vicinity of the DNA-bound non-histone protein. Alternatively, an array of several regularly spaced nucleosomes could form adjacent to one sequence-positioned nucleosome. These mechanisms have been called statistical positioning (6). Statistical positioning would be most effective in yeast where the DNA linkers between adjacent nucleosomes are on average very short, only 18 bp for a 165-bp nucleosome repeat length (NRL) (7), and therefore not much statistical variation in nucleosome linker lengths can occur. Nucleosome positioning could also result from the boundary effect provided by the attachment of DNA regions to nuclear structures (8), chromatin remodeling (5,9) or as a result of DNA replication from a bidirectional replication origin (7). In addition, nucleosome positioning could result from the DNA sequence preferences of the histones themselves. It has been known for some time that in vitro nucleosomes form with high preference on certain DNA sequences (10–13) and tend to avoid other sequences (14–21).
Evidence was provided that there is a genomic code for nucleosome positioning, and that ∼50% of the nucleosome positions in yeast (±35 bp) result from histone preferences for certain DNA motifs (22). A different, complementary approach reported similar results for computationally predicting the positions of positioned yeast nucleosomes based on the genomic DNA sequence (±35 bp), but concluded that only ∼25% of the positioned nucleosomes can be attributed to the preferences of certain DNA sequence motifs for histones (23), a value considered to be too low for the existence of a nucleosome positioning code. A critical evaluation of the statistics used by Segal et al. (22) in 2006 also suggested that the performance of the proposed nucleosome positioning code is more modest than claimed (24). In 2007, Lee et al. (3) reported that there was a poor correlation between their microarray-determined genome-wide nucleosome occupancy values and the predictions by Segal et al. (22) in 2006. However, they found that there was a moderate correlation (R = 0.44) between their measured nucleosome occupancy values and a collection of DNA structural or sequence parameters. This degree of correlation suggests that only about 19% (R2 × 100%) of their nucleosome occupancy values are represented by the DNA structure/sequence parameters that they used in their model. Consistent with these findings, it was suggested that statistical positioning (discussed earlier), rather than intrinsic positioning, largely accounts for the nucleosome positioning observed in S. cerevisiae (25). In addition, it was reported in a genome-wide study, that Caenorhabditis elegans, which has (on average) longer nucleosome linkers than yeast, generally lacks sequence-dictated nucleosome positioning (26).
Recently, a direct genome-wide comparison of in vivo and in vitro nucleosome positioning in yeast was performed using the massively parallel Illumina sequencing system (27). The number of reads overlying each base pair for DNA sequences extracted from nucleosomes that were excised from native or reconstituted chromatin by micrococcal nuclease was used to assess the nucleosome occupancy at each base pair. This same sequencing approach had been used earlier to assess the nucleosome occupancy per base pair of the much smaller SV40 virus genome, where it was found that unique nucleosome positions did not occur (28). Control experiments were also performed by Kaplan et al. (27), using ∼40 000 synthesized 150-bp DNA sequences to validate their yeast genomic DNA results. In these experiments, competitive reconstitution and microarray analysis were used to assess the affinities of each synthetic DNA sequence for histones to show that 5-mers contained in genomic sequences that had high (or low) affinities had corresponding affinities in the synthetic DNAs. Kaplan et al. (27) concluded from their direct genome-scale experiment that intrinsic nucleosome sequence preferences do have a dominant role in determining the nucleosome organization in vivo.
Shortly after the Kaplan et al. (27) study in 2009, Zhang et al. (29) reported a similar parallel sequencing study for yeast chromatin in vivo and in vitro, but using somewhat different methodology. They concluded that intrinsic histone–DNA interactions are not the major determinant of nucleosome positioning.
It is clear that there are apparent conflicts in the current literature on the question: are nucleosome positions in vivo primarily determined by histone–DNA sequence preferences?
In an attempt to resolve these apparent conflicts, in this study we first examined the degree of correlation between nucleosome occupancies from the yeast in vitro parallel sequencing data and those from the in vivo microarray data of Lee et al. (3). We found that nucleosome occupancies in vitro and in vivo correlate less well when the data from the two different studies are compared than when the parallel sequencing data of Kaplan et al. (27) in vitro and in vivo are compared. We discuss a potential problem with correlation analysis using scatter plots, when large numbers of superimposed points are present. We then analyzed the synthetic DNA nucleosome occupancy data provided by Kaplan et al. (27) in a more direct way than the authors reported and found that there is not a very good correlation between their parallel sequencing data and their microarray data for these sequences. We suggest possible causes for the apparent discrepancies between the Illumina-Solexa parallel sequencing data and the microarray data, and between the two recent genome-wide parallel sequencing studies (27,29). We precisely calculate the effect of ‘statistical positioning’ in yeast. Furthermore, we examine what it really means to say that genomes encode an intrinsic nucleosome organization that can explain approximately half of the in vivo nucleosome positions.
SOME CHARACTERISTICS OF YEAST CHROMATIN AND NUCLEOSOME ARRAYS
Nucleosomes are generally taken to contain 147 bp of DNA that is tightly wrapped 1.7 times around the histone core. In chromosomal DNA, the nucleosomes are connected by relatively short and variable lengths of histone-free linker DNA (2). The NRL is the average distance between the midpoints of the two linkers flanking a nucleosome. The NRL is usually measured by limited nuclease digestion of the chromatin, and analysis of the resulting periodic purified DNA lengths on an agarose gel, arising from the nucleosome oligomers excised. Yeast has an unusually short NRL of only 165 bp, compared to the more typical 190 ± 5 bp of metazoans (7). Thus, most nucleosomes in yeast are very close to each other, and the DNA linkers are short. The average yeast nucleosome linker length calculated from the bulk chromatin NRL is 165 bp−147 bp = 18 bp. Consistent with the NRL value, the core histone to DNA weight ratio in yeast is 1.0 ± 0.2 (30), which corresponds to one nucleosome for every 167 bp. From a high-resolution genome-wide microarray study (3), the average length of non-nucleosomal DNA was estimated to be 32.4 bp. This value reflects the presence of occasional nucleosome-free regions (with lengths larger than average linkers) in addition to the linker DNA. Therefore, biochemical analysis, NRL analysis and genome-wide microarray analysis all indicate that the proportion of the genome contained in nucleosomes is very high, with an average occupancy value (see definition below) >0.80 [147 bp/(147 bp + 32.4 bp)]. Nucleosomes in arrays can be, but do not have to be, spaced regularly with respect to each other. In addition, the arrangement of nucleosomes on the DNA molecules in the different cells of a sample does not necessarily have to be the same.
A positioned nucleosome is defined as one for which the nucleosome center (or a nucleosome boundary) occurs at the same nucleotide coordinates, ± x bp, in all of the DNA molecules in the sample (7). Segal et al. (22) used an x-value of 35 bp to compare predicted nucleosome positions with measured nucleosome positions. Lee et al. (3) have identified over 70 000 ‘positioned nucleosomes’ in yeast using high-resolution microarray analysis. They define a ‘well-positioned’ nucleosome as one that hybridizes to 31–38 consecutive probes of length 25 bp that were separated by 4 bp. This definition would correspond to an x-value of ∼13 bp. Zhang et al. (29) used an x-value of 10 bp. Nucleosomes are sometimes referred to as being uniquely positioned, if they occupy the same position (±x bp) on all of the DNA molecules in the sample (7,31,32). It is sometimes stated that nucleosomes occupy alternative positions, or arrays have multiple positioning frames (on different DNA molecules), when the measured positions indicate that two (or more) distinct nucleosomes appear to overlap with one another if they were present on the same DNA molecule (7,32,33), which cannot happen.
Recently, in 2009, Segal and Widom (34) have precisely defined nucleosome positioning and nucleosome occupancy. They define nucleosome positioning at a base pair as the probability that a nucleosome center (or a nucleosome start) is at that base pair. The value of this probability can be estimated approximately by dividing the number of nucleosome centers found at the base pair in question by the number of nucleosome centers found in the region ±83 bp from the base pair in question. For a uniquely positioned nucleosome, with center positioned precisely on the base pair in question on all molecules in the sample, the probability value at that base pair would be one. Segal and Widom define nucleosome occupancy at a given base pair to be the total probability with which that base pair is covered by any of the nucleosomes that could potentially cover it. They point out that the occupancy at base pair i is the sum of the probabilities of all of the (mutually exclusive) nucleosomes starting from base pair i-146 to base pair i. They further point out that occupancies, like probabilities, vary between 0 and 1. More simply, the occupancy at position i can be thought of as the fraction of molecules in the sample that have any part of a nucleosome covering position i. It is worth noting that for a random nucleosome arrangement, the probability that a nucleosome, in chromatin having 20-bp linkers (but no nucleosome-free regions), starts at any position is 1 bp/167 bp. Following Segal and Widom, the sum of this constant probability over the 147 positions from i-146 to i, leads to an occupancy value of 147 bp/167 bp = 0.88, consistent with Kornberg and Stryer [(6), and Figure 4A, upper curve]. Moreover, the average nucleosome occupancy value for any region of chromatin having a core histone to DNA weight ratio of 1.0 is ∼147 bp/167 bp = 0.88 (6). Including the experimentally determined nucleosome-free regions (3), reduces the genome-average occupancy value to 0.82 (see above).
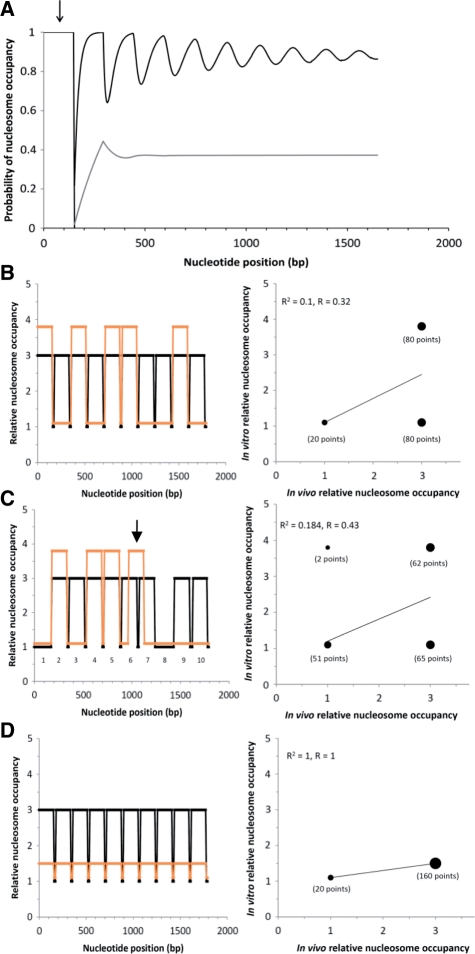
Figure 4.
Effects on nucleosome positioning and on the in vitro–in vivo occupancy correlation for chromatin reconstituted at less than or equal to half the average in vivo occupancy value. (A) The probability that a base pair is contained in a nucleosome was calculated for in vivo yeast chromatin (average linker length, L = 20 bp) according to Kornberg and Stryer’s 1988 equation (5) (black curve), and for in vitro reconstituted chromatin at a histone to DNA ratio value of 0.42 times the physiological value (L = 250 bp, see text) (gray curve). The diameter of the nucleosome core (d) is 147 bp. A well-positioned nucleosome, directed by the DNA sequence, is placed on base pairs 0–146; the other nucleosomes have random positions. The DNA molecules are assumed to be very long; only 10 nucleosomes are shown. (B) Simulated variation of relative nucleosome occupancy values with nucleotide position for one possible arrangement of five intrinsically positioned nucleosomes in vitro (orange curve) out of 10 well-positioned nucleosomes in vivo (black curve). Nucleosome cores are 150 bp; linkers are 20 bp. Points were taken every 10 bp. The scatter plot comparing the in vitro and in vivo occupancy values for all nucleotide positions is shown at the right. The number of points in each of the three clusters is indicated. R2 and R values are given. (C) Simulated variation of relative nucleosome occupancy values for 10 potential nucleosome positions in vivo (black curve), two of which (1 and 8) are nucleosome-free because the sequences disfavor nucleosome formation, and three (2, 4 and 5) have sequences that favor nucleosome formation. In vitro (orange curve), there are four nucleosomes present, compared to the eight in vivo; these nucleosomes avoid the two nucleosome-free regions, and occupy the three regions that favor nucleosomes; the fourth nucleosome (arrow) is not positioned. Other features of the simulation are as described for (B). (D) Simulated variation of relative nucleosome occupancy values for 10 nucleosome positions, all of which are intrinsically positioned on some fraction (<0.5) of the DNA molecules in the sample or for some fraction (<0.5) of the time. Other features of the simulation are as described for (B).
COMPARISON OF THE PARALLEL SEQUENCING IN VITRO NUCLEOSOME OCCUPANCIES OF KAPLAN ET AL. WITH THE MICROARRAY IN VIVO NUCLEOSOME OCCUPANCIES OF LEE ET AL.
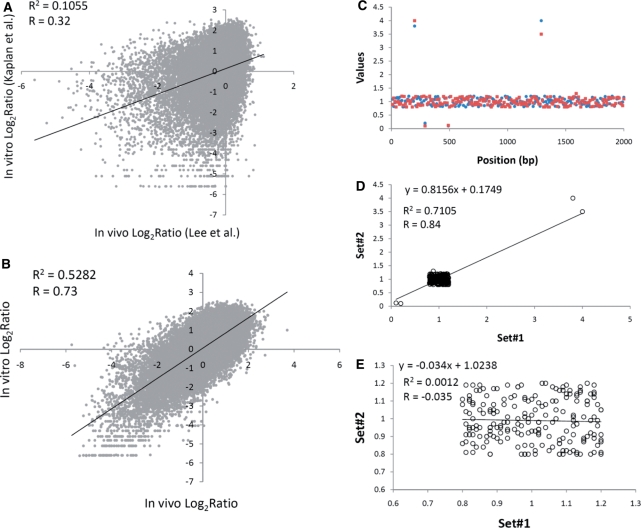
Kaplan et al. (27) reported that a high degree of similarity exists between the in vivo and in vitro nucleosome organizations in yeast, with a correlation coefficient R = 0.74 between the nucleosome occupancy values. We decided to see whether the in vitro nucleosome occupancy data of Kaplan et al. (27) correlate as well with the high-resolution in vivo microarray data, previously reported by Lee et al. (3). We chose to represent the data (Figure 1A) using only chromosome 4, the largest yeast chromosome, because the scatter plot repeatedly piles tens of thousands of points on top of each other for the whole genome, making it difficult to interpret the data. Results similar to Figure 1A for the whole genome are shown in Supplementary Figure S1A. We also performed the same analysis in Figure 1B, comparing the Kaplan et al. (27) parallel sequencing in vitro data to the Kaplan et al. 2009 (27) parallel sequencing in vivo data. Whereas the chromosome 4 parallel sequencing in vitro versus in vivo data (Figure 1B) gives a very similar correlation coefficient (R = 0.73) to the Kaplan et al. (27) genome wide result (R = 0.74), the correlation coefficient for the microarray (in vivo) data (Figure 1A) is significantly less (R = 0.32). In both cases, the P-values are very small, due to the large numbers of data points present. Thus, the in vitro parallel sequencing data do not correlate nearly as well with the microarray in vivo data as they do with the parallel sequencing in vivo data.
Figure 1.
Scatter plots and correlation coefficients for yeast chromosome 4 and a simulation to illustrate the ‘influential point effect’. (A) Comparison of the in vitro nucleosome occupancy ratios of Kaplan et al. (27) by Illumina–Solexa sequencing and the in vivo nucleosome occupancy ratios of Lee et al. (3) by microarray analysis. Points were taken every 48 bp. (B) Comparison of the in vitro nucleosome occupancy ratios of Kaplan et al. (27) by Illumina–Solexa sequencing and the in vivo nucleosome occupancy ratios of Kaplan et al. (27) by Illumina–Solexa sequencing. Points were taken every 50 bp. (C) Simulated nucleosome occupancy data for the case where 196 points from each of two data sets (set #1: blue circles; set #2: red squares) exhibit small random deviations from y = 1, but four pairs of points exhibit larger correlating deviations. Points were plotted every 10 bp. (D) Scatter plot for the simulated nucleosome occupancy data shown in (C). (E) Scatter plot for the nucleosome occupancy data shown in (C) omitting the four influential correlating points.
Actually, even the weak apparent correlation (R = 0.32) of the data points in Figure 1A is overestimated because of the well-known ‘influential point effect’, which occurs when there are a large number of points present in a small region of the scatter plot plus a small number of more spread out correlated points (35). For example, the points in the lower left quadrant of Figure 1A are unusually influential. Omitting just some of these points, <1% of the total points plotted, lowers the value of R to 0.26 (data not shown). The ‘influential point effect’ is clearly illustrated by the simulated data shown in Figure 1C–E. Here, there are 200 total points in each of two data sets (over 2000 bp), 196 of which deviate randomly by small amounts in their y-values from y = 1. Additionally, there are four correlated pairs of points, which deviate by greater values. The scatter plot for these data is shown in Figure 1D. The correlation coefficient appears to be high and significant (R = 0.84, P < 10−10). However, when the four actually correlating points are omitted, the correlation vanishes (R = −0.035), as shown in Figure 1E. Thus, the scatter plot and Pearson correlation coefficient overestimates the number of points that really correlate, when large numbers of points are superimposed in a small region of the scatter plot. The color-coded, whole-genome, plot (Supplementary Figure S1A) indicates that there are very high densities of poorly correlating points clustered near the origin of the scatter plot, and suggests that, at best, a weak correlation exists.
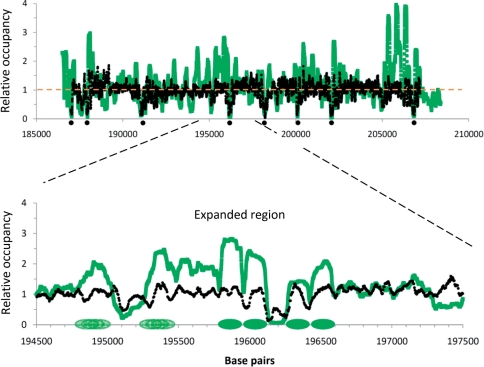
We also compared the relative nucleosome occupancies for the Kaplan et al. (27) in vivo parallel sequencing data with the Lee et al. (3) in vivo data across the 20 000-bp region of chromosome 14 (187 000–207 000) used in Figure 1 of Kaplan et al. (27). Figure 2 shows the data of Kaplan et al. (27) for a 20-Kb region of chromosome 14, reported to be a ‘typical’ genomic DNA region. The green curve is nearly identical to the green curve displayed in Figure 1 of Kaplan et al. (27) (labeled YPD, in vivo). Here, their data is plotted as connected points, rather than as a histogram, and a sliding 4-bp average was applied to compare the results with the 4-bp resolution data of Lee et al. (3). Superimposed on this curve is the data reported by Lee et al. (3) (black curve). It is apparent that the Kaplan et al. (27) relative occupancies have more values that are significantly greater than the genome average value (orange dashed line at y = 1) than the Lee et al. (3) data. The high-occupancy regions of Kaplan et al. (27) do not appear to correlate very well with the corresponding regions of Lee et al. (3). However, eight low-occupancy regions (indicated by dots) do correspond in this 20-Kb region, which should contain ∼120 nucleosomes [20 000 bp/165 bp nucleosome−1, where 165 bp is the bulk NRL (7)].
Figure 2.
In vivo relative nucleosome occupancy for yeast chromosome 14: 187 000–207 000 (upper curve), as reported by Kaplan et al. (27) (Figure 1). The green curve is the YPD (in vivo) data of Kaplan et al. (27) from Illumina–Solexa sequencing; the dashed orange line at y = 1 represents the genome-wide average; the black curve is the (in vivo) data of Lee et al. (3) by microarray analysis. Black dots indicate eight regions where both studies found low nucleosome occupancies. The inset (below) shows an expanded view of the 3000-bp region (194 500–197 500). Four well-positioned nucleosomes (dark green ovals) and two ‘fuzzily’ positioned nucleosomes (overlapping light green ovals) assigned on the basis of the green curve are shown on the x-axis.
To confirm that the 20 000-bp region is really a typical region, we examined all of chromosome 14 for the presence of nucleosome-enriched regions (NERs) and nucleosome-depleted regions (NDRs). These regions were defined as DNA regions of sizes at least 100 bp that had relative occupancies >1 SD (NER) or <1 SD (NDR) from the average value. For the NERs, there were 454, with mean separation of 1676 bp ± 2133 bp for the microarray data; and 559, with mean separation of 1363 bp ± 1536 bp for the parallel sequencing data. Approximately 31% of the microarray-determined NERs overlapped with parallel sequencing NERs. For the NDRs, there were 520, with mean separation of 1469 bp ± 1537 bp for the microarray data; and 510, with mean separation of 1496 ± 1416 bp for the Illumina–Solexa sequencing data. Approximately 60% of the microarray-determined NDRs overlapped with parallel sequencing NDRs. Thus, the 20 000-bp region shown in Figure 2 seems to be representative of the whole chromosome, and indicates that there is a poor agreement (30%) between the NERs determined by the two experimental methods, but a better agreement (60% match) for NDRs.
It is clear from the microarray data (black curve) that the small deviations of the relative nucleosome occupancies from the control value (y = 1) preclude the precise identification of nucleosome positions over most of this 20-Kb region. Even for the parallel sequencing data, coordinates of positioned nucleosomes are difficult to assign. An expanded 3-Kb region is shown below, as was also provided by Kaplan et al. (27). Of the ∼18 nucleosomes expected to be present in this 3-Kb region, we would only feel somewhat confident in assigning unique positions to four nucleosomes (Figure 2, dark green ovals) and several possible positions each to two nucleosomes (light green overlapping ovals), based upon the parallel sequencing nucleosome occupancy values (green curve). Only in these regions are there elevated occupancy values flanked by significantly lower occupancy values that are separated by ±150 bp. The curve resulting from the unaveraged sequencing data is essentially identical to the curve shown here (data not shown). Thus, the nucleosome occupancy data from neither study is really consistent with 80% (14 of the 18) of the nucleosome positions in this ‘typical’ region being well-defined.
Whereas the Lee et al. (3) relative occupancy values (Figure 2, upper, black curve) are roughly consistent with actual occupancy values that can occur in yeast chromatin, the Kaplan et al. (27) values (green curve) are not. As described earlier in the section ‘Some characteristics of yeast chromatin and nucleosome arrays’, nucleosome occupancy values, by definition, vary from 0 to 1, and the average nucleosome occupancy for a typical region of yeast chromatin should be ∼0.80 (see above). To convert the relative nucleosome occupancy in Figure 2 to actual nucleosome occupancy, we can adjust the y-axis scale slightly in Figure 2, upper graph (and in Figure 1 of Kaplan et al. (27), upper graph), so that the genome average occupancy is at 0.80 instead of the arbitrarily set value of 1. Then, the relative occupancy value of 2 becomes actual occupancy of 1.6; the relative occupancy value of 3 becomes actual occupancy 2.4; etc. However, there are many actual nucleosome occupancy values in the Kaplan et al. data (27) that significantly exceed 1 (1.25 on the relative occupancy scale of Figure 2), the maximum possible value. Thus, the large increases in read numbers reported by Kaplan et al. (27) over the genome average cannot solely reflect nucleosome occupancy variations.
DIRECT COMPARISON OF THE HISTONE AFFINITIES OF THE SAME DNA SEQUENCES BY THE PARALLEL SEQUENCING AND THE MICROARRAY TECHNIQUES
Recently, the three major second-generation DNA sequencers were evaluated for potential sequence-dependent read-number variation (36). Long-range polymerase chain reaction (PCR) was used to ensure DNA fragmentation into equal numbers of short DNA fragments for sequencing. It was found that all of the second-generation sequencers gave read-number variations in excess of 100-fold. Typically, DNA regions extending more than 100 bp gave anomalously high or low read numbers, on average approximately every 1000 bp. The Illumina GA (Solexa) machine gave considerably more variation than the Roche-454 machine. These large, machine-dependent, read-number variations would seem to pose a problem in experiments for which conclusions are based on read numbers.
To address concerns regarding possible biases caused by the sequence preferences of micrococcal nuclease or by possible biases in parallel sequencing, Kaplan et al. (27) prepared ∼40 000 unique synthetic double-stranded 150-bp DNA sequences, thereby avoiding micrococcal nuclease digestion. The synthetic DNAs were of various types: 9907 concatenated 10-mers, 5782 random sequences, 3680 sequences consisting of dinucleotides having a range of periodicities, 22 236 yeast chr III sequences, and 1873 mouse sequences. They devised an experiment to measure the affinities of these synthetic DNA sequences without the use of parallel sequencing. Briefly, the combined pool of synthetic DNA sequences was reconstituted in vitro with limiting amounts of histones, thereby allowing nucleosomes to form on preferred sequences. The DNA that formed nucleosomes was separated from the DNA that did not by native gel electrophoresis. The relative abundance of each synthetic DNA in the nucleosome-forming fraction was then quantified by microarray analysis. These results provided a measure of the affinity of each synthetic DNA sequence; sequences that preferentially form nucleosomes would be more abundant than those that have low preferences for nucleosome formation.
Kaplan et al. (27) reported that the nucleosome-forming sequence preferences of the various 5-mers (1024 possibilities) contained in the synthetic DNA sequences was in excellent agreement with 5-mers contained in yeast genomic DNA reconstituted in vitro (R = 0.83). In turn, they concluded that this experiment confirms that the sequence specificities that they found through parallel sequencing were due to intrinsic nucleosome preferences, rather than being an artifact of their experimental approach. In addition, they measured the affinities of the same synthetic DNA sequences by parallel sequencing but did not directly compare the affinities obtained by their parallel sequencing with those obtained by their microarray analysis.
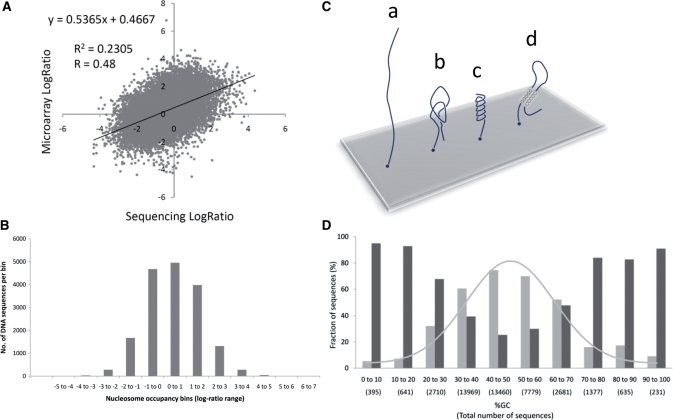
Figure 3A shows this comparison for 26 627 sequences out of the 43 879. Kaplan et al. (27) report (http://genie.weizmann.ac.il/pubs/nucleosomes08/nucleosomes08_data.html, Synthetic Oligonucleotides) that the remaining 17 252 sequences out of the 43 879 gave either zero reads or >500 reads (considered to be too high a number compared with the other sequences) by parallel sequencing; these were omitted from the Supplementary Data spreadsheet. Nucleosome occupancy log-ratios of all 43 879 sequences measured by microarray analysis were reported. Figure 3A shows only a modest correlation (R = 0.48) between the parallel sequencing results and the microarray results. The R2 value of 0.23 indicates that only 23% of the data are represented by the linear relation shown. Moreover, it can be seen that in the log-ratio range of 0 to −1 by parallel sequencing axis (x-axis), corresponding to only a slightly lower nucleosome-forming preference than the average value, some of the lowest and the highest log-ratios are found by microarray analysis (y-axis).
Figure 3.
Analysis of the nucleosome occupancy ratios, a possible problem with Illumina–Solexa read number comparisons, and %GC for the synthetic 150-bp DNAs examined by both microarray analysis and Illumina–Solexa sequencing by Kaplan et al. (27). (A) Scatter plot and correlation coefficient for the 26 627 DNAs for which log ratios were reported from both microarray analysis and Illumina–Solexa sequencing. (B) Microarray-determined nucleosome occupancy distribution for the 17 251 DNAs eliminated by Kaplan et al. (27) because the number of Illumina–Solexa reads was either zero or >500. (C) Illustration of a possible problem arising during Illumina–Solexa bridge amplification due to the formation of base sequence dependent single-stranded DNA structures. (a) The unstructured free DNA can loop down so that the free end can find a support-bound primer, as intended. (b) Local structure facilitates priming by placing the free DNA end closer to the support-bound primers. (c) Stacking interactions hinder priming by stiffening the DNA. (d) Hairpin formation of GC-rich homopolymer regions interfere with priming. (D) GC-content distribution of the total set of ∼40 000 synthetic DNAs (light grey bars and fitted Gaussian curve) and the ∼17 000 omitted (anomalous read number) synthetic DNAs (dark grey bars).
It is possible that the 17 252 omitted sequences with zero or very high (>500) sequence read numbers reflect the very low or very high affinities of these DNAs for histones, and that sequencing is just more sensitive than microarray analysis. To see whether these sequences have a bimodal distribution of log-ratios by microarray analysis that were higher-than-average or lower-than-average, we determined this distribution from the Supplementary Data reported. The histogram plot shown in Figure 3B is nearly normally distributed, rather than bimodal, indicating that nucleosome occupancies for the excluded synthetic DNA sequences grossly disagree when determined by parallel sequencing or by microarray analysis.
The Illumina–Solexa solid-phase DNA sequencing system is a powerful massively parallel system whose ability to sequence DNA has been demonstrated (37). In this system, the single-stranded DNA attached to the solid support must base pair to a nearby primer that is also attached to the support for amplification. However, this ‘bridge amplification’ step necessary to amplify the signal might be influenced by DNA sequence effects. It has been appreciated for some time that single strands of DNA are not disordered polymers. Their local structures seem to be well defined and dependent on the base composition and base sequence (38). Several possibilities are illustrated in Figure 3C. In (a), the unstructured free DNA can loop down for the end to find a bound primer, as intended. In (b), local structure facilitates priming by placing the free DNA end closer to the support-bound primers; whereas in (c), stacking interactions hinder priming by stiffening the DNA. In (d), hairpin formation of GC-rich homopolymer regions interferes with priming.
It is difficult to identify the sequence motifs which might possibly affect priming and read numbers from the data available. As high G/C content sequences might potentially promote problem (d) in Figure 3C, and oligo dA runs (low G/C content) are consistent with problem (c) (38), we simply tabulated the fractions of sequences present in bins of increasing %GC for both the total set of ∼40 000 synthetic DNAs, about 45% of which were yeast or mouse sequences, and the ∼17 000 sequences that had either anomalously low (zero) or anomalously high (>500) read numbers. These data are plotted in Figure 3D. Figure 3D shows that while the total set of synthetic DNA sequences (light shaded bars) is approximately normally distributed with respect to %GC, the distribution of the anomalous read sequences (dark shaded bars) are quite biased toward both low and high %GC, consistent with potential problems (c) and (d) in Figure 3C. We cannot obtain information about sequence motifs that might possibly give high reads (b) from the data provided.
The fact that more than 17 000 synthetic DNA sequences gave anomalous read numbers by parallel sequencing, while having nearly normally distributed nucleosome occupancy values by microarray analysis, and our analysis that the anomalous-read sequences tended to have unusually high or low GC-contents, suggests that Illumina–Solexa read numbers may be influenced by sequence in the 2009 Kaplan et al. study (27). This study would have benefited from a genomic DNA control, as used by others (3,29), such as randomly sheared purified genomic DNA to normalize each nucleosomal DNA value from the same region, to assess the background read variations across the genome. If the background Illumina–Solexa read numbers across the genome were known, it would have been possible to correct nucleosome occupancy ratios for any sequence-dependent read number influence. It is not possible to assess the extent to which possible sequence-dependent read number influenced the correlation analyses of Kaplan et al. (27) because of influential point effects on the scatter plots (discussed earlier), and because read numbers from parallel sequencing were also involved in the computational model for nucleosome positioning.
WHAT DOES IT MEAN TO SAY THAT GENOMES ENCODE AN INTRINSIC NUCLEOSOME ORGANIZATION THAT CAN EXPLAIN APPROXIMATELY HALF OF THE IN VIVO-DETERMINED NUCLEOSOME POSITIONS?
In 2006, Segal et al. (22) stated that genomes encode an intrinsic nucleosome organization that can explain approximately half of the in vivo nucleosome positions. The meaning of this statement is not entirely obvious, and here we wish to examine what it could mean. First, we consider the case that nearly all of the nucleosomes in the yeast genome have readily identifiable positions, obtained from some type of experiment. Then, we consider the case where some positions are only slightly more likely than other positions. Of course, if nucleosome occupancy values do not vary appreciably from the background levels, and lack distinct nucleosome period modulations, the correlation analysis will not be very reliable because of the influential point effect, as discussed in Figure 1C–E.
We have mentioned the phenomenon of statistical positioning (6) in the Introduction. Through statistical positioning, a single intrinsically positioned nucleosome could induce the positioning of adjacent nucleosomes on sequences which do not have any preferences for positioning nucleosomes. In Figure 4A (upper curve), we use Kornberg and Stryer’s (6) statistical mechanical equation 5 to precisely calculate the probability of nucleosome occupancy near a boundary, for example, a single intrinsically positioned nucleosome. We used parameters (d = 147 bp, L = 20 bp) reflecting yeast chromatin. The parameter d is the number of base pairs contained in a nucleosome, and L is the average linker length, consistent with the NRL of yeast chromatin. The calculation takes into account all possible non-overlapping ways of placing nucleosomes, in addition to the positioned one, on a very long DNA. The intrinsically positioned nucleosome (arrow) begins at 0 bp; the other nucleosomes can form anywhere. It can be seen that a substantial amount of nucleosome positioning is induced on DNA that has no preference for one position over another by a single intrinsically positioned nucleosome. This result does not preclude the possibility that genomic DNA sequences evolved to position most nucleosomes through a ‘DNA code’. It just shows that one of 10 nucleosomes being positioned by the DNA sequence is more than sufficient to generate the extent of positioning found by experiment. However, to address the question of whether half of the nucleosomes are intrinsically positioned, some way of distinguishing between the intrinsically positioned nucleosomes and the statistically positioned nucleosomes is required.
An excellent way of eliminating statistical positioning effects, but maintaining intrinsically positioned nucleosomes on the DNA, is to reconstitute the chromatin in vitro at a low (for example, 40% of the physiological) histone to DNA ratio. This method is precisely what Kaplan et al. (27) did. Figure 4A, gray lower curve, shows that under these conditions (d = 147 bp, L = 250 bp), statistical positioning does not occur to an appreciable extent. There is simply too low a nucleosome density for the nucleosomes to influence each other. The mean linker length, L = 250 bp, rather than 20 bp, is consistent with in vitro reconstitution at 0.42 times the in vivo histone octamer to DNA ratio: (20 + 147)-bp octamer−1/(250 + 147)-bp octamer−1. Under these conditions, only the intrinsically positioned nucleosomes have high probabilities of forming on specific 147-bp regions. However, there are some consequences of using a low histone to DNA ratio.
To illustrate these consequences, let us assume that there are 5 out of 10 nucleosomes that can adopt intrinsic (DNA-sequence-defined) positions, in accord with the statement that 50% of the nucleosomes are intrinsically positioned. In vivo (black curves in Figure 4B) all 10 nucleosomes are well positioned, but only 50% are intrinsically positioned. The particular arrangement of the intrinsically positioned nucleosomes does not matter. However, the arrangement is specified by the DNA sequence; hence, it must be the same on all DNA molecules. Otherwise, it would not be consistent with intrinsic nucleosome positioning. In vitro (orange curve), there are only five nucleosomes present, on average, and they will tend to occupy the five DNA-sequence-defined positions. The positioned nucleosomes can vary around their preferred positions, leading to some overlap (on different molecules) and, consequently, the nonzero backgrounds shown. The scatter plot for comparing how well the in vitro nucleosome positions agree with the in vivo positions in Figure 4B is shown alongside. The correlation coefficient, R, is only 0.32. R cannot really be much higher for the assumptions made here. Thus, a consequence of in vitro chromatin assembly at a low histone-to-DNA ratio, for the case that nearly all of the nucleosomes in the yeast genome have readily identifiable positions, and that only about 50% of them are intrinsically positioned, is that the in vivo and in vitro nucleosome positions cannot correlate well. This interpretation is not consistent with the R value obtained by Kaplan et al. (27) of 0.74.
Including nucleosome-free regions does not improve the correlation much. Figure 4C (black curve) shows 10 potential nucleosome positions in vivo; two of them are nucleosome-free because the sequences (numbered 1 and 8) disfavor nucleosome formation, and three (2, 4 and 5) have sequences that favor nucleosome formation. Thus, 5 of the 10 potential nucleosome positions are specified by the DNA sequence, as in Figure 4B. To obtain a histone to DNA weight ratio of 0.50 for the in vitro chromatin (orange curve), as in Figure 4B, we add another nucleosome (arrow) that is not positioned (because the three intrinsic positioning sites: 2, 4, and 5 are occupied). The scatter plot alongside Figure 4C shows that for this situation R = 0.43, still well short of 0.74.
Alternatively, the statement that genomes encode an intrinsic nucleosome organization that can explain approximately half of the in vivo nucleosome positions could be taken to mean that essentially ‘all’ of the nucleosomes are intrinsically positioned only about half of the time, or somehow, on only half of the copies of the identical DNA sequences in the sample. This interpretation would permit the in vitro and in vivo nucleosome positions to correlate strongly even at low histone to DNA ratios, as shown in Figure 4D. Here, the in vivo nucleosome arrangement is the same as in Figure 4B, but all of the in vivo sites tend to be occupied preferentially for about 50% of the in vitro nucleosomes. The degree of nucleosome occupancy over the background is necessarily low. For this interpretation, nucleosomes are not ‘well positioned’ if they only occupy their positions (±x bp) half of the time or on half of the identical copies of the DNA sequences. Moreover, if x were too large, the nucleosomes would hardly be positioned at all. One would have to wonder what the biological function of such an encoding might be.
CONCLUSIONS AND THE POSSIBLE FUNCTIONS OF NUCLEOSOME POSITIONING
We conclude that, on close examination, the data of Kaplan et al. (27) do not support the claim that nucleosome sequence preferences have a dominant role in determining in vivo nucleosome organization. A similar conclusion has been reached by Zhang et al. in 2009, based upon their experiments using recombinant Drosophila chromatin assembly factors, and different methodology (29). Consistent with the data of others (3,23–26), it appears that only a relatively small fraction of the nucleosomes in S. cerevisiae are ‘positioned’ as a consequence of histone preferences for DNA sequence motifs. For much of genomic DNA, nucleosome occupancy values exhibit only small deviations from the average value, with occasional low values (for example, Figure 2, black curve). However, there is good evidence and general agreement that low nucleosome occupancy value regions correlate with promoter regions (3,16–18,20,21,23,25,26,29).
Although both Kaplan et al. (27) and Zhang et al. (29) used parallel sequencing, which appears to be subject to considerable DNA sequence-dependent read number variation (36), a critical difference in methodology between the two groups makes the Zhang et al. (29) work largely immune to the problem. Kaplan et al. (27) assessed nucleosome occupancy, whereas Zhang et al. assessed the degree of nucleosome positioning. As genomic DNA regions that exhibit anomalously high read numbers, due to sequence, generally extend over more than 100 bp (36), a base pair within this region will appear to have a high nucleosome occupancy; a large number of (147 bp) reads will span the base pair in question. In contrast, the fraction of (147 bp) read centers that are within a 20-bp window (29) will be low, scoring as a base pair having a low or average degree of nucleosome positioning. Therefore, in addition to the theoretical advantage of examining the degree of (translational) nucleosome positioning rather than occupancy (29), there is a practical advantage as well. We suggest that this methodological difference is primarily responsible for the apparent discrepancy between the two parallel sequencing studies.
There is no doubt that nucleosome positioning in genomes is not random. Small inherent sequence preferences for nucleosome formation appear to exist throughout genomic DNA. These sequence preferences are not strong enough to precisely position individual nucleosomes. However, it is plausible that these preferences conspire with each other over thousands or tens of thousands of base pairs (or more) to influence nucleosome array formation, which could in turn influence chromatin higher order structure (39,40). Evidence for the influence of long-range periodic DNA sequence oscillations on nucleosome array formation in the mouse genome (41), and on chromosome function in the human genome (42) has been reported.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Department of Biological Sciences, Purdue University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 4.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 5.Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Holde KE. Chromatin. New York: Springer Verlag; 1989. [Google Scholar]
- 8.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 9.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpson RT, Stafford DW. Structural features of a phased nucleosome core particle. Proc. Natl Acad. Sci. USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald DJ, Anderson JN. Unique translational positioning of nucleosomes on synthetic DNAs. Nucleic Acids Res. 1998;26:2526–2535. doi: 10.1093/nar/26.11.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 13.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 14.Simpson RT, Kunzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979;6:1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT) EMBO J. 1982;1:173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl Acad. Sci. USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 18.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segal MR. Re-cracking the nucleosome positioning code. Stat. Appl. Genet. Mol. Biol. 2008;7:Article14. doi: 10.2202/1544-6115.1367. [DOI] [PubMed] [Google Scholar]
- 25.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambrose C, Lowman H, Rajadhyaksha A, Blasquez V, Bina M. Location of nucleosomes in simian virus 40 chromatin. J. Mol. Biol. 1990;214:875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wintersberger U, Smith P, Letnansky K. Yeast chromatin. Preparation from isolated nuclei, histone composition and transcription capacity. Eur. J. Biochem. 1973;33:123–130. doi: 10.1111/j.1432-1033.1973.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 31.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 32.Zachau HG, Igo-Kemenes T. Face to phase with nucleosomes. Cell. 1981;24:597–598. doi: 10.1016/0092-8674(81)90084-2. [DOI] [PubMed] [Google Scholar]
- 33.Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- 34.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuels ML, Witmer JA. Statistics for the Life Sciences. 3rd edn. NJ, USA: Pearson Education, Inc.; 2003. [Google Scholar]
- 36.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloomfield VA, Crothers DM, Tinoco I., Jr . Physical Chemistry of Nucleic Acids. New York: Harper & Row; 1974. p. 100. [Google Scholar]
- 39.Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl Acad. Sci. USA. 1993;90:9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelhardt M. Choreography for nucleosomes: the conformational freedom of the nucleosomal filament and its limitations. Nucleic Acids Res. 2007;35:e106. doi: 10.1093/nar/gkm560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cioffi A, Fleury TJ, Stein A. Aspects of large-scale chromatin structures in mouse liver nuclei can be predicted from the DNA sequence. Nucleic Acids Res. 2006;34:1974–1981. doi: 10.1093/nar/gkl078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takasuka TE, Cioffi A, Stein A. Sequence information encoded in DNA that may influence long-range chromatin structure correlates with human chromosome functions. PLoS ONE. 2008;3:e2643. doi: 10.1371/journal.pone.0002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.