Abstract
The supramolecular assembly of a novel cyanine dye, 3,3′-di(3-sulfopropyl)-4,5,4′,5′-dibenzo-9-ethyl-thiacarbocyanine triethylammonium salt (ETC) was designed to verify specific intramolecular G-quadruplexes from duplex and single-strand DNAs. Spectral results have shown that ETC presented two major distinct signatures with specific intramolecular G-quadruplexes in vitro: (i) dramatic changes in the absorption spectra (including disappearance of absorption peak around 660 nm and appearance of independent new peak around 584 nm); (ii) ∼70 times enhancement of fluorescence signal at 600 nm. Furthermore, based on 1H-nuclear magnetic resonance and circular dichroism results, the preferring binding of ETC to specific intramolecular G-quadruplexes probably result from end-stacking, and the loop structure nearby also plays an important role.
INTRODUCTION
Telomeres, the ends of chromosomes, are essential for genome integrity and chromosome replication (1). Telomeres normally contain repeats of guanine-rich (G-rich) motifs, for example, the hexameric repeats of TTAGGG/CCCTAA in vertebrate telomeres. Of special interest is that the 3′-overhang G-rich single strand with 50–200 bases could adopt special structures under physiological condition, termed ‘G-quadruplex’. They may play important physiological functions in vivo, such as facilitating chromosome association and alignment during meiosis (2). Recently, quadruplex-folded telomeric DNA has been found to perturb telomere function and inhibit the activity of telomerase, an enzyme overexpressed in >85% of human cancers, hence opening up a novel avenue for cancer therapy in G-quadruplex stabilizing agents (3–7).
In addition, bioinformatics sequence analysis indicates that G-rich tracts capable of G-quadruplex formation are prevalent in human genome (8–10). For example, promoter regions spanning 1 kb upstream of transcription start sites of genes are significantly enriched in putative G-quadruplex-forming motifs and these putative promoter G-quadruplex-forming regions strongly associate with nuclease hypersensitivity sites (11). Such promoter-based G-quadruplexes may be directly involved in gene regulation at the level of transcription (12), which leads to extensive investigations of the structure and the role of promoter-mediated G-quadruplex in the promoters of many oncogenes, such as c-myc (13–15), c-kit (16) and bcl-2 (17).
G-quadruplex structure has been characterized in vitro (18), which is stabilized by Hoogsteen hydrogen bonding among four guanine bases arranged in a square planar configuration. However, the DNA strands of G-quadruplexes can assemble into either intramolecular (a single strand folds upon itself) or intermolecular (formed by two or more strands) configuration in vitro. Furthermore, G-quadruplexes exhibit extensive structural polymorphism; the DNA-strand orientation may be either parallel or antiparallel, even both conformations (termed hybrid) in some cases, for example, the (3 + 1) G-quadruplex motif in human telomeres (19,20). Depending on DNA sequence and extrinsic cation, an oligonucleotide even can exist as a mixture of several different quadruplex forms (21). Therefore, identifying particular quadruplex structure in human telomeres both in vitro and in vivo are still complicated tasks, which are important in the study of cell proliferation, cancer research and drug development.
So far, several direct evidences for the presence of G-quadruplex structure both in vitro (22) and in vivo (23,24) have been reported. However, these methods are still limited in biochemical aspect, such as single-chain antibody synthesis (23) and G-quadruplex interacting proteins discovery (25). Meanwhile, some distinct G-quadruplex structures could be verified by using organic ligands (26–28) in vitro. Besides these strategies, supramolecular assemblies, which are considered as intermediates between small molecules and macromolecules, may be another potential class of probe. Due to the noncovalent interaction among the components in the supramolecular assembly, the interaction forces among them are relatively weak, making spectral properties of the assembly to be easily manipulated by varying the environments. Obviously, this special feature would enable supramolecular assemblies to be potential excellent probe.
In previous research, we had found that supramolecular assembly of a cyanine dye could reflect specific features (such as unique chirality or molecular arrangement) in the presence of some biomacromolecules (29–31), which may be caused by the sensibility of dye assemblies to environmental change. Furthermore, we had successfully recognized mixed G-quadruplex in human telomeres from other DNA motifs by the supramolecular assembly of a novel cyanine dye 3,3′-di(3-sulfopropyl)-4,5,4′,5′-dibenzo-9-ethyl- thiacarbocyanine triethylammonium salt (ETC, shown in Figure 1) (32). Here, we discussed the interaction between ETC and more DNA sequences (derived from both human telomeres and oncogenic promoters) and specific G-quadruplex structures (including hybrid, parallel and antiparallel motifs). It has been found that ETC J-aggregates could recognize specific intramolecular G-quadruplexes from duplex and single-strand DNAs based on structural features, no matter where the sequences derived from, accompanying with notable spectral changes. This recognition probably results from the binding mode (end-staking and loop interaction) between ETC and specific G-quadruplex.
Figure 1.
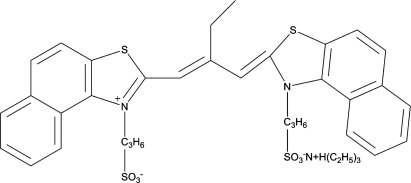
The molecular formula of cyanine dye ETC.
MATERIALS AND METHODS
Materials and sample preparation
The cyanine dye (ETC) was synthesized according to Hamer (33) and Fichen’s (34) methods, and the purity was proved by mass spectrometry and nuclear magnetic resonance (NMR) (shown in Supplementary Data). Calf thymus DNA (D4522) were purchased from Sigma-Aldrich Co. and used without further purification. All oligonucleotides were purchased from Invitrogen Co. (Beijing, China) and purified by PAGE (purity 98%). Analytical grade methanol, KH2PO4, K2HPO4, NaH2PO4 and Na2HPO4 were purchased from Beijing Chem. Co. Ultrapure water prepared by Milli-Q Gradient ultrapure water system (Millipore) was used throughout the experiments.
The stock solution of ETC was prepared by dissolving ETC in methanol. The stock solution of CT was prepared by dissolving calf thymus DNA directly into phosphate-buffered saline (PBS) (K+) (10 mM KH2PO4/K2HPO4, 1 mM EDTA, pH 7.4). The stock solutions of the oligonucleotides S24, S22, S17, H24, A24, A22, c-myc 2345, c-kit1, bcl-2 2345 and TBA were prepared by dissolving oligonucleotides directly into PBS (K+) or PBS (Na+) (10 mM NaH2PO4/Na2HPO4, 1 mM EDTA, pH 7.4), in terms of Table 1. The stock solutions of double-strand samples were prepared by heating the two complementary oligonucleotides (D24, D22 and D17) or the self-complementary oligonucleotide (D12 and D26) at 85°C for 15 min in PBS (K+) followed by a slow cooling (>6 h) to room temperature. The stock solutions of the oligonucleotides bcl-2 2345 C5 and bcl-2 2345 C21 were prepared by dissolving oligonucleotides directly into PBS (K+). All DNA samples had been stored for more than 24 h at 4°C and then structurally identified by the circular dichroism (CD) spectra.
Table 1.
DNA samples with different sequences and their abbreviations
| Abbr. | Sequences | Motifs | |
|---|---|---|---|
| CT | Calf thymus DNA | Double strand | |
| D24 | [5′-(TTAGGG)4-3′]/[5′-(CCCTAA)4-3′] | ||
| D22 | [5′-AGGG(TTAGGG)3-3′]/[5′-(CCCTAA)3CCCT-3′] | ||
| D17 | [5′-CCAGTTCGTAGTAACCC-3′]/ | ||
| [5′-GGGTTACTACGAACTGG-3′] | |||
| D26 | [5′-CAATCGGATCGAATTCGATCCGATTG-3′] | ||
| D12 | [5′-CGCGAATTCGCG-3′] | ||
| S24 | [5′-(CCCTAA)4-3′] | Single strand | |
| S22 | [5′-(CCCTAA)3CCCT-3′] | ||
| S17 | [5′-CCAGTTCGTAGTAACCC-3′] | ||
| H24 | [5′-TTGGG(TTAGGG)3A-3′] | Hybrid G4 | |
| A24a | [5′-(TTAGGG)4-3′] | Antiparallel G4 | |
| A22a | [5′-AGGG(TTAGGG)3-3′] | Antiparallel G4 | |
| c-myc 2345 | [5′-TGAGGGTGGGGAGGGTGGGGAA-3′] | From oncogenic promoters | Parallel G4 |
| c-kit1 | [5′-AGGGAGGGCGCTGGGAGGAGGG-3′] | Parallel G4 | |
| bcl-2 2345 | [5′-GGGCGCGGGAGGAATTGGGCGGG-3′] | Hybrid G4 | |
| TBA | [5′-GGTTGGTGTGGTTGG-3′] | From thrombin binding DNA aptamer | Antiparallel G4 |
aThese oligonucleotides were dissolved in PBS (Na+) (10 mM NaH2PO4/Na2HPO4, 1 mM EDTA, pH 7.4).
The measured sample was prepared by mixing a quantity of ETC solution with DNA solution, staying at 4°C for 1 h, and then being diluted by corresponding buffer solution. The concentration of methanol was 2% (v/v). Then the samples were kept in darkness overnight at 4°C before measurement in order to realize the full complexation.
Spectral measurement
Absorption, fluorescence and CD spectra were taken on a UV-1601PC spectrophotometer, a Hitachi F-4500 spectrophotometer and a JASCO J-815 spectrophotometer, respectively, in 10-mm quartz cells at room temperature. Xenon arc lamp was used in the excitation light source in fluorescence measurement. The excitation wavelength was 530 nm. Both excitation and emission slits were 5 nm and the scan speed was 240 nm/min. All CD spectra were collected at 1000 nm/min, with five scans averaged.
Polyacrylamide gel electrophoresis experiment
The polyacrylamide gel electrophoresis (PAGE) was conducted in 1× TBE (Tris base-boric acid-EDTA) buffer solution with 20% native gels. The gels were run at 100 V for 2 h at room temperature. Then the gels were incubated in 1× SYBR Gold and 20 μM ETC PBS (K+) solution for 30 min, respectively, rinsed with ultrapure water, and then photographed in GE Typhoon Trio. The excitation wavelength was 532 nm. Fluorescence images were recorded under the emission filters of 526 nm (for SYBR Gold) and 610 nm (for ETC), respectively.
NMR experiment
The stock solution of ETC was prepared by dissolving ETC in CD3OD. The stock solution of oligonucleotides was prepared by samples directly into NMR buffer solution [10 mM PBS (K+), 1 mM EDTA, 1 μM TSP, 90% H2O/10% CD3OD (v/v)]. All DNA samples had been stored for >24 h at 4°C and then identified by the CD spectra. The measured sample was prepared by mixing a quantity of ETC solution with DNA solution, and the final concentrations of DNAs were 200 μM. All NMR spectra were recorded on a Bruker Avance 600 spectrometer which is equipped with a 5-mm BBI probe capable of delivering z-field gradients up to 50 G cm–1. The experiments were carried out at 25°C (bcl-2 2345, H22 and c-kit1) and 30°C (TBA), respectively.
Molecular modeling
All the molecular modeling works and simulations were performed using the Insight II 2005 software (Accelrys Inc., San Diego, CA, USA) on a DELL 5300 workstation under CHARMM force field. The structure of ETC was first built using the sketching module and then minimized for 200 steps by the steepest descent algorithm. The structures of the bcl-2 2345, H24, c-kit1 and TBA G-quadruplex were obtained from the RCSB Protein Data Bank and the PDB IDs were 2F8U, 2GKU, 2O3M and 148D, respectively. The binding sites of the receptors were concluded and defined by the NMR titration results. Then, ETC was arranged to the binding site of the receptor and 5000 steps of minimization procedure of the receptor–ligand complex were performed using the steepest descent algorithm. During the minimization process, the DNA bases which form the binding site were flexible while other bases were fixed.
RESULTS
Recognizing intramolecular hybrid/parallel G-quadruplexes from duplex and single-strand DNAs by significant spectral changes of ETC
Due to the extended planar π-electron conjugated system, ETC tends to self-assembly in PBS (K+) and exhibits only a predominant absorption band at 660 nm assigned to J-aggregates (35) (as shown in supporting information).
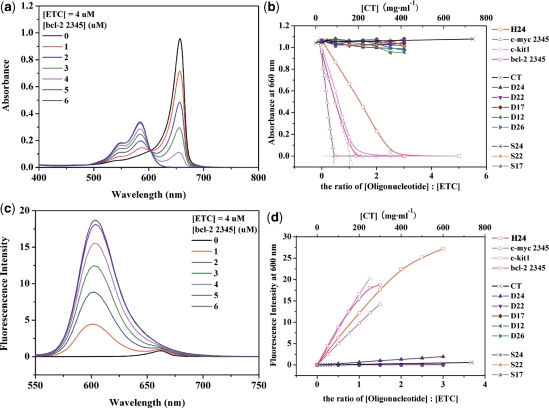
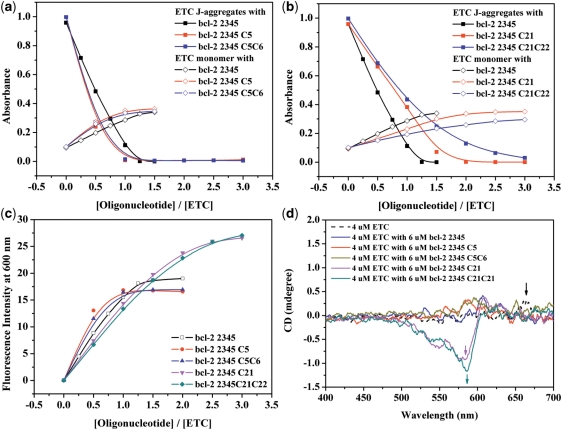
Figure 2a shows the absorption spectra of 4-μM ETC with different concentrations of bcl-2 2345 [hybrid G-quadruplex (17)]. Clearly, addition of bcl-2 2345 resulted in a gradual decrease, and eventually disappearance in the absorbance of ETC J-aggregates, accompanying with the appearance of a new peak located at 584.5 nm, which could be assigned to ETC monomer (35). When the ratio [bcl-2 2345] : [ETC] is 1.5 : 1, bcl-2 2345 could completely disassemble ETC J-aggregates to monomer. According to the method suggested by Walwick and co-workers (36), the Job curves (dashed lines in Figure 2b) intercrossed at about 1.02, indicating that the binding ratio of ETC and bcl-2 2345 is 1: 1. Similarly, G-quadruplex in human telomeres H24 [hybrid structure (20)], and G-quadruplex derived from oncogenic promoters c-myc 2345 [parallel structure (15)] and c-kit1 [parallel structure (16)] could also induce the disassembly of ETC J-aggregates and the appearance of the ETC monomer peak. On the other hand, the DNAs with other motifs, including linear duplex CT, D24, D22 and D17; hairpin duplex D26 and D12; and single-strand S24, S22 and S17 could hardly decrease the absorbance at 660 nm nor induced the new peak around 584.5 nm under similar conditions. Obviously, the DNAs with intramolecular hybrid/parallel G-quadruplex structures could completely disassemble ETC J-aggregates to monomer, while duplex and single-strand DNAs could not.
Figure 2.
The absorption (a) and fluorescence (c) spectra of 4-μM ETC with different concentrations of bcl-2 2345. The changes of 4 μM ETC J-aggregates absorbance (b) and monomer fluorescence intensity (d) against the ratio of [DNAs] : [ETC] and the concentration of CT (μg ml−1), respectively.
Therefore, ETC can recognize intramolecular hybrid/parallel G-quadruplex structure (whether it is derived from human telomeres or oncogenic promoters) from duplex and single-strand DNAs simply by measuring the absorption of monomer. The appearance of ETC monomer peak around 584.5 nm, about 80 nm apart from that of J-aggregates, can be considered as a unique signature.
In order to provide more significant feature, the fluorescence properties of ETC with various DNAs were also examined simultaneously. ETC monomer and J-aggregate have weak but unique fluorescence peaks at 600 and 662 nm, respectively (as shown in Supplementary Data). As shown in Figure 2d, the fluorescence intensity of ETC monomer could be strongly enhanced by adding bcl-2 2345, H24, c-myc 2345 or c-kit1. The fluorescence intensity increased simultaneously with increasing the ratios of [DNA] : [ETC]. This behavior could be interpreted by the competition between radiation transition and radiationless transition caused by facile rotation around methine bridge (37). When ETC monomer is bound to G-quadruplexes with specific motifs, the rotation is hindered and radiationless transition is inhibited, thus resulting in the enhancement of ETC monomer’s fluorescence intensity. The strong enhancement of ETC monomer’s fluorescence intensity (∼70 times stronger than that in methanol) indicates strong interaction between ETC monomer and hybrid/parallel G-quadruplexes. In the case of duplex and single-strand DNAs, on the contrary, the enhancement of ETC monomer’s fluorescence intensity is so weak that it will not interfere with the recognition. Therefore, the strong enhancement of ETC monomer fluorescence intensity can be considered as another unique signature to distinguish intramolecular hybrid/parallel G-quadruplex structure from duplex and single-strand DNAs.
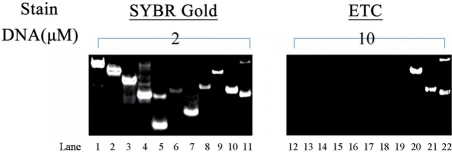
This dramatic fluorescence signature also suggested ETC assembly could be applied as structural probe or disease monitor vastly. As an example, a recognition experiment was performed on PAGE (as shown in Figure 3). Under the concentration of 10 μM, only bcl-2 2345, c-myc 2345 and c-kit1 could be stained by 20 μM ETC in PBS (K+) (lanes 20–22) while duplex and single-strand DNAs did not give rise to any interference (lanes 13–20). As a control, the positions of all DNAs were located by SYBR Gold (lanes 1–11). Further, the concentration limits of bcl-2 2345, c-myc 2345 and c-kit1 stained by 20-μM ETC were also detected. As shown in Supplementary Data, when [bcl-2 2345] > 1.5 μM, [c-myc 2345] > 0.5 μM and [c-kit1] > 3 μM, the G-quadruplexes could be recognized by ETC on PAGE.
Figure 3.
Recognition experiments on PAGE. Two-micromolar DNAs stained by SYBR Gold (lanes 1–11) and 10-μM DNAs by 20-μM ETC in PBS (K+) (lanes 12–22), respectively. Lanes 1–11 and 12–22 correspond to D24, D22, D17, D26, D12, S22, S17, S24, c-myc 2345, c-kit1 and bcl-2 2345, successively.
Binding characterization of ETC to intramolecular hybrid G-quadruplex
In order to understand the mechanism of ETC recognizing intramolecular hybrid G-quadruplex structure, CD measurements of both ETC J-aggregates and monomer with various DNAs were carried out. As shown in Figure 4, in PBS (K+) without any DNAs, ETC itself presented a weak positive CD signal around 660–670 nm (dashed lines), assigned to ETC J-aggregates. Adding DNAs with hybrid G-quadruplex structure to ETC induced the complicated changes of CD signals around 480–620 nm, which could be assigned to ETC monomer, and CD signals assigned to J-aggregates vanished (solid lines). Concretely, bcl-2 2345 induced a relatively weak positive signal at 607 nm, while H24 induced a relatively weak negative signal around 550–590 nm. The intricate CD signals indicate that these hybrid G-quadruplexes could interact with ETC in the form of monomer and twist the molecular frame of ETC. Probably, binding and twisting by hybrid G-quadruplex disturbs the exciton transition among ETC molecules in J-aggregates, and consequently leads to the disaggregation of the supramolecular assembly.
Figure 4.
The CD spectra of 4-μM ETC (dashed lines) and 4-μM ETC with different concentrations of (a) bcl-2 2345; (b) H24; (c) c-kit1; and (d) c-myc 2345, respectively.
As expected, on the other hand, duplex and single-strand DNAs could not induce monomer CD signal, but translated CD signals of J-aggregates to bisignate signals with a negative first Cotton Effect at longer wavelength and a positive second one at shorter wavelength centered around 650 nm, which are also assigned to ETC J-aggregates (as shown in Supplementary Data). The results indicated that the interactions between ETC and these DNAs are not strong enough to ‘snatch’ ETC monomer from J-aggregates, nor disassemble J-aggregates. They can only interact with ETC mainly in the form of J-aggregates.
ETC monomer end-stacking on hybrid G-quadruplex
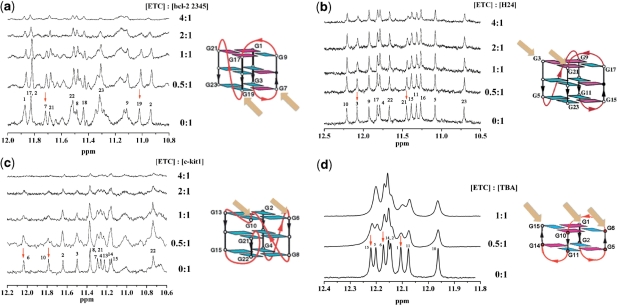
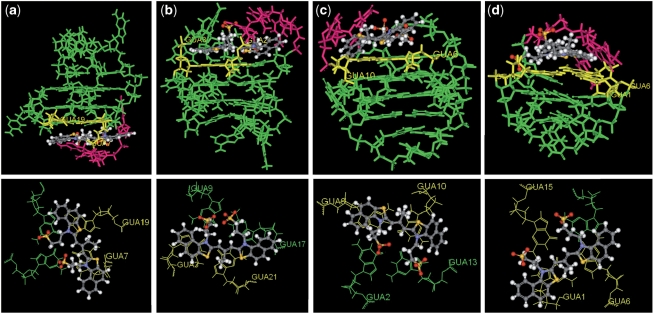
Proton NMR spectroscopy has been widely used to study the interaction between small molecule ligands and G-quadruplexes (38). In order to investigate the binding characterization of ETC and specific G-quadruplex in detail, the 1H-NMR spectra of ETC with hybrid G-quadruplex samples (bcl-2 2345 derived from oncogenic promoters and H24 from human telomeres) were investigated. In the 1H-NMR spectra of bcl-2 2345 and H24, the guanine imino proton signals were well resolved in the downfield region (about 10–12 p.p.m.) (20,39). As shown in Figure 5, adding ETC to bcl-2 2345 or H24 caused dramatic line-broadening and decreasing of these signals’ intensities. Furthermore, with the increasing [ETC], obviously the G7 and G19 imino proton signals of bcl-2 2345 exhibited much larger changes in half-width and intensity (marked by red arrows), suggesting that the binding of ETC to bcl-22345 are mostly located near the G7 and G19 region (14). The NMR-based folding topology of bcl-22345 is also shown in the right side, and the orange arrows point out the locations of G7 and G19 bases. In the case of H24, the larger changes of imino proton signals assigned to G3 and G21 of H24 also indicate the similar end-staking mode. Figure 8 shows the top projection of the ETC–DNA complex based on molecular modeling results. In order to get a clear view, only the interaction G-tetrad was shown. Obviously, ETC binds onto the end G-quartet of hybrid G-quadruplexes in the form of monomer. The end-stacking interaction is probably a main reason why hybrid G-quadruplexes could induce disassembly of ETC J-aggregates and strong enhancement of ETC monomer fluorescence intensity, while duplex and single-strand DNAs could not.
Figure 5.
The 1H-NMR spectra of 200 μM (a) bcl-2 2345; (b) H24; (c) c-kit1; (d) TBA with different concentration of ETC (on the left side) and the NMR-based folding topologies of the DNAs (on the right side). The red arrows show the signals changed strongest and the orange ones show the corresponding binding sites.
Figure 8.
The plots of the structure of (a) ETC-bcl-2 2345; (b) ETC-H24; (c) ETC-c-kit1 and (d) ETC-TBA complex from molecular mechanics simulation (on the top), and the top projections of the locations of ETC stacking onto the end G-tetrad (on the button). The binding sites based on 1H-NMR results are yellow and the lateral loops involved in the interaction are pink.
Loop interaction
Obviously, both the G7 and G9 bases of bcl-22345 are in the same end G-quartet, indicating that ETC molecule most probably stacks on one specific end of bcl-2 2345. In the case of H24, the NMR spectral changes also lead to similar results. However, if end-stacking interaction was concerned only, ETC monomer should bind to both ends of G-quadruplex. The binding ratio of ETC and hybrid G-quadruplex would be 2 : 1. However, it is not the case.
It is known that loops in G-quadruplex may exhibit various conformations and they play important roles in the interaction between G-quadruplex and the ligands (40–42). In order to discuss the role of G-quadruplex loop structure, the interaction between ETC and hybrid G-quadruplex samples with different length of certain loops (the conformations were identified by CD, and the data were shown in Supplementary Data) have been investigated.
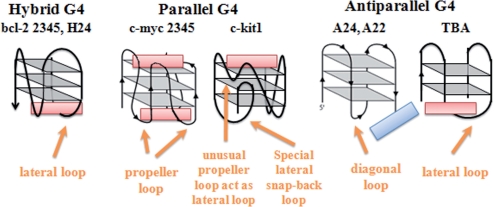
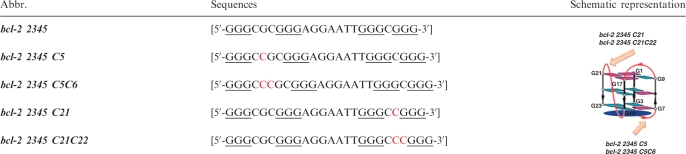
Figure 6 is the topologies of hybrid G-quadruplex structures used in this work. It is shown that hybrid G-quadruplex has one propeller loop and two lateral loops (opposite to and next to the propeller one). Therefore, two groups of bcl-2 2345 derivatives with certain loop extension had been designed. As shown in Table 2, compared with bcl-2 2345, bcl-2 2345 C5 and bcl-2 2345 C5C6 have longer lateral loops opposite to the propeller one, while bcl-2 2345 C21 and bcl-2 2345 C21C22 have longer propeller loops.
Figure 6.
The topologies of all kinds of G-quadruplexes, as well as the interaction between ETC molecule and them.
Table 2.
Hybrid G-quadruplex samples with different loops
 |
In the case of the former group, bcl-2 2345 C5 can induce the changes of the absorption and fluorescence spectra more sharply than bcl-2 2345 do under the same condition (as shown in Figure 7), indicating it has higher affinity to ETC. Hence, it is believed that the longer lateral loop in G-quadruplex opposite to the propeller one has less steric hindrance for the binding of ETC monomer, which would facilitate ETC binding to hybrid G-quadruplex. Figure 8 are the plots of the structures of ETC–DNA complex by using the Insight II 2005 software. Clearly, the lateral loops opposite to the propeller ones (pink bases) result in a cavity and ‘snatch’ part of ETC molecule, and, consequently, facilitate ETC stacking on the end G-quartet (yellow bases). In this interaction mode, a longer and more random lateral loop may not help reducing (probably on the contrary, increasing) the steric hindrance nearby the end G-quartet. As expected, the affinity of ETC and bcl-2 2345 C5C6 is almost the same (even somewhat weaker) as that of ETC and bcl-2 2345 C5, indicating excessive long lateral loop opposite to the propeller one would block, rather than facilitate ETC stacking on hybrid G-quadruplex. On the other end, the lateral loop next to the propeller loop is not fit for ‘snatching’ ETC molecule, but block ETC accessing to the G-quartet. So ETC can stack on only one end of hybrid G-quadruplex.
Figure 7.
The changes of 4-μM ETC J-aggregates and monomer absorbance against the ratio of [bcl-2 2345 derivatives] : [ETC]: (a) bcl-2 2345, bcl-2 2345 C5 and bcl-2 2345 C5C6; (b) bcl-2 2345, bcl-2 2345 C21 and bcl-2 2345 C21C22. (c) The changes of 4-μM ETC monomer fluorescence intensity against the ratio of [bcl-2 2345 derivatives] : [ETC]. (d) The CD spectra of 4-μM ETC (dashed line) and 4-μM ETC with 6 μM various bcl-2 2345 derivatives.
For the latter group of derivatives, as shown in Figure 7, bcl-2 2345 C21 has weaker and bcl-2 2345 C21C22 has the weakest ability to disassembly ETC J-aggregates, inferring longer propeller loop weaken the binding force. At the same time, bcl-2 2345 C21 and bcl-2 2345 C21C22 caused stronger fluorescence intensity and larger induced CD signal of ETC monomer, indicating that the influences of ETC molecular frame by binding bcl-2 2345 C21 and bcl-2 2345 C21C22 increase with the extension of the propeller loop. Probably, the propeller loop twists ETC molecular frame too strongly to weaken the stacking of ETC to the end G-quartet. And the longer the propeller loop is, the weaker the binding force of ETC to hybrid G-quadruplex is.
Binding characterization of ETC to intramolecular parallel G-quadruplex
Besides hybrid G-quadruplex, dramatic spectral changes of ETC could be induced by some intramolecular parallel G-quadruplexes. Therefore, the interaction characterizations of ETC to two intramolecular G-quadruplexes c-myc 2345 and c-kit1 were also discussed.
In the case of c-myc 2345 which folds into typical parallel G-quadruplex motif in PBS (K+), it can induce the completely transformation of ETC J-aggregates to monomer in the ratio [c-myc 2345]:[ETC] = 0.5 : 1, inferring stronger interaction between c-myc 2345 and ETC. As shown in Figure 2, the Job curve (dashed lines) of c-myc 2345 intercrossed at 0.45, indicating that the binding ratio of ETC and c-myc 2345 is 2: 1, which is different from those of ETC and hybrid G-quadruplexes. That is probably owing to the loop structure of c-myc 2345. As shown in Figure 6, c-myc 2345 has three propeller loops and no lateral or diagonal loop, the steric hindrance is very small in both ends, and consequently, ETC could stack on both ends of c-myc 2345.
Besides normal parallel G-quadruplex structure, the interaction between ETC and special parallel G-quadruplex c-kit1 (with two unusual loops, as shown in Figure 6) was also investigated. As shown in Figure 2, both the curve of absorption and fluorescence intensity against the ratio of [ETC]:[c-kit1] are similar with those of bcl-2 2345, indicating the interaction between ETC and c-kit1 is similar with that between ETC and bcl-2 2345. The 1H-NMR results also show that the binding of ETC to c-kit1 are mostly located near the G7 and G19 region (as shown in Figure 5), inferring ETC could stack on only one specific end (G2-G6-G10-G13) of c-kit1. In order to discuss the functions of the two unusual loops of c-kit1, the structure of ETC-c-kit1 complex were calculated by using the Insight II 2005 software on the basis of 1H-NMR results. As shown in Figure 8, the unusual propeller loop (C11-T12, pink bases), which results in a cavity and acts as the lateral loop opposite to the propeller one in hybrid G-quadruplex, could ‘snatch’ part of ETC molecule and facilitate ETC stacking on the end G-quartet. On the other hand, the five-membered lateral snap-back loop (A16-G17-G18-A19-G20) is long and in random motif. It blocks ETC accessing to the other G-quartet, acting as a diagonal loop.
Binding characterization of ETC to intramolecular antiparallel G-quadruplex
Besides hybrid and parallel G-quadruple structure, specific G-rich oligonucleotides also could fold into other G-quadruplex structures under certain conditions. For example, the DNA oligonucleotide d(TTAGGG)4 and d[AGGG(TTAGGG)3] in human telomeres can fold into both mixed/hybrid G-quadruplex in the presence of K+ and antiparallel G-quadruplex in the presence of Na+ (termed as A24 and A22). Therefore, in order to deepen the recognition property of ETC under different conditions, the interaction between ETC and A24, A22 in PBS (Na+) also have been discussed.
As in PBS (K+), ETC also tends to self-assembly in PBS (Na+) and exhibits only a predominant absorption band at 660 nm assigned to J-aggregates. And based on the UV-melting results, the J-aggregates show almost the same stability in both PBS (K+) and PBS (Na+) (as shown in Supplementary Data).
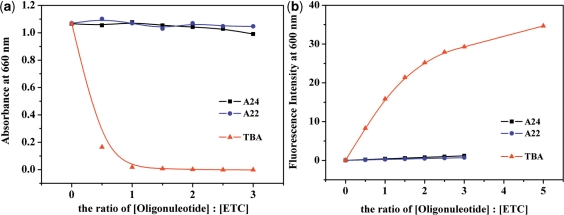
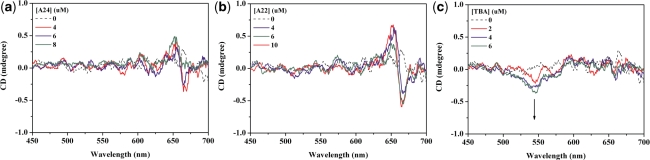
The absorption and fluorescence spectral results (as shown in Figure 9) shown that A24 and A22 [which would be in the motif of intramolecular antiparallel G-quadruplex in PBS (Na+) (43,44)] could induce neither the disassembly of ETC J-aggregates nor obvious enhancement of ETC monomer fluorescence intensity. At the same time, the CD spectra (as shown in Figure 10) also indicated that addition of A24 and A22 could not induce monomer CD signal, but translated J-aggregates positive CD signals to bisignate signals of ETC J-aggregates. Clearly, the interactions between ETC and A24, A22 are too weak to disassemble ETC J-aggregates, just like duplex and single-strand DNAs.
Figure 9.
The changes of 4-μM ETC J-aggregates monomer absorbance (a) and monomer fluorescence intensity (b) against the ratio of [DNAs]: [ETC].
Figure 10.
The CD spectra of 4-μM ETC (dashed lines) and 4-μM ETC with different concentrations of (a) A24; (b) A22; and (c) TBA, respectively.
Considering the end-stacking mode of ETC to specific G-quadruplex, it is reasonable that the steric hindrance provided by the diagonal loop or the two lateral loops opposite to each other in A24 and A22 probably is the key factor for the weak binding affinities between ETC and two G-quadruplexes.
Besides regular antiparallel G-quadruplex structure, some unique antiparallel G-quadruplexes with distinct loop structure also have been found in vitro (45). For examples, the DNA oligonucleotide d(GGTTGGTGTGGTTGG) (termed as TBA), which could bind to thrombin and inhibit its enzymatic activity in the chain of reactions that lead to blood clotting (46), would be in the motif of a special intramolecular antiparallel G-quadruplex in the presence of K+ (47). In order to examine whether ETC could recognize this special antiparallel G-quadruplex structure, the interaction between ETC and TBA was also investigated. As shown in Figures 9 and 10, TBA could induce all of the unique signatures in the interaction with ETC, as hybrid/parallel G-quadruplexes do: appearance of new absorption peak assigned to ETC monomer; strong enhanced ETC monomer fluorescence intensity; and the induced CD signals assigned to ETC monomer. As shown in Figure 6, compared with A24 and A22, TBA has a lateral loop, rather than diagonal loop on one end. Therefore, at the end of TBA, there has enough interspace to allow ETC molecule to stack on the G-quartet. As expected, both the 1H-NMR (Figure 5) and molecular modeling (Figure 8) results proved that the binding of ETC to TBA are located the G1-G6-G10-G15 G-quartet.
DISCUSSION
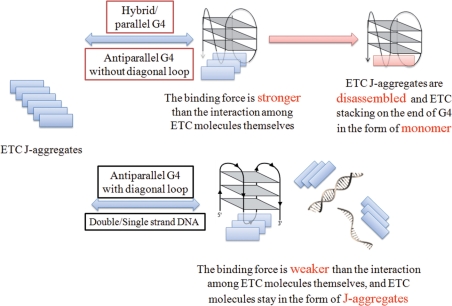
Based on the results, it is believed that the recognition of G-quadruplex by ETC supramolecular assembly is quite different from that by organic probes, which depends on distinct property changes in the transition of the balance of probe ↔ probe–DNA complex. Owing to the unique properties of supramolecular assembly, the recognition depends on the distinct properties changes in the transition of the balance of ETC(J-aggregates) ↔ ETC(J-aggregates)-DNA complex ↔ ETC(monomer)-DNA complex. As shown in Figure 11, the binding force (including end-stacking and loop interaction) between ETC and specific G-quadruplex is stronger than that among ETC molecules themselves. ETC molecule would be ‘snatched’ from J-aggregate and bound on the end of G-quadruplex in the form of monomer. Consequently, ETC J-aggregates are disassembled and the unique spectral signatures appear. On the other hand, owing to large steric hindrance or mismatching binding site, the binding force between ETC and other motifs (including duplex, single-strand DNA and specific antiparallel G-quadruplex) is so weak that it could not disassemble ETC J-aggregates. They could only interact with ETC in the form of J-aggregates weakly, and induce invisible spectral changes.
Figure 11.
The recognition mechanism of specific G-quadruplex by ETC supramolecular assembly compared with other DNA motifs.
In this strategy, since the recognition signatures come from the distinct spectral properties of supramolecular arrangements, spectral changes by recognition would be more visible and clearer than those from different states of the same molecule. First, the absorption peak could be blue-shifted about 80 nm in the presence of specific G-quadruplex. Chen et al. (48) have reported a carbocyanine dye DODC, as G-quadruplex ligand, which could give rise to a new absorption peak when specifically binding to dimeric hairpin G-quadruplexes. However, the small shoulder peak of DODC, which is blue-shifted by about 30 nm from primary peak, is relatively hard to be resolved and could not be a clear signature in recognizing certain G-quadruplex structure. In the case of ETC, however, the well-resolved new independent peak is a much clearer signature. Furthermore, the fluorescence intensity of ETC monomer could be enhanced >70 times by hybrid/parallel G-quadruplex, which is also ∼25 times than that of ETC J-aggregates. Compared with the known molecule used for the G-quadruplex fluorescence probes, such as BMVC, which can recognize and verify antiparallel G-quadruplex structure in human telomeres from linear duplex DNA through only small shift of fluorescence peak (∼30 nm) (49), clearly, ETC molecule which presents strong fluorescence enhancement (∼70 times) may have better features as a potential probe.
However, as a kind of supramolecular assembly, each ETC J-aggregate contains a number of dye molecules. Only the concentration of all ETC molecules (in the form of both J-aggregate and monomer) could be calculated under experimental conditions. The actual number of ETC J-aggregates and monomers are still a mystery. It is an unsolved problem in supramolecular assembly research. Therefore, the further exploration of the quantitative affinity and dynamics of ETC for various DNA motifs is necessary.
CONCLUSION
A novel cyanine dye ETC J-aggregates has been shown to change its spectral properties upon interaction with various DNA motifs. Owing to the unique properties of supramolecular assembly, intramolecular hybrid, several kinds of parallel G-quadruplex, and some specific antiparallel G-quadruplex without diagonal loop could strongly interact with ETC monomer and disassemble ETC J-aggregates, and consequently induce two indispensable signatures: (i) dramatic absorption changes (including disappearance of absorption peak around 660 nm and appearance of independent new peak around 584 nm); (ii) the strong enhancement (∼70 times) of fluorescence signal at 600 nm.
Further, the binding characterizations of ETC to the specific G-quadruplexes were discussed. It is proved that ETC stacks on one specific end of hybrid, some specific parallel and some specific antiparallel G-quadruplexes, or on both ends of normal parallel G-quadruplex. As shown in Table 3, the loops nearby the end G-quartet are also involved in the interaction. Some specific lateral or unusual propeller loops could ‘snatch’ part of ETC molecule and facilitates stacking on the end G-quartet, while diagonal or special lateral snap-back loop would block the access of ETC molecule to the G-quadruplex frame.
Table 3.
The influences of loop structures in the interaction between ETC and various G-quadruplexes
| Loop structural features | Examples | Influences in the interaction with ETC |
|---|---|---|
| Propeller loop | ||
| Normal | c-myc 2345 | Allow ETC to end-stacking |
| Unusual | c-kit1 | Snatch ETC and strengthen the interaction |
| Lateral loop | ||
| Opposite to propeller one | bcl-2 2345, H24 | Snatch ETC and strengthen the interaction |
| Next to propeller one | bcl-2 2345, H24 | Block ETC to access |
| Opposite to each other | A22, A24, TBA | Block ETC to access |
| Opposite to groove | TBA | Allow ETC to end-stacking |
| Snap-back | c-kit1 | Block ETC to access |
| Diagonal loop | A22, A24 | Block ETC to access |
Unlike some biochemical strategy, recognizing specific G-quadruplex by ETC supramolecular assembly is only based on structure, no matter derived from human telomeres, non-telomeric oncogenic promoters or other part of genome. Compared with organic probes, the spectral signatures come from transition of molecular arrangement are more visible and clearer than those from transition of states of the same molecule. This recognizing strategy by using ETC supramolecular assembly may offer a new approach for identifying and probing specific DNA motifs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for Open Access Charge: Author's personal funds.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi E. Genetics – DNA's; molecular gymnastics. Science. 2006;312:1467–1468. doi: 10.1126/science.312.5779.1467. [DOI] [PubMed] [Google Scholar]
- 4.Fu BQ, Huang J, Ren L, Weng XC, Zhou YY, Du YH, Wu XJ, Zhou X, Yang GF. Cationic corrole derivatives: a new family of G-quadruplex inducing and stabilizing ligands. Chem. Commun. 2007:3264–3266. doi: 10.1039/b704599a. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Li L, Xiang J, Tang Y, Zhang H, Yang S, Li Q, Yang Q, Xu G. Screening potential antitumor agents from natural plant extracts by G-quadruplex recognition and NMR methods. Angew. Chem.-Int. Edit. 2008;47:5590–5592. doi: 10.1002/anie.200800913. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Li L, Xiang J, Sun H, Tang Y. Fast screening and structural elucidation of G-quadruplex ligands from a mixture via G-quadruplex recognition and NMR methods. Biochimie. 2009;91:304–308. doi: 10.1016/j.biochi.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, Xiang J, Li X, Chen L, Xu X, Tang Y, Zhou Q, Li L, Zhang H, Sun H, et al. Stabilizing parallel G-quadruplex DNA by a new class of ligands: two non-planar alkaloids through interaction in lateral grooves. Biochimie. 2009;91:811–819. doi: 10.1016/j.biochi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dexheimer TS, Fry M, Hurley LH. In: Quadruplex Nucleic Acids. Neidle S, Balasubramanian S, editors. Cambridge, UK: RSC Publishing; 2006. pp. 180–207. [Google Scholar]
- 13.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 14.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded g-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomolec. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 19.Neidle S, Parkinson GN. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 20.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Frantz JD, Gilbert W, Tye BK. Identification and characterization of a nuclease activity specific for G4 tetrastranded DNA. Proc. Natl Acad. Sci. USA. 1993;90:3157–3161. doi: 10.1073/pnas.90.8.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang GW, Cech TR. The beta-subunit of oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 26.Dash J, Shirude PS, Balasubramanian S. G-quadruplex recognition by bis-indole carboxamides. Chem. Commun. 2008:3055–3057. doi: 10.1039/b806042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun HX, Xiang JF, Tang YL, Xu GZ. Regulation and recognization of the extended G-quadruplex by rutin. Biochem. Biophys. Res. Commun. 2007;352:942–946. doi: 10.1016/j.bbrc.2006.11.125. [DOI] [PubMed] [Google Scholar]
- 28.Waller ZAE, Shirude PS, Rodriguez R, Balasubramanian S. Triarylpyridines: a versatile small molecule scaffold for G-quadruplex recognition. Chem. Commun. 2008:1467–1469. doi: 10.1039/b718854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang YZ, Xiang JF, Tang YL, Xu GZ, Yang WP. Transition of H- and J-aggregate of a cyanine dye based on cation embedded in aggregation. Chem. Lett. 2006;35:1316–1317. [Google Scholar]
- 30.Zhang Y, Xiang J, Tang Y, Xu G, Yan W. Chiral transformation of achiral J-aggregates of a cyanine dye templated by human serum albumin. ChemPhysChem. 2007;8:224–226. doi: 10.1002/cphc.200600548. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, Xiang J, Li Q, Yan W, Zhou Q, Tang Y, Xu G. Chiral transformation of cyanine dye aggregates induced by small peptides. J. Phys. Chem. B. 2008;112:8783–8787. doi: 10.1021/jp803076d. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Xiang J, Yang S, Zhou Q, Li Q, Tang Y, Xu G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. Recognizing mixed G-quadruplex in human telomeres. Chem. Commun. 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- 33.Hamer FM. The Chemistry of Heterocyclic Compounds. New York: Interscience; 1964. [Google Scholar]
- 34.Ficken GE. The Chemistry of Synthetic Dyes. New York: Academic Press; 1971. [Google Scholar]
- 35.Herz AH. Aggregation of sensitizing dyes in solution and their adsorption onto silver halides. Adv. Colloid Interface Sci. 1977;8:237–298. [Google Scholar]
- 36.Bean RC, Shepherd WC, Kay RE, Walwick ER. Spectral changes in a cationic dye due to interaction with macromolecules. III. Stoichiometry and mechanism of the complexing reaction1. J. Phys. Chem. 1965;69:4368–4379. [Google Scholar]
- 37.Guo CN, Xiang JF, Feng J, Tang YL, Chen CP, Xu GZ. Effect of TiO2 colloids on the fluorescence behavior of two cyanine dyes. J. Colloid Interface Sci. 2002;246:401–409. doi: 10.1006/jcis.2001.7912. [DOI] [PubMed] [Google Scholar]
- 38.Gavathiotis E, Heald RA, Stevens MFG, Searle MS. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)(4) containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Dai JX, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guedin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Campbell NH, Patel M, Tofa AB, Ghosh R, Parkinson GN, Neidle S. Selectivity in ligand recognition of G-quadruplex loops. Biochemistry. 2009;48:1675–1680. doi: 10.1021/bi802233v. [DOI] [PubMed] [Google Scholar]
- 43.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 44.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 45.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 47.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 48.Chen Q, Kuntz ID, Shafer RH. Spectroscopic recognition of guanine dimeric hairpin quadruplexes by a carbocyanine dye. Proc. Natl Acad. Sci. USA. 1996;93:2635–2639. doi: 10.1073/pnas.93.7.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.