Abstract
Human sirtuin 1 (SIRT1) is a NAD+-dependent deacetylase that participates in cell death/survival, senescence and metabolism. Although its substrates are well characterized, no direct regulators have been defined. Here, we show that SIRT1 associates with SKI-interacting protein (SKIP) and modulates its activity as a coactivator of retinoic acid receptor (RAR). Binding assays indicated that SKIP interacts with RAR in a RA-dependent manner, through a region that overlaps the binding site for SIRT1. SKIP augmented the transcriptional activation activity of RAR by cooperating with SRC-1, and SIRT1 suppressed SKIP/SRC-1-enhanced RAR transactivation activity. The suppression was dependent on the deacetylase activity of SIRT1 and was enhanced by a SIRT1 activator, resveratrol. In contrast, the suppression was relieved by SIRT1 knockdown, overexpression of SKIP and treatment with a SIRT1 inhibitor, splitomicin. Upon SKIP overexpression, the recruitment of SIRT1 to the endogenous RARβ2 promoter was severely impaired, and SKIP was recruited to the promoter instead. Finally, resveratrol treatment inhibited RA-induced neuronal differentiation of P19 cells, accompanied by reductions in the neuronal marker nestin and a RAR target gene, RARβ2. This inhibition was relieved by either knockdown of SIRT1 or overexpression of SKIP. These data suggest that SIRT1 and SKIP play reciprocal roles in the regulation of RAR activity, which is implicated in the regulation of RA-induced neuronal differentiation of P19 cells.
INTRODUCTION
Human SIRT1 (sirtuin1), one of the seven mammalian SIRT homologs, is closely related to yeast Sir2, which possesses NAD+-dependent class III histone deacetylase activity. Recent studies strongly suggest that mammalian SIRT1, like yeast Sir2, is involved in transcriptional silencing of integrated reporter genes by chromatin modification via histone deacetylation, DNA damage responses (1,2),and life span extension following caloric restriction (3). SIRTI also deacetylates non-histone proteins, including various transcription factors that are involved in growth regulation, the stress response and endocrine signaling. For example, SIRT1 negatively regulates p53-dependent apoptosis by deacetylating p53 in response to cellular damage (4–6). Other substrates of SIRT1, including DNA repair protein Ku70 (7), FOXO family proteins (8–9) and NF-κB (10), are involved in the stress response. In energy metabolism and insulin signaling, SIRT1 activates gluconeogenesis and represses glycolysis in the liver via deacetylation of PGC-1α (11). Increasing levels of SIRT1 in the pancreatic β cells of mice resulted in repressed UCP2 transcription and enhanced glucose-stimulated insulin secretion (12). Upon binding, SIRT1 deacetylates the androgen receptor (AR) and represses DHT-induced AR signaling in human prostate cancer (13). Thus, it has been speculated that SIRT1 is widely involved in mammalian physiology, with roles in metabolism, senescence, apoptosis and tumorigenesis. In addition, recent studies have suggested a role for SIRT1 in cellular differentiation. SIRT1 represses nuclear receptor PPARγ by docking with its co-repressors, NcoR and SMRT, and attenuates adipogenesis in 3T3-L1 cells (14). SIRT1 also regulates muscle gene expression and differentiation by deacetylating MyoD and PCAF (15). Moreover, SIRT1 participates in the growth and maturation of the embryo and in gametogenesis (16). Nevertheless, a role of SIRT1 in neuronal differentiation, specially derived from embryonic stem cells, remains largely unknown.
To identify additional targets of SIRT1, we first performed a yeast two-hybrid screen of a human complementary DNA (cDNA) library using SIRT1 as bait, and found that SIRT1 is functionally associated with SKI-interacting protein (SKIP), which has been described as a transcriptional coactivator of nuclear receptors and other transcription regulators (17,18). Here, we provide the first evidence for a retinoic acid receptor (RAR) regulatory pathway controlled reciprocally by SIRT1 and SKIP.
MATERIALS AND METHODS
Plasmids and cloning
All cDNAs were made according to standard methods and verified by sequencing. The full-length SKIP cDNA was amplified by PCR from a HeLa cDNA library and was inserted into the 5′-XhoI and 3′-BamHI or 3′-BglII sites of each vector. Expression plasmids for the SIRT1 wild type and point mutant (SIRT1 HY) are described elsewhere (19). For yeast two-hybrid screening, the full-length SIRT1 cDNA was inserted into the bait plasmid pBTM116 (LexA DBD vector). SIRT1 deletion mutants were created by PCR amplification and were also subcloned into pBTM116. For transient transfections, the Flag-tagged SIRT1 or SKIP gene was placed in the pcDNA3 vector. For the localization assay, GFP-tagged recombinant constructs were created in pEGFP-C3 (BD Biosciences, Palo Alto, CA, USA), respectively. For GST-fused proteins, pGEX4T-1 (Amersham Pharmacia Biotech, Piscataway, NJ, USA) was used.
Cell culture and differentiation
The H1299 cells derived from a non-small-cell lung tumor were grown and maintained in RPMI 1640 medium, and COS-1, P19, and HEK293 cells were grown and maintained in Dulbecco’s Modified Eagle Medium (DMEM) medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and an antibiotic-antimycotic mix (all from Gibco BRL, Gaithersburg, MD, USA) in a 5% CO2 atmosphere at 37°C. For differentiation, P19 cells were aggregated in bacterial Petri plates at a density of 105 cells/ml and treated with 1 µM RA and/or 120 µM resveratrol (RES) for 96 h, with subculturing in fresh medium after 48 h with each treatment. On day 4, the aggregates were transferred to cell culture plates, and RA was eliminated from the medium. Cells were then seeded on 10-cm-diameter plates, at a density of 3 × 105 cells/plate. After 12 h, the medium was changed to 0.5% FBS medium, and the cells were allowed to differentiate for another 6 days.
Yeast two-hybrid screening and assay
A HeLa cDNA library in the prey plasmid pGAD10 (BD Biosciences) was screened for proteins that interacted with SIRT1, using the yeast reporter strain L40. Screening was performed as reported previously, using LexA-fused SIRT1 as the bait (19). To map the SKIP interaction domain of SIRT1, deletion derivatives of SIRT1 were fused with LexA DBD by subcloning into pBTM116. The deletion mutants of SKIP were fused with VP16 AD by subcloning into the pASV3 vector. The level of interaction was determined by quantitative β-galactosidase assays.
GST pull-down assay
A GST-fusion protein of SKIP (amino acids 174–373) or SIRT1 (amino acids 114–217) was expressed in Escherichia coli and purified on glutathione-Sepharose beads (Amersham Pharmacia Biotech) by standard methods. An approximately equal amount of GST or GST-SIRT1 was mixed with in vitro translated Flag-SIRT1, SKIP or RARα using a TNT reticulocyte system (Promega, Madison, WI, USA). Bound proteins were detected on western blots with antibodies against Flag (for SIRT1 and SKIP) and RARα.
Western blotting, immunoprecipitation and immunofluorescence microscopy
For western blotting (WB), cells were lysed in lysis buffer (20) supplemented with a protease inhibitor cocktail (Roche, Basel, Switzerland). Proteins were separated by electrophoresis on 8–12% sodium dodecyl sulfate (SDS)-polyacrylamide gels, transferred to nitrocellulose, and incubated with primary antibodies. The commercially available primary antibodies used were rabbit polyclonal antibodies (SIRT1, sc-15404; RARα, sc-551; RARβ, sc-552; GFP, sc-8334: Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-Flag M2 antibody (F3165; Sigma, St Louis, MO, USA) and mouse monoclonal anti-β-actin antibody (A1978; Sigma). The blots were next incubated with peroxidase-conjugated mouse or rabbit IgG secondary antibodies (Amersham Pharmacia Biotech). The protein bands were detected with an ECL system (Amersham Pharmacia Biotech).
For the immunoprecipitation (IP) assay, cells treated under various conditions were washed with ice-cold phosphate-buffered saline (PBS) and lysed in RIPA buffer supplemented with a protease inhibitor cocktail (Roche) (20). The lysates were incubated overnight at 4°C with a 1 : 200 dilution of the indicated antibodies. After 2 h of incubation at 4°C with A/G-agarose beads (Santa Cruz Biotechnology), the beads were washed three times with RIPA buffer. The immune complexes were released from the beads by boiling and analyzed by WB using the indicated antibodies.
For co-localization, H1299 cells were co-transfected with GFP-SKIP and Flag-SIRT1 or Flag-RARα, washed with PBS, fixed with 4% paraformaldehyde in PBS for 1 min at room temperature, and then permeabilized for 4 min in PBS containing 0.5% Triton X-100. After washing, the cells were incubated with anti-SIRT1 or anti-RARα antibody (1 : 100 dilution) in blocking buffer (PBS and 2% BSA) for 1 h and then incubated with Texas Red-conjugated anti-rabbit IgG (1 : 200 dilution; Amersham Pharmacia Biotech). After the final washing, the cells (either GFP- or Texas Red-labeled) were visualized under a fluorescence microscope (AX70; Olympus Optical Co, Tokyo, Japan). Hoechst (Sigma) staining was used to localize chromosomal DNA in the nucleus.
Transient transfection and luciferase reporter assay
HEK293 cells were seeded in a 12-well culture plate and transiently transfected with RARE-luciferase reporter enzyme, and SV40-driven β-galactosidase (β-gal) expression vector as an internal control. Depending on the experimental conditions, the SIRT1, SIRT1 HY or SKIP expression vector was co-transfected using Lipofectamine Plus reagent (Invitrogen). The luciferase activity was measured by adding 20 µl of luciferin into 30 µl of cell lysate and using an analytical luminescence luminometer, according to the manufacturer’s instructions (Promega). The β-gal activity was determined in 96-well plates using a microplate reader at 405 nm. The luciferase activity was normalized to the β-gal activity.
Real-time-reverse transcriptase
Total RNA from undifferentiated or differentiated P19 cells was extracted using TRIzol reagent (Gibco-BRL) according to the manufacturer's; instructions. RNA (5 μg) was reverse-transcribed using Superscript II reverse transcriptase (RT; Invitrogen) and oligo(dT) primer (New England Biolabs, Beverly, MA, USA). The primers used for PCR were as follows: nestin, forward, 5′-CAGATGTGGGAGCTCAATCG-3′, and reverse, 5′-GCCTCCTCGATGGTCCGCTC-3′; glyceraldehydes-3-phosphate dehydrogenase (GAPDH), forward, 5′-GTGGATATTGTTGCCATCA-3′ and reverse, 5′-GACTCCACGACGTACTCA-3′; Hoxa1, forward, 5′-TGGAGGAAGTGAGAAAGTTGG C-3′, and reverse, 5′-ATGGGAGTCGAGAGGTTTCC-3′; Hoxb1, forward, 5′-CCATATCCTCCGCCGCAG-3′, and reverse, 5′-CGGACTGGTCAGAGGCATC-3′.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously (20). H1299 cells were transfected with empty Flag vector or Flag-SKIP vector and treated with or without 1 µM AtRA for 4 h. Cross-linked, immunoprecipitated chromatin complexes were obtained by IP with the indicated antibodies, and the cross-linking was reversed according to Upstate’s protocol (Upstate, Chicago, IL, USA). The DNA pellets were recovered and analyzed by PCR, using a primer pair that encompasses the RARβ2 promoter region: forward, 5′-AAGCTCTGTGAGAATCCTG-3′, and reverse, 5′-GGATCCTACCCCGACGGTG-3′.
RNA interference
The sequences of the custom siRNA duplex for SIRT1 and the control have been described (19). The transfection of siRNA was performed with Lipofectamine 2000 (Gibco BRL) in Opti-MEM I reduced-serum medium (Gibco BRL), according to the manufacturer's; instructions. SIRT1 knockdown was verified by WB using anti-SIRT1 antibody.
RESULTS
SIRT1 interacts specifically with SKIP through distinct domains
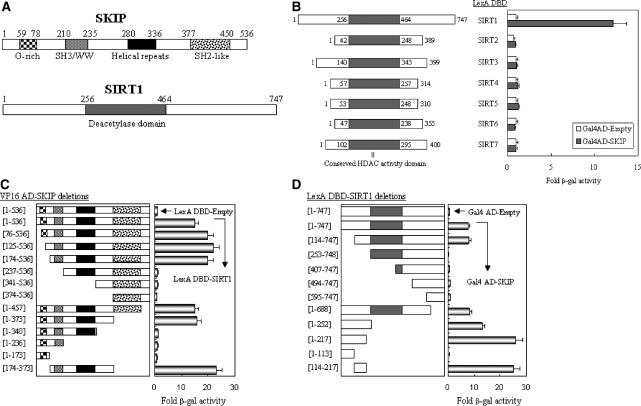
To identify cellular regulator(s) of SIRT1, we performed a yeast two-hybrid genetic screening of a HeLa cDNA library fused to GAL4 AD (activation domain), with LexA-fused SIRT1 as bait. We identified a SKIP/NCoA-62 protein from several rounds of screening and selection. SKIP, a transcriptional co-regulator, modulates the transcriptional activities of various transcription factors, including nuclear receptors such as vitamin D receptor (VDR) (17,18). The structural features of SIRT1 and SKIP are depicted in Figure 1A. Yeast two-hybrid assays using LexA DBD-fused SIRT family members and Gal4 AD-fused SKIP constructs revealed that, of the seven SIRT members, SKIP selectively associates with SIRT1 (Figure 1B). Further domain-mapping assays indicated that amino acid residues 174–373 of SKIP (Figure 1C) and 114–217 of SIRT1 (Figure 1D) are responsible for the interaction between SKIP and SIRT1. Amino acid residues 114–217 of SIRT1 are distinct from other SIRT family members, supporting a specific interaction of SKIP with SIRT1 among the SIRT family.
Figure 1.
Mapping of the interaction domains between SKIP and SIRT1. (A) Structural features of SKIP and SIRT1. (B) Specific interaction of SKIP with SIRT1 among the SIRT family. Yeast two-hybrid assays were performed using LexA DBD-fused SIRT family members (SIRT 1–7) and Gal4 AD-fused SKIP (originally recovered from a yeast two-hybrid screening). The interaction was evaluated by β-gal assays. Fold-activity indicates the value relative to the value of the Gal4 AD empty control. (C) Mapping of the SKIP domain responsible for SIRT1 binding. SKIP deletions were fused to the Gal4 AD vector and introduced into yeast L40, together with LexA DBD-fused SIRT1. Fold-activity indicates the β-gal value relative to that of the VP16 AD-SKIP plus LexA DBD empty control. (D) Mapping of the SIRT1 domain responsible for SKIP binding. SIRT1 deletions were fused to the LexA DBD vector and introduced into yeast L40, together with Gal4 AD-fused SKIP. Fold-activity indicates the β-gal value relative to that of the LexA DBD-SIRT1 plus Gal4 AD empty control. All the results are the means of three independent experiments ± SD.
SIRT1 interacts with SKIP in vitro and in vivo
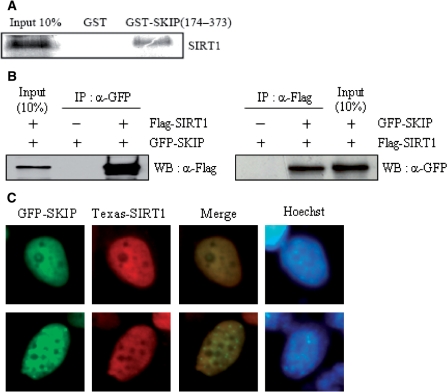
Using a GST pull-down assay, we determined that SIRT1 directly interacts with amino acids 174–373 of SKIP in vitro (Figure 2A). The physical interaction of SIRT1 and SKIP in mammalian cells was confirmed by performing IP assays. Flag-tagged SIRT1 was co-transfected along with an empty GFP or GFP-tagged SKIP construct in COS-1 cells. IP with an anti-GFP antibody and WB with an anti-Flag antibody indicated that SIRT1 interacted with SKIP in vivo (Figure 2B, left). This result was verified by reverse co-transfection, followed by IP with an anti-Flag antibody and WB with an anti-GFP antibody (Figure 2B, right). To further substantiate our proposed mode of interaction, we determined the subcellular distribution of SIRT1 and SKIP in H1299 cells. Immunofluorescence microscopy showed that Flag-SIRT1 and GFP-SKIP were co-localized in speckle-like nuclear substructures (Figure 2C).
Figure 2.
Interaction between SKIP and SIRT1 in vitro and in vivo. (A) GST pull-down assay. In vitro translated Flag-tagged SIRT1 was incubated with GST or GST-SKIP (amino acid residues 174–373). The bound proteins were visualized by SDS–PAGE and subsequent western blotting using anti-Flag antibody (for SIRT1). (B) Immunoprecipitation (IP) analysis. H1299 cells were co-transfected with Flag-tagged SIRT1 and GFP-tagged SKIP, and vice versa. Lysates were subjected to IP using anti-GFP or anti-Flag antibodies. Bound protein was identified by western blotting anti-Flag or anti-GFP antibodies. (C) Co-localization of SKIP and SIRT1. H1299 cells were transfected with GFP-SKIP and Flag-SIRT1. The cellular location of SIRT1 was determined using Texas Red-conjugated anti-rabbit IgG. Images were visualized by fluorescence microscopy. Hoechst staining was used to locate the nucleus.
SKIP interacts with RARα through an overlapping binding domain for SIRT1
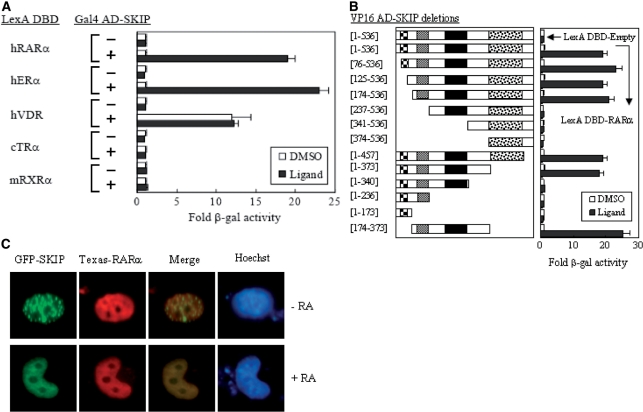
SKIP was originally discovered as a nuclear protein that interacts with the SKI oncoprotein (21), and was later found to interact with VDR, augmenting VDR-activated transcription (22). To understand the role of SKIP in other nuclear receptor-mediated transcriptional activations, we first investigated whether SKIP could interact with other nuclear receptors and the ligand-dependency requirement for the interaction. As shown by the yeast two-hybrid assays, SKIP interacts with RARα and ERα in a ligand-dependent manner, whereas no ligand dependency was observed for the interaction between SKIP and VDR, and no interactions were detected between SKIP and TRα or RXRα (Figure 3A). We selected RARα for further studies because of its ligand-dependent interaction with SKIP. Subsequent domain-mapping assays using a series of N- and C-terminal deletions of SKIP indicated that RARα can bind to the central region of SKIP, covering amino acids 174–373, which overlaps with the SIRT1 binding region (Figure 3B). This observation was further confirmed by immunofluorescence microscopy. When GFP-SKIP and Flag-RARα were co-expressed in the absence of RA, SKIP displayed speckle-like nuclear substructures, as shown in Figure 3C, while RARα localized in the nucleoplasm. However, RA treatment caused the two proteins to merge in the nucleoplasm, supporting an RA-dependent interaction between RARα and SKIP.
Figure 3.
Interaction between SKIP and RARα. (A) Interaction of SKIP with nuclear receptors (NRs). Yeast two-hybrid assays were performed with the indicated NRs in the presence of their cognate ligand. (B) Mapping of the SKIP domain responsible for the interaction with RARα in the presence of the ligand all-trans retinoic acid (AtRA). SKIP deletions were fused to the VP AD vector and introduced into yeast L40, together with LexA DBD-fused RARα. Fold-activity indicates the β-gal value relative to that of the VP16 AD-SKIP plus LexA DBD empty control. (C) Immunofluorescence microscopy. H1299 cells were transfected with GFP-SKIP and Flag-tagged RAR. Transfections were performed in the absence and presence of AtRA. Hoechst staining was used to locate the nucleus.
SKIP competes with SIRT1 for RARα binding
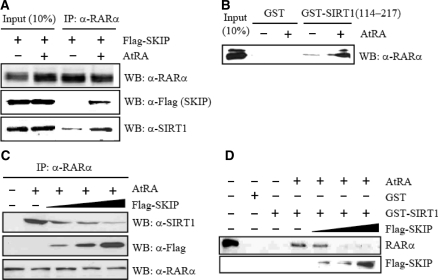
Given that SKIP can interact with both SIRT1 and RA-bound RARα, we probed whether SIRT1 interacts with RARα in the presence of RA. IP and subsequent WB assays using Flag-SKIP overexpression cellular extracts indicated that RARα forms a complex with both SKIP and SIRT1. Complex formation with SKIP was RA dependent, and complex formation with SIRT1 was RA enhanced (Figure 4A). The RA-enhanced interaction of SIRT1 with RARα was confirmed with a GST pull-down assay using purified GST-SIRT1 (amino acids 114–217) and in vitro translated RARα (Figure 4B). This portion of SIRT1 is also responsible for SKIP binding, as described in Figure 1D, suggesting that SKIP and SIRT1 compete with each other for RA-dependent interactions with RARα. To analyze the competition between SIRT1 and SKIP for RARα binding in vivo, H1299 cells were transfected with increasing amounts of Flag-tagged SKIP expression vector. Cellular extracts were then subjected to IP with anti-RARα antibody and WB with anti-Flag (for SKIP) and anti-SIRT1 antibody. The SIRT1 binding to RARα decreased inversely with SKIP expression (Figure 4C). Similarly, the GST pull-down assays revealed that RARα binding to GST-SIRT1 decreased gradually with increasing levels of SKIP (Figure 4D). Overall, our in vivo and in vitro assays revealed that SKIP forms a complex with SIRT1 and RARα at its low level but compete with SIRT1 for RARα binding at high level, suggesting that SKIP and SIRT1 may play reciprocal roles in regulating RARα activity.
Figure 4.
Competition between SKIP and SIRT1 for RARα binding. (A) Ternary complex among RARα, SKIP and SIRT1. H1299 cells were transfected with Flag-tagged SKIP in the absence and presence of RA. Cell lysates were prepared and immunoprecipitated with anti-RARα antibody. Precipitated proteins were identified on western blots using anti-Flag (for SKIP) and anti-SIRT1 antibodies. (B) Direct interaction between SIRT1 and RARα. In vitro translated RARα (in pSG5 vector) was incubated with GST or GST-SIRT1 (amino acids 114–217). The bound proteins were visualized by SDS–PAGE and subsequent western blotting using anti-RARα antibody. (C) Competition in vivo. H1299 cells were transfected with increasing amounts of Flag-SKIP expression vector in the presence of RA. Lysates were subjected to IP with anti-RARα antibody, followed by western blotting with anti-SIRT1 antibody. Protein levels of endogenous RARα and overexpressed SKIP were visualized on western blots. (D) Direct competition analysis. In vitro translated RARα was incubated with GST or GST-SIRT1 (1–217) and further reacted with increasing amounts of in vitro translated Flag-tagged SKIP in the presence of RA. The bound proteins were visualized by SDS–PAGE and subsequent western blotting with anti-Flag antibody.
SKIP enhances and SIRT1 represses the transcriptional activity of RARα
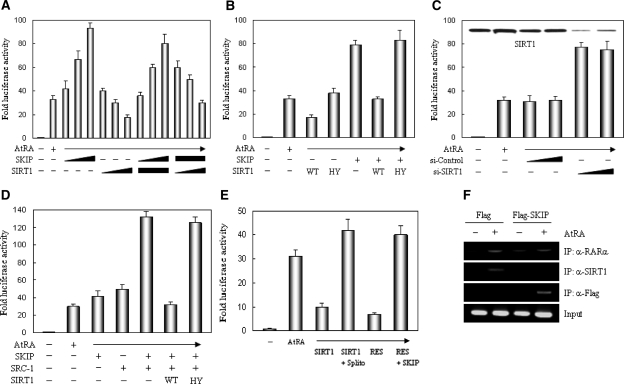
To evaluate whether the RARα binding observed above is functionally relevant, we determined the effects of SIRT1 or SKIP on the transcriptional activity of RARα, using the RA-responsive RARE-luciferase reporter gene. As shown in Figure 5A, SKIP enhanced and SIRT1 repressed the RA-dependent transcriptional activation of RARα in a dose-dependent manner. Further assays using a deacetylase-defective SIRT1 mutant (HY) demonstrated that the deacetylase activity of SIRT1 is required for RARα repression (Figure 5B). When SIRT1 was depleted by siRNA treatment, RARα activity increased markedly, supporting that SIRT1 itself mediates RARα repression (Figure 5C). As expected from another report (23), SKIP and a steroid receptor coactivator, SRC-1, cooperated for RA-induced RARα transcriptional activation, but this cooperation was again diminished by SIRT1 in a deacetylase activity-dependent manner (Figure 5D). To confirm the deacetylase activity-dependent RARα repression mediated by SIRT1, a known SIRT1 inhibitor, splitomicin (Splito) and an activator, RES, were utilized. RARα repression by SIRT1 was impaired completely by splitomicin. However, RARα repression was achieved by RES without SIRT1 over-expression, and this repression was released by SKIP over-expression (Figure 5E). We next investigated whether this competition for RARα binding occurs in vivo, by ChIP assays. Under normal conditions, SIRT1 was recruited to the chromatinized RARβ2 promoter in the presence of RA, but upon SKIP overexpression, SKIP replaced SIRT1 on the promoter (Figure 5F). Overall, these results suggest that SIRT1 represses and SKIP augments RARα transcriptional activation by competing for both RARα binding at the protein level and RA-dependent chromatin binding at the RARβ2 promoter when they were overexpressed.
Figure 5.
Reciprocal effect of SKIP and SIRT1 on the transcriptional activity of RARα. (A) SKIP enhances and SIRT1 suppresses RARα transactivation. H1299 cells were transfected with increasing amounts (0, 0.1, 0.3 and 0.5 µg) of Flag-SKIP or Flag-SIRT1, in combination with the RARα expression vector and RAR-responsive RARE-luciferase reporter. Luciferase activity was determined after normalizing to the observed β-gal activity. All data are represented as means ± SD from triplicate experiments. Fold-luciferase activity indicates the relative ratio obtained in the presence and absence of AtRA. (B) Requirement of deacetylase activity of SIRT1 for RARα repression. H1299 cells were transfected with combinations of SIRT1 wild type (WT), SIRT1 HY mutant (deacetylase-defective), and SKIP expression vector. Cell lysates were then subjected to luciferase assays. (C) Depletion of SIRT1 abrogated RARα repression. H1299 cells were transfected with SIRT1 siRNA or control siRNA. Cell lysates were subjected to luciferase assays, and the expression of SIRT1 was monitored by western blotting with anti-SIRT1 antibody. (D) SIRT1 impairs RARα transactivation induced by SKIP-SRC-1 cooperation in a deacetylase-dependent manner. H1299 cells were transfected with combinations of SKIP, SRC-1 and SIRT1 WT or SIRT1 HY expression vectors. Cell lysates were subjected to luciferase assays. (E) Effect of SIRT1 inhibitor or activator on RARα activity. H1299 cells were transfected with SIRT1 or SKIP expression vector and treated with 240 µM SIRT1 inhibitor, splitomicin (Splito) or 120 µM SIRT1 activator, resveratrol (RES), for 12 h. Cell lysates were subjected to luciferase assays. (F) SKIP blocks SIRT1 binding to RA-responsive RARβ2 promoter in vivo. H1299 cells were transfected with empty Flag vector or Flag-SKIP expression vector and treated with or without 1 µM AtRA for 4 h. Soluble chromatin was prepared and immunoprecipitated with the indicated antibodies. The final DNA pellets were PCR-amplified using a primer pair that covers the RARβ2 promoter region.
SIRT1 and SKIP oppositely regulate RA-induced differentiation of P19 cells
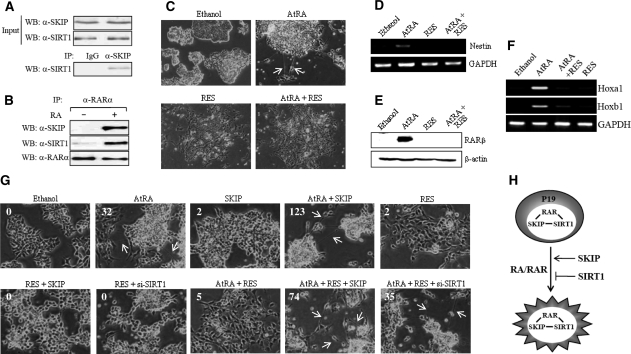
To address the biological significance of the reciprocal regulation of RARα transcriptional activation as described above, we first determined the endogenous interactions between SKIP, SIRT1 and RARα, and next analyzed the effects of SIRT1 and SKIP on RA-induced cellular differentiation in P19 cells. These cells represent a murine embryonic carcinoma cell line that possesses pluripotency, like embryonic stem cells (24), and efficiently differentiates to neuron and glial cells upon RA treatment (25). IP with anti-SKIP antibody and WB with anti-SIRT1 antibody exhibited the endogenous interaction between SKIP and SIRT1 (Figure 6A). Subsequent IP assays using anti-RARα antibody indicated that RARα, SKIP and SIRT1 form a ternary complex in the presence of RA (Figure 6B). As shown in Figure 6C, upon RA treatment, the P19 cells began to extrude neurite-like structures from an embryonic body, indicative of neuronal differentiation. However, additional treatment with the SIRT1 activator RES prevented the P19 cells from producing neurite-like structures. To support this morphological change, we analyzed the mRNA expression of nestin, a known neuronal marker (26), by RT-PCR. RA induced the expression of nestin during differentiation, but RA-induced nestin expression was abolished with RES treatment (Figure 6D). Similar results were obtained when the protein and mRNA expression of RARβ and Hox genes, a known RA target genes, was analyzed by WB and RT-PCR, respectively (Figure 6E and F). Overall, these findings suggest that SIRT1 interferes with RA-induced neuronal differentiation by repressing the expression of a neuronal marker and RAR target genes.
Figure 6.
Reciprocal effect of SKIP and SIRT1 on the RA-induced differentiation of P19 cells. (A) Endogenous SKIP interacts with SIRT1. P19 cell lysates were prepared and immunoprecipitated with pre-immune serum (IgG) or anti-SKIP antibody. Precipitated proteins were revealed by WB using anti-SIRT1 antibody. (B) RARα forms a ternary complex with SKIP and SIRT1 in the presence of RA. (C) Resveratrol blocks RA-mediated neural differentiation in P19 cells. Cells were aggregated in bacterial Petri plates and treated with 1 µM RA and/or 120 µM resveratrol (RES) for 96 h. The aggregates were transferred to cell culture plates and were allowed to differentiate for another 6 days without RA or resveratrol. Neurite-like structures are shown by arrows. (D) Effect of resveratrol on the mRNA expression level of a neural differentiation marker, nestin. RT-PCR was performed using total RNA from neurally differentiated P19 cells, treated as above, using primers specific for the nestin coding sequence. GAPDH was used as an internal control. (E, F) Effect of resveratrol on RA-regulated gene expression. Western blotting was performed to monitor protein expression, using P19 cell extracts and antibodies against RARβ and β-actin (for control) (E). To analyze the expression of Hoxa1 and Hoxb1, RT-PCR was employed (F). (G) Repression of RA-induced neural differentiation by resveratrol is released by SKIP. P19 cells transfected with Flag-SKIP or SIRT1 siRNA and control siRNA were aggregated in bacterial Petri plates and treated with 1 µM RA and/or 120 µM resveratrol for 96 h. Subsequently, differentiation was allowed for 6 days, as described above. The number of neurite-like structures counted per 1000 P19 cells was shown on each figure. (H) Proposed model of the reciprocal roles of SIRT1 and SKIP in RAR signaling. The molecular interplay among RAR, SKIP and SIRT1 may precisely regulate the transcriptional activity of RAR either by repression or enhancement during RA-induced neuronal differentiation.
To determine the reciprocal roles of SIRT1 and SKIP in P19 cell differentiation, we used SKIP overexpression, RES and SIRT1 siRNA. As shown in Figure 6G, SKIP overexpression significantly increased the formation of neurite-like structures in the presence of RA, although the expression vector transfection efficiency was <10% in P19 cells. The number of neurite-like structures counted per 1000 P19 cells was shown on each figure. RES blocked SKIP-induced neuronal differentiation, and its effect was reversed by SKIP overexpression. In addition, the depletion of SIRT1 reversed the negative effect of RES on the RA-induced neuronal differentiation, suggesting that SIRT1 is required for RES to be negatively functional. Consistent with the transcriptional data shown in Figure 5, these results suggest that the reciprocal roles of SIRT1 and SKIP in the regulation of the transcriptional activity of RARα are responsible for their different roles in modulating RA-induced neuronal differentiation of P19 cells (Figure 6H).
DISCUSSION
In this study, we demonstrated reciprocal roles of SIRT1 and SKIP in the regulation of RARα-dependent gene transcription and RA-dependent neuronal differentiation. We identified SKIP as a novel binding partner of SIRT1. Extensive binding assays revealed that in addition to binding to SIRT1, SKIP interacts with RARα through a binding domain that overlaps that for SIRT1. We consistently found that SIRT1 competes with SKIP for RARα binding at the protein level and at the chromatin-associated RARβ2 promoter. This overexpression based competition resulted in differential regulation of RARα transactivation, with SKIP enhancing and SIRT1 repressing the transcriptional activity of RARα. Finally, we provided evidence to suggest that the reciprocal regulation of RARα-activated transcription by SIRT1 and SKIP may account for the differential regulation of RARα during RA-induced neuronal differentiation.
Our investigation raises questions regarding how SIRT1 represses but SKIP stimulates RARα-dependent transcription. Recent studies have provided evidence of SIRT1 repression of transcriptional activation mediated by other nuclear receptors such as PPAR and AR. SIRT1 represses PPARγ activity by binding to its cofactors, NCoR and SMRT, although it was uncertain whether SIRT1 deacetylates PPARγ or histones, or both, at target genes (27). In the case of AR, SIRT1 binds and deacetylates AR to repress AR signaling (13). Another study on AR reported that AR antagonism requires SIRT1 binding and deacetylation of histone H3 locally at the AR-responsive promoter (28). The SIRT1-mediated hypoacetylation of histone H3 has also been implicated in the suppression of MyoD-dependent transcription (15). In a study on the role of SIRT1 in histone binding and histone modification, when SIRT1 was forced to interact with a synthetic Gal4-responsive promoter, it deacetylated histones H3 (at lysine 9) and H4 (at lysine 16), and recruited histone H1 to the promoter through an interaction with the N-terminal portion of SIRT1, thereby reducing the expression of the reporter gene (2). The involvement of SIRT1 in other histone modifications such as methylation is indicated by the finding that SIRT1 interacts directly with and deacetylates histone methyltransferase SUV39H1, specific for H3K9. The deacetylation of SUV39H1 at its active site increases SUV39H1 activity, thereby elevating the levels of H3K9me3 to mediate transcriptional silencing (29). Our data revealed that SIRT1 repression of RARα transactivation requires the deacetylase activity of SIRT1. Transient transfection assays also revealed that Gal4-SIRT1 could repress the expression of a reporter gene (data not shown). It remains to be determined whether the deacetylation of RARα, histone or SUV39H1 is responsible for SIRT1 repression of RARα-dependent transcription. Although the mechanism of SIRT1-mediated repression is interesting, here we focused on the switching mechanism from SIRT1 repression to SKIP activation, which provides another level of SIRT1 regulation. We demonstrated that overexpressed SKIP competes with and displaces SIRT1 from a repression complex at the RARα-activated promoter, resulting in transcriptional activation.
SKIP has been described as a coactivator of Notch (30) and nuclear receptors, including VDR (22,23), as well as a co-repressor for Checkpoint Suppressor 1 (31), MAGE-A1 (32) and Smad2/3 (33) in association with the co-repressors NCoR/SMRT and/or HDAC. Interestingly, SKIP plays a dual role in CBF1- and nuclear receptor-mediated transcription. SKIP represses CBF1-dependent transcription by associating with SMRT. To activate the CBF1-repressed promoter, NotchIC displaces SMRT from the repressor complex and interacts with SKIP and CBF1. SKIP also regulates nuclear receptor-mediated transcription by switching its association with co-repressor NCoR/SMRT and coactivator p300 (34). In this study, we provide evidence for another role of SKIP in regulation: activated or overexpressed SIRT1 competes with and displaces SKIP from the activation complex at the RARα-activated promoter, resulting in transcriptional repression.
Our study provides the first evidence for a RAR regulatory pathway controlled reciprocally by SIRT1 and SKIP, although the biological significance of the reciprocal regulation remains unknown. Retinoic acid (RA) is used to differentiate stem cells into neurons by inducing neuron-specific genes (35,36), but its detailed mechanism is still poorly understood. In this study, we used P19 EC cells to delineate the molecular mechanism underlying RA-induced neuronal differentiation and presented the reciprocal functions of SIRT1 and SKIP. SIRT1 activation by RES repressed RA-induced differentiation, probably through the targeted repression of the nestin and other RA target genes, whereas SIRT1 knockdown or SKIP overexpression stimulated RA-induced differentiation. SIRT1 has been implicated in the regulation of differentiation through adipogenesis (14,37), spermatogenesis (38) and neurogenesis (39,40). During the neurogenesis of neuronal stem cells in response to environmental redox, SIRT1 represses Mash1 transcription by forming a complex with Hes1, but SIRT1 displacement allows Hes1 to recruit CBP to activate Mash1 transcription: SIRT1 drives astrocyte fate, whereas the release of SIRT1 drives neuronal fate. SIRT1 is highly expressed in mouse embryonic stem cells (41), where it may cause the cells to die after exposure to ROS, leaving healthy cells to proliferate (42). Moreover, in embryonic cells, the SIRT1 inhibits the p53-mediated suppression of Nanog expression, thus allowing the cells to express Nanog and maintain their self-renewal potential. However, the role of SIRT1 in the RA-induced differentiation of ES cells has not been determined. Based on the present study using ES-like P19 cells, we propose that SIRT1 may associate with RAR and repress RAR-activated transcription in undifferentiated P19 cells, and that SIRT1 disappears or SKIP displace SIRT1 from RAR to enhance RAR-mediated transcription during the neuronal differentiation in response to RA, although the precise molecular interplay among RAR, SKIP and SIRT1 remains to be determined.
FUNDING
The National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (KRF-2008-313-C00864 and R01-2008-000-10902-0 to E.-J.K.), and (R01-2007-000-10308-0 to S.-J.U.). Funding for open access charge: Brain Korea 21 Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 3.Chen WY, Wang DW, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 5.Vaziri H, Dessain SK, Eaton EN, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 6.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 8.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, LinY, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by theSIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 9.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 10.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 12.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Eric Ford E, Cras-Méneur C, Permutt MA, Imai SI. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Powell M, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, et al. The Hormonal Control of Androgen Receptor Function through SIRT1. Mol. Cell. Biol. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, de Oliveira RM, Leid M, Michael W, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 16.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald PN, Dowd DR, Zhang C, Gu C. Emerging insights into the coactivator role of NCoA62/SKIP in vitamin D-mediated transcription. J. Steroid Biochem. Mol. Biol. 2004;89:179–186. doi: 10.1016/j.jsbmb.2004.03.097. [DOI] [PubMed] [Google Scholar]
- 18.Folk P, Půta F, Skruzný M. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell. Mol. Life Sci. 2004;61:629–640. doi: 10.1007/s00018-003-3215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Lee HK, Park UH, Kim EJ, Um SJ. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl R, Wani B, Hayman MJ. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene. 1998;16:1579–1586. doi: 10.1038/sj.onc.1201687. [DOI] [PubMed] [Google Scholar]
- 22.Baudino TA, Kraichely DM, Jefcoat SC, Jr, Winchester SK, Partridge NC, MacDonald PN. Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription. J. Biol. Chem. 1998;273:16434–16441. doi: 10.1074/jbc.273.26.16434. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 2001;276:40614–40620. doi: 10.1074/jbc.M106263200. [DOI] [PubMed] [Google Scholar]
- 24.van der Heyden MA, Defize LH. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc. Res. 2003;58:292–302. doi: 10.1016/s0008-6363(02)00771-x. [DOI] [PubMed] [Google Scholar]
- 25.Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan Y, Yang J, Bian W, Jing N. Mouse nestin protein localizes in growth cones of P19 neurons and cerebellar granule cells. Neurosci. Lett. 2001;302:89–92. doi: 10.1016/s0304-3940(01)01664-0. [DOI] [PubMed] [Google Scholar]
- 27.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol. Endocrinol. 2007;21:1807–1821. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 30.Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol. Cell. Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott KL, Plon SE. CHES1/FOXN3 interacts with Ski-interacting protein and acts as a transcriptional repressor. Gene. 2005;359:119–126. doi: 10.1016/j.gene.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Laduron S, Deplus R, Zhou S, Kholmanskikh O, Godelaine D, De Smet C, Hayward SD, Fuks F, Boon T, De Plaen E. MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription. Nucleic Acids Res. 2004;32:4340–4350. doi: 10.1093/nar/gkh735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figueroa JD, Hayman MJ. Differential effects of the Ski-interacting protein (SKIP) on differentiation induced by transforming growth factor-beta1 and bone morphogenetic protein-2 in C2C12 cells. Exp. Cell. Res. 2004;296:163–172. doi: 10.1016/j.yexcr.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Leong GM, Subramaniam N, Issa LL, Barry JB, Kino T, Driggers PH, Hayman MJ, Eisman JA, Gardiner EM. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem. Biophys. Res. Commun. 2004;315:1070–1076. doi: 10.1016/j.bbrc.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Glaser T, Brüstle O. Retinoic acid induction of ES-cell-derived neurons: the radial glia connection. Trends Neurosci. 2005;28:397–400. doi: 10.1016/j.tins.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 37.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 38.Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS ONE. 2008;3:e1571. doi: 10.1371/journal.pone.0001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hisahara S, Chiba S, Matsumoto H, Horio Y. Transcriptional regulation of neuronal genes and its effect on neural functions: NAD-dependent histone deacetylase SIRT1 (Sir2alpha) J. Pharmacol. Sci. 2005;98:200–204. doi: 10.1254/jphs.fmj05001x2. [DOI] [PubMed] [Google Scholar]
- 40.Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schröter F, Ninnemann O, Siegert E, Bendix I, Brüstle O, Nitsch R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell. Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 41.McBurney MW, Yang X, Jardine K, Bieman M, Th'n;g J, Lemieux M. The absence of SIR2α protein has no effect on global gene silencing in mouse embryonic stem cells. Mol. Cancer Res. 2003;1:402–409. [PubMed] [Google Scholar]
- 42.Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]