Abstract
N7-methylguanine at position 46 (m7G46) in tRNA is produced by tRNA (m7G46) methyltransferase (TrmB). To clarify the role of this modification, we made a trmB gene disruptant (ΔtrmB) of Thermus thermophilus, an extreme thermophilic eubacterium. The absence of TrmB activity in cell extract from the ΔtrmB strain and the lack of the m7G46 modification in tRNAPhe were confirmed by enzyme assay, nucleoside analysis and RNA sequencing. When the ΔtrmB strain was cultured at high temperatures, several modified nucleotides in tRNA were hypo-modified in addition to the lack of the m7G46 modification. Assays with tRNA modification enzymes revealed hypo-modifications of Gm18 and m1G37, suggesting that the m7G46 positively affects their formations. Although the lack of the m7G46 modification and the hypo-modifications do not affect the Phe charging activity of tRNAPhe, they cause a decrease in melting temperature of class I tRNA and degradation of tRNAPhe and tRNAIle. 35S-Met incorporation into proteins revealed that protein synthesis in ΔtrmB cells is depressed above 70°C. At 80°C, the ΔtrmB strain exhibits a severe growth defect. Thus, the m7G46 modification is required for cell viability at high temperatures via a tRNA modification network, in which the m7G46 modification supports introduction of other modifications.
INTRODUCTION
To date, more than 100 modified nucleosides have been found in various RNA species (1,2). In particular, tRNA contains numerous modified nucleosides (3). All of these modified nucleosides are produced post-transcriptionally by specific tRNA modification enzymes or guide RNA systems (1). Among them, N7-methylguanine at position 46 (m7G46) in the variable region is one of the common modifications in tRNA and forms a tertiary base pair with the C13–G22 base pair (4,5).
The m7G46 modification is generated by tRNA (m7G46) methyltransferase [tRNA (guanine-N7-)-methyltransferase, EC 2. 1. 1. 33; TrMet (m7G46)] (1,6). It has been reported that the yeast enzyme is composed of two protein subunits (Trm8 and Trm82) and their genes have been identified (7). Bacterial genes have also been experimentally identified and named trmB (classical name, yggH) from Escherichia coli (8), Bacillus subtilus (9) and Aquifex aeolicus (10). There is a clear structural difference between eukaryotic and bacterial tRNA (m7G46) methyltransferases: the eukaryotic enzyme is a heterodimer (7,11,12), while the bacterial enzyme is a monomer (8) or homodimer (9). Recently, crystal structures of the eukaryotic (13) and bacterial (9) enzymes have been reported. These structural studies strongly suggest that the substrate RNA recognition mechanisms differ considerably between the eukaryotic and bacterial enzymes, although both enzymes have a similar catalytic domain. In our previous study, we reported that the A. aeolicus TrmB recognizes the G46 base from the T-stem side in tRNA (10). Recently, we have reported that the C-terminal region, which is found in thermophilic bacterial enzymes, is required for protein stability at high temperatures and contributes to the selection of the precise guanine nucleotide (i.e. G46) to be modified (14). Furthermore, we investigated the RNA recognition mechanism of the yeast enzyme (Trm8–Trm82 complex) and found that yeast Trm8–Trm82 has more stringent recognition requirements for the tRNA molecule than A. aeolicus TrmB (15). Thus, protein structure–function relationship studies have been useful in the elucidation of the RNA recognition mechanism.
Functional studies of the m7G46 modification have also recently been performed. Gene disruption mutants of yeast (7) or E. coli (8) have revealed that the tRNA (m7G46) methyltransferase activity in yeast and E. coli is not essential for cell viability. However, it has been reported that a yeast double mutant strain lacking both the trm8 and trm4 genes showed rapid degradation of tRNAVal (16) [trm4 encodes yeast tRNA (m5C34, 40, 48, 49) methyltransferase (17)]. Thus, the m7G46 modification in yeast contributes to the stability of tRNA in conjunction with the other modified nucleotide(s) around the variable region in tRNA. Moreover, recently, we have reported that a gene involved in m7G modification of tRNA is required for infection by the phytopathogenic fungus Colletotrichum lagenarium (18). In comparison with these eukaryotic enzymes, there is limited information about the bacterial enzyme. In the current study, we have focused on characterization of a trmB gene disruptant (ΔtrmB) strain of Thermus thermophilus HB8, an extreme thermophilic eubacterium. We report the importance of the m7G46 modification for growth at high temperatures and propose a tRNA modification network, in which the m7G46 modification has a positive effect on formation of other modifications in tRNA.
MATERIALS AND METHODS
Materials
[Methyl-14C]-S-adenosyl-l-methionine (AdoMet) (1.95 GBq/mmol) was purchased from Perkin Elmer. Non-radioisotope labeled AdoMet was obtained from Sigma. DE52 is a product of Whatman. Q-Sepharose Fast Flow was bought from GE Healthcare. DNA oligomers were purchased from Invitrogen, and T7 RNA polymerase was from Toyobo. Other chemical reagents were of analytical grade.
Strain and media
The culture source of T. thermophilus HB8 was a kind gift from Dr Tairo Oshima (Tokyo University of Pharmacy and Life Science). The cells were grown in rich medium [0.8% polypeptone, 0.4% yeast extract, and 0.2% NaCl, pH 7.5 (adjusted with NaOH)]. The medium was supplemented with 0.35 mM CaCl2 and 0.17 mM MgCl2 after autoclaving. To make plates, gellan gum (Wako Pure Chemicals) was added to the medium (final concentration, 1.5%).
Selection of the target gene for disruption
We searched for the target gene in the T. thermophilus HB8 genome by BLAST-search using the amino acid sequence of the E. coli TrmB (classical name, YggH) (8). One target gene (TTHA1619), which was annotated as a gene encoding a methyltransferase of unknown function, was found to be a candidate for T. thermophilus trmB. The expected amino acid sequence of TTHA1619 shares high homology with E. coli and A. aeolicus TrmB proteins. The TTHA1619 gene was amplified by polymerase chain reaction (PCR) using the following primers: Tth TrmBN, 5′-GGG GCA TAT GCT GGT CGT GCC CGC CCG CCT CCA C-3′; Tth TrmBC, 5′-GGG GGA ATT CTT AGG TGT GGT CCT GGA CCA CCT C-3′. Underlined regions show restriction enzyme sites (Nde I and Eco RI). The amplified DNA was digested with Nde I and Eco RI, and ligated into the multi-cloning linker of pET30a E. coli expression vector. The expression of TTHA1619 protein in E. coli BL21 (DE3) Rosetta 2 strain was performed according to the manufacturer’s manual. The protein was partially purified by heat-treatment and DE52 column chromatography according to the purification procedure for A. aeolicus TrmB (10). The enzymatic activity and modified nucleotide analysis were performed using yeast and T. thermophilus tRNAPhe transcripts as described in our previous report (14).
Construction of ΔtrmB (ΔTTHA1619) strain
The TTHA1619 gene was disrupted by replacement with the highly thermostable kanamycin nucleotidyltransferase (HTK) gene (19,20). The plasmid vector containing the TTHA1619 region disrupted by the HTK gene was purchased from RIKEN Biological Resource Center (Tsukuba, Japan) (21). Thermus thermophilus cells in late-log phase were transformed by the vector according to the report (22) and mutant colonies were selected on a plate containing 500 µg/ml kanamycin at 70°C. The genomic DNA from each colony was isolated, analyzed by PCR and Southern hybridization, and then the sequence of the replaced region was determined on ABI PRISM 310 DNA sequencers. Southern hybridization was performed at 55°C as reported previously (23). An alkaline phosphatase-labeled probe was prepared using the AlkPhos Direct Labeling system (GE Healthcare) and the hybridized bands were detected by monitoring the alkaline phosphatase activity consuming ECF substrate with a Typhoon 9400 Variable Mode Imager (Amersham Biosciences).
Measurement of tRNA methyltransferase activities in S-100 and P-100 wash fractions
In this study, we used yeast and T. thermophilus tRNAPhe transcripts as standard substrates. The transcripts were prepared by using T7 RNA polymerase and purified by Q-Sepharose column chromatography and 10% polyacrylamide gel electrophoresis (PAGE) (7 M urea).
Cell extracts from the wild type and ΔTTHA1619 were prepared from late-log phase cells cultivated at 67°C for in vitro methyl-transfer assay. Wet cells (0.3g) were suspended in 2 ml of buffer A [50 mM Tris–HCl (pH 7.6), 5 mM MgCl2, 6 mM 2-mercaptoethanol, 50 mM KCl]. The cells were ground in a mortar with 0.15 g aluminum oxide and then the suspension was centrifuged at 8000g for 20 min. The supernatant fraction was further centrifuged at 100 000g for 2 h. The resultant supernatant was used as the S-100 fraction. Glycerol was added to the S-100 fraction to a final concentration of 50% v/v and stored at −30°C. The P-100 wash fraction was prepared from the precipitate of the centrifugation at 100 000g. The precipitate was homogenized with 200 µl of buffer A containing 1 M ammonium chloride and then centrifuged at 100 000g for 2 h. Subsequently, the supernatant was dialyzed against buffer A containing glycerol (final concentration 50% v/v) and used as the P-100 wash fraction.
Transfer RNA methyltransferase activities in the S-100 and P-100 wash fractions were analyzed as follows: 30 µg protein from the S-100 or P-100 wash fraction, 0.2 A260 unit yeast tRNAPhe transcript and 0.78 nmol [methyl-14C]-AdoMet were incubated in 400 µl of buffer A at 60°C for 1 h. The RNA was extracted with phenol-chloroform and then recovered by ethanol precipitation. The RNA pellet was dissolved in 3 µl of 50 mM sodium acetate (pH 5.0), and digested with 2.5 units of nuclease P1 (Wako Pure Chemicals). The sample was separated using two dimensional thin layer chromatography (2D-TLC) as described previously (25). The 14C-methylated nucleotides were monitored with a Fuji Photo Film BAS2000 imaging analyzer.
Nucleosides analysis by HPLC
Class I tRNA fractions were purified from the wild-type and ΔTTHA1619 cells in late-log phase cultured at 67°C. Briefly, total RNA fraction was prepared by phenol-chloroform extraction. Subsequently, the total RNA fraction was loaded on a Q-Sepharose column equilibrated with buffer B [20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 400 mM NaCl] and then a small RNA fraction (mainly tRNA and 5S rRNA) was eluted with buffer B containing 600 mM NaCl. The class I tRNA fraction was further purified by 10% PAGE (7 M urea). The class I tRNA fraction (0.2 A260 unit) was digested with 2 µg snake venom phosphodiesterase (Sigma), 20 µg RNaseA (Invitrogen), and 0.125 U bacterial alkaline phosphatase (Takara) in 20 µl of 50 mM Tris–HCl (pH 8.0) at 37°C overnight. Nucleosides were analyzed on an HPLC (Hitachi L-2000 system) equipped with a reverse phase C18 column (NUCLEOSIL 100 C18; 25 cm × 4.6 mm, 7 µm; GL Sciences, Inc). The solvent system consisted of buffer C [50 mM sodium phosphate (pH 5.1)] and buffer D (buffer C containing 70% methanol). The nucleosides (20 µl) were chromatographed using a flow rate of 1 ml/min with a multistep linear gradient as follows: 3% buffer D from 0 to 10 min, 3–35% D from 10 to 50 min, 35–98% B from 50 to 65 min, 98–100% B from 65 to 75 min and 100% buffer C from 75 to 85 min. Standard modified nucleosides [1-methyladenosine (m1A), 5-methylcytidine (m5C), 2′-O-methyladenosine (Am), 2′-O-methylcytidine (Cm) and N6-methyladenosine-5′-monophosphate (pm6A)] were purchased from Sigma. The pm6A nucleotide was dephosphorylated with bacterial alkaline phosphatase before use. 5-Methyl-2-thiouridine (m5s2U) was received as a kind gift from Dr Naoki Shigi (National Institute of Advanced Industrial Science and Technology, Japan). The elution points of 1-methylguanosine (m1G), 2-methylguanosine (m2G), 2′-O-methylguanosine (Gm), 7-methylguanosine (m7G), 5-methyluridine (m5U) and pseudouridine (Ψ) were determined by enzymatic formations using the tRNA modification enzymes, TrmD, Trm1, TrmH, TrmB, TrmA and TruB, respectively. The nucleoside contents quoted in Figure 5 were calculated as follows: the peak areas of all nucleosides were integrated and the ratio of each modified nucleoside was calculated. Subsequently, the ratio of each modified nucleoside in the ΔtrmB sample was divided by that in the wild-type sample.
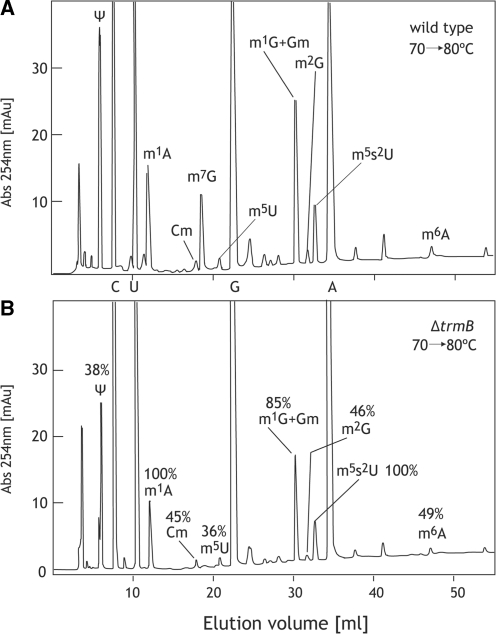
Figure 5.
The contents of modified nucleosides in class I tRNA from the ΔtrmB cells cultured at 70–80°C decrease. The modified nucleosides in class I tRNA from the wild-type (A) and the ΔtrmB (B) cells cultured at 70–80°C were compared. The content of each modified nucleosides was calculated as described in the ‘Materials and methods’ section, and is depicted in the figure.
Purification of native tRNAPhe by solid-phase DNA probe
3′-Biotinylated DNA oligomer (5′-TTC AGT CGC ATG CTC TAC CAA CT–biotin 3′) was used as a hybridization probe. The probe sequence is complementary from A36 to A14 of T. thermophilus tRNAPhe. Purification of tRNAPhe by solid-phase DNA probe was performed as described in our previous report (25). The eluted tRNAPhe was further purified by 10% PAGE (7 M urea).
RNA sequencing
RNA sequences of the purified tRNAPhe (0.02 A260 unit) from the wild-type and ΔTTHA1619 strains were determined by Kuchino’s post labeling method (26) with slight modifications as follows. Limited cleavage by formamide was performed at 90°C for 90 s, because the structure of T. thermophilus tRNAPhe is more stable as compared with tRNA species from mesophiles. For rapid detection of the modified nucleotides, we initially performed TLC using a single solvent system (isobutylic acid: conc. ammonia: water, 66:1:33, v/v/v). Later, we identified the modified nucleotides by 2D-TLC (24). 5′-32P-labeled nucleotides were monitored with a Fuji Photo Film BAS2000 imaging analyzer. Standard nucleotides were marked by UV260nm irradiation.
Growth phenotype analyses by plate culture
Thermus thermophilus wild-type and ΔtrmB strains were cultivated in rich medium at 67°C overnight. The diluted culture medium containing cells (A600 = 0.1) and sequential dilutions (10–1, 10–2, 10–3 and 10–4) (5 µl each) were spotted on to rich medium plates. These plates were incubated at 50°C for 50 h, at 60°C for 33 h, at 70°C for 13 h, at 75°C for 13 h and at 80°C for 15 h.
Translation activity analyses by 35S-Met incorporation
The wild-type and ΔtrmB strains were cultured at 70°C. When the cell density (A600 nm) had reached 0.5, the culture medium (20 ml) was pre-incubated for 20 min at 80°C and then supplemented with 4.8 MBq of 35S-Met. Culture medium (1 ml each) was sampled at various timepoints and the cells collected by centrifugation at 3500g for 2 min. The cells were washed with 500 µl of the medium and collected by centrifugation at 3500g for 2 min. The cells were resuspended in 20 µl of buffer A after which 20 µl of sodium dodecyl sulfate (SDS) loading buffer [100 mM Tris–HCl (pH 6.8), 200 mM dithiothreitol, 2.5% SDS, 0.2% bromophenol blue, 20% glycerol] was added. The sample was boiled for 10 min and centrifuged at 21 500g for 5 min. The supernatants were analyzed by 15% SDS–PAGE. The gels were stained with Coomassie brilliant blue and 35S-Met incorporation was monitored with a Fuji Photo Film BAS2000 imaging analyzer.
Melting profile analyses of class I tRNA and tRNAPhe
The purified tRNAPhe and class I tRNA fractions from the wild-type and ΔtrmB strains were prepared as described above. The tRNAPhe transcript containing the m7G46 modification was prepared by methylation of the tRNAPhe transcript by A. aeolicus TrmB. Before the melting point measurement the tRNA fraction was annealed in buffer E [50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl] from 80 to 40°C for 60 min and then the melting curve was recorded on a spectrophotometer, UV-1650PC (Shimadzu). The temperature was increased from 20 to 95°C for 75 min. The melting profiles were obtained by averaging the two scans. The melting temperatures were calculated from first derivative plots.
Preparation of tRNA modification enzymes
Aquifex aeolicus TrmB was purified according to our previous report (10,14). Thermus thermophilus tRNA (Gm18) methyltransferase (TrmH) (23,27–30) and tRNA (m1A58) methyltransferase (TrmI) (31) were kindly provided by Anna Ochi (Ehime University) and Masayuki Minoji (Ehime University), respectively. Escherichia coli tRNA m5U54 methyltransferase (TrmA) (32–34) was a gift from Chikako Iwashita (Ehime University). Aquifex aeolicus tRNA (m1G37) methyltransferase (TrmD) (35–37) was a gift from Mr Takashi Toyooka (Ehime University).
Aminoacylation of tRNAPhe
The phenylalanine–tRNA synthetase (Phe-RS) fraction was prepared as follows. Briefly, the S-100 fraction (20 ml) of the wild-type strain was loaded onto a DE52 column (column volume, 10 ml) and Phe-RS was eluted by a KCl linear gradient (50–350 mM). The fractions containing Phe-RS were identified by Phe charging activity. The aminoacylation assay was performed using 14C-Phe (18.4 GBq/mmol, Perkinelmer) as described in the reference (38). Phe charging activity was measured using 0.03A260 units of purified tRNAPhe using a filter assay.
Northern hybridization
The small RNA fraction was purified by Q-Sepharose column chromatography as described above. The samples were separated by 10% PAGE (7 M urea), transferred to a membrane (Hybond-N+, GE Healthcare) by electro blotting, and fixed by UV254 nm irradiation. Northern hybridization was performed with hybridization buffer (GE Healthcare) and 5′-32P-labeled DNA probe at 52°C or 59°C [in the case of tRNAVal (CAC)]. The DNA probe sequences are as follows: tRNAPhe (UUC), 5′-TCA GTC GCA TGC TCT ACC AAC-3′; tRNAval (CAC), 5′-AAC CGT GTG AGG CGA GCG CTC TT-3′; tRNAArg (CGG), 5′-CGG AGG CCG ACG CTC TAT C-3′; tRNATyr (UAC), 5′-TAC AGA CCG TCC CCT TTG GC-3′; tRNAIle (AUC), 5′-ATC AGG CGT GCG CTC TAA CC-3′. The hybridized bands were monitored with a Fuji Photo Film BAS2000 imaging analyzer.
RESULTS
Construction of a potential trmB disruptant
In order to investigate the function of the m7G46 modification in tRNA, we constructed a trmB disruptant strain of T. thermophilus HB8. We searched for the target gene in the T. thermophilus genome database by BLAST search using the amino acid sequence of E. coli TrmB. We identified the TTHA1619 gene as a candidate for T. thermophilus trmB. The expected amino acid sequence of TTHA1619 gene product shares high homology with both E. coli and A. aeolicus TrmB proteins (data not shown). The protein has a distinct basic amino acid rich region at its C-terminus, which is common to thermophilic TrmB proteins (14). To check the tRNA methyltransferase activity of the TTHA1619 gene product, we performed PCR cloning and inserted the amplified DNA into the pET30a E. coli expression vector. The recombinant protein was partially purified by heat treatment, and subsequently by DE52 column chromatography (data not shown). We confirmed tRNA methyltransferase activity by 14C-methyl transfer assay using yeast and T. thermophilus tRNAPhe transcripts, and 14C-pm7G formation activity by 2D-TLC (data not shown). On the basis of these experimental results, we selected the TTHA1619 gene as the target for gene disruption.
The plasmid vector for replacement of TTHA1619 by the HTK gene was purchased from RIKEN Biological Resource Center (21) and homologous recombination was performed according to the ref. (22). Colonies were isolated on a kanamycin plate and their genomic DNAs were prepared. The location of the HTK gene in the genome was analyzed by Southern hybridization. As a result, we were able to isolate a candidate ΔTTHA1619 strain (data not shown). We performed PCR cloning of the homologous recombination region and confirmed the DNA sequence (data not shown). These results confirmed that we had successfully selected a ΔTTHA1619 strain.
Absence of tRNA (m7G) methyltransferase activity in cell extract from the ΔTTHA1619 strain
To confirm the absence of tRNA (m7G) methyltransferase activity in the ΔTTHA1619 strain, we prepared S-100 and P-100 wash fractions from the wild-type and ΔTTHA1619 strains. The S-100 fraction from the wild-type or ΔTTHA1619 strain, yeast tRNAPhe transcript, and 14C-AdoMet were incubated at 60°C for 1 h. The RNA was recovered by ethanol precipitation and digested with nuclease P1. The 14C-labeled mononucleotides were analyzed by 2D-TLC (Figure 1A). Left panel of Figure 1A shows the result from the wild-type strain. A 14C-pm7G spot was clearly detected in addition to spots representing 14C-pm1G and 14C-pGm (14C-pm1G and 14C-pGm were probably derived from TrmD and TrmH activities, respectively). It should be mentioned that TrmI was mainly present in the P-100 wash fraction (data not shown) and its activity was barely detected in the S-100 fraction (Figure 1A). Furthermore, in the case of T. thermophilus, the m5U54 modification is generated by the 5,10-methylenetetrahydrofolate-dependent tRNA methyltransferase, TrmFO (39,40). Thus, 14C-pm5U was not detected in this experiment, whilst pm7G, pm1G and pGm were identified as 14C-labeled mononucleotides in the wild-type sample. In contrast, the 14C-pm7G spot was completely absent from the ΔTTHA1619 sample (Figure 1, right panel), demonstrating that the tRNA (m7G) methyltransferase activity was not present in the S-100 fraction from the ΔTTHA1619 strain. Furthermore, analysis of the AdoMet-dependent tRNA methyltransferase activities in the P-100 wash fractions confirmed that the tRNA (m7G) methyltransferase activity was not observed in the fraction from the ΔTTHA1619 strain (data not shown). Taking these experimental results together, we concluded that the tRNA (m7G) methyltransferase activity was absent from the cell extract of the ΔTTHA1619 strain.
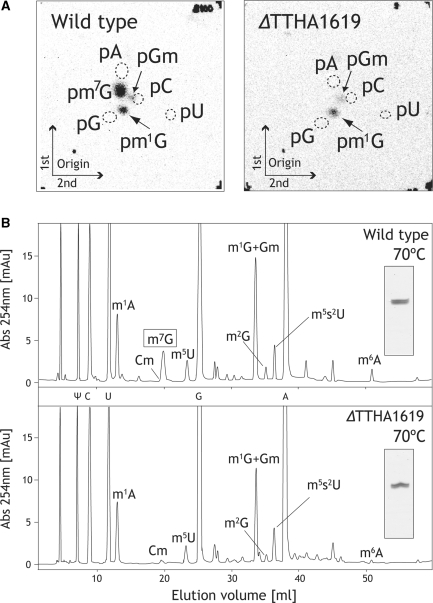
Figure 1.
Absence of m7G formation activity and lack of m7G nucleoside in extract and tRNA from the ΔTTHA1619 strain. (A) yeast tRNAPhe transcript, [methyl-14C]-AdoMet and S-100 fraction of the wild-type (left) or ΔTTHA1619 (right) strain were incubated at 60°C for 1 h, and 14C-methylated nucleotides were analyzed by 2D-TLC. Positions of standard markers (pA, pG, pC and pU) are enclosed by dotted circles. (B) nucleoside analyses of the class I tRNA fractions from the wild-type (upper) and ΔTTHA1619 (lower) strains. 0.03 A260 units of the purified class I tRNA fractions were analyzed by 10% PAGE (7 M urea) and the gel was stained with toluidine blue (insets).
Absence of the m7G modification in the Class I tRNA fraction from the ΔTTHA1619 strain
Next, we performed nucleoside analysis of the class I tRNA fraction, because the m7G46 modification is not found in class II tRNA species. The class I tRNA fractions from the wild-type and ΔTTHA1619 strains were purified as shown in Figure 1B insets. Figure 1B upper panel shows the result of the nucleoside analysis from the wild-type strain. All labeled modified nucleosides in Figure 1B were experimentally identified by comparison with standard markers or enzymatic formation as described in experimental procedures (data not shown). The m7G nucleoside eluted at 20.0 ml. In contrast, the peak representing m7G was missing from the ΔTTHA1619 sample (Figure 1B, lower). Thus, we confirmed the absence of the m7G46 modification in class I tRNA from the ΔTTHA1619 strain.
As well as the alteration described above we also noticed changes in other modifications of class I tRNA from the ΔTTHA1619 strain. As shown in Figure 1B, the content of the m1G + Gm and m6A seemed to be decreased. Because m1G and Gm eluted at the same point (32.4 min) on our HPLC system, they could not be distinguished. The m6A modification is generally produced in two ways. One is derivation from the m1A58 modification produced by TrmI, through non-enzymatic conversion of m1A to m6A. Another is the m6A37 modification, for which the responsible enzyme has not been identified. Moreover, at least, three modifications (Gm18, m5s2U54 and m1A58) in T. thermophilus tRNA are often not complete and the contents of these modifications in tRNA change according to the culture conditions (especially culture temperature) (41–44). Therefore, we carefully analyzed modification rates of these changes in ΔtrmB strain. The detailed analyses of these modifications are described in a later section.
Absence of the m7G46 modification in purified tRNAPhe
The sequence of tRNAPhe has been reported (Figure 2A) (41). In T. thermophilus tRNAPhe, the m7G modification exists only at position 46. To confirm the absence of this modification in tRNAPhe from the ΔTTHA1619 strain, we purified tRNAPhe by the solid phase DNA probe method, which was recently reported (25). The DNA probe sequence was designed to be complementary to the RNA sequence from the D-loop to the anti-codon loop (Figure 2A), because the sequence in this region of tRNAPhe is quite different from those of the other T. thermophilus tRNA species. To elute the RNA efficiently, tetramethylammonium chloride was selected as the salt in the hybridization buffer. This approach was successful in purifying tRNAPhe from tRNA mixtures of the wild-type and ΔTTHA1619 strains (Figure 2B). We further purified the tRNAPhe by 10% PAGE (7 M urea).
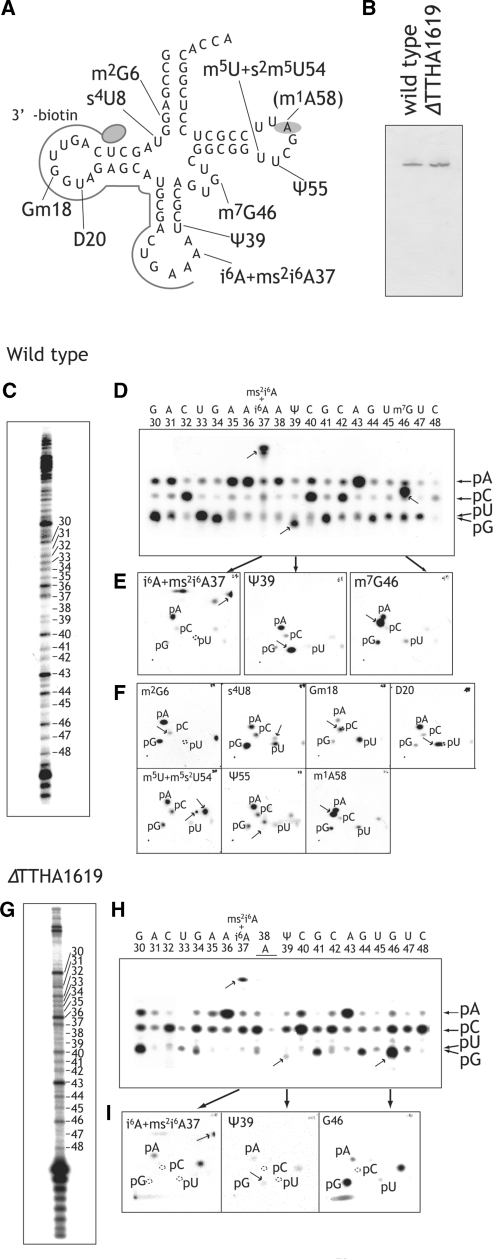
Figure 2.
Sequence analyses of purified tRNAPhe from the wild-type and ΔTTHA1619 strains by Kuchino’s post labeling method. (A) Nucleotide sequence of T. thermophilus tRNAPhe is depicted as a cloverleaf structure. The 3′-biotin DNA probe is illustrated. The m1A58 modification was identified in this work. (B) 0.01 A260 units of purified tRNAPhe from the wild-type (left) and ΔTTHA1619 (right) strains were analyzed by 10% PAGE (7 M urea). The gel was stained by toluidine blue. The purified tRNAPhe from the wild-type (C) and ΔTTHA1619 (G) strains was partially cleaved by formamide. Then the 5′-end of each fragment was labeled with γ-32P-ATP and T4 polynucleotide kinase. The RNA fragments were separated by 15% PAGE (7M urea). Numbers correspond to the nucleotide positions in tRNAPhe. The tRNAPhe fragments of the wild-type (D) and ΔTTHA1619 (H) strains were cut off from the gels in panels C and G, respectively. The fragments were digested with nuclease P1 and their 5′-nt were analyzed by TLC. In panels D and H, nucleotides at positions from 30 to 48 are shown. Positions of standard markers (pA, pG, pC and pU) are indicated by arrows at the right side of the thin layer plates. (E, I) Modified nucleotides of panels D and H were analyzed by 2D-TLC. The arrows indicate spots of modified nucleotides. (F) TLC patterns of the all modified nucleotides identified in tRNAPhe from the wild-type strain are shown.
To determine precise positions of the modified nucleosides, we performed RNA sequencing. In these experiments, we selected Kuchino’s post label method (26) to visualize the modifications. Briefly, the purified tRNAPhe was partially cleaved by formamide, and then the 5′-end of each fragment was labeled with γ-32P-ATP and T4 polynucleotide kinase. The RNA fragments were separated by 15% PAGE (7 M urea) (Figure 2C and G), cut off, extracted and recovered by ethanol precipitation. The recovered RNA was completely digested with nuclease P1 and 32P-labeled nucleotides were analyzed by TLC. Because the RNA sequence of T. thermophilus tRNAPhe has been reported (41), we initially performed one-dimensional TLC to search for modified nucleotides rapidly (Figure 2D and H), and then modified nucleotides were analyzed by the standard 2D-TLC method (Figure 2E, F and I). Figure 2C–F shows the results obtained from the tRNAPhe of the wild-type strain. In Figure 2C and D, nucleotide positions from 30 to 48 can be seen. In this region, the presence of three modified bases [i6A37 (partially modified to ms2i6A37), Ψ39, and m7G46] has been reported (Figure 2A). In fact, these modifications could be detected in our purified tRNAPhe from the wild-type strain (Figure 2D and E). In the same way, we analyzed the other positions of the purified tRNAPhe from the wild-type strain and we could detect from m2G6 to A73 in the tRNAPhe (data not shown). As a result, seven additional modifications [m2G6, s4U8, Gm18, D20, m5U54 (partially modified to m5s2U54), Ψ55 and m1A58] could be detected (Figure 2F), consistent with nucleosides analysis of the purified tRNAPhe by HPLC (data not shown). It should be mentioned that the m1A58 modification in T. thermophilus tRNAPhe has not been previously reported (41). As described above, the content of the m1A58 modification in tRNA changes according to the culture conditions (41–44). In the current study, we purified tRNAPhe from cells cultured at 70°C under 1 l per 1 min air supply into 1 L rich medium. The difference in the m1A58 modification between previous studies and those described here may be caused by differences in the culture conditions and/or purification procedures.
Next, we analyzed the tRNAPhe from the ΔTTHA1619 strain (Figure 2G–I). As expected, position 46 was found to be unmodified G46 (Figure 2H and I). These experimental results clearly showed that the TTHA1619 gene is the T. thermophilus trmB. Hereafter, we describe ΔTTHA1619 as ΔtrmB.
Growth defect of the ΔtrmB strain at high temperatures
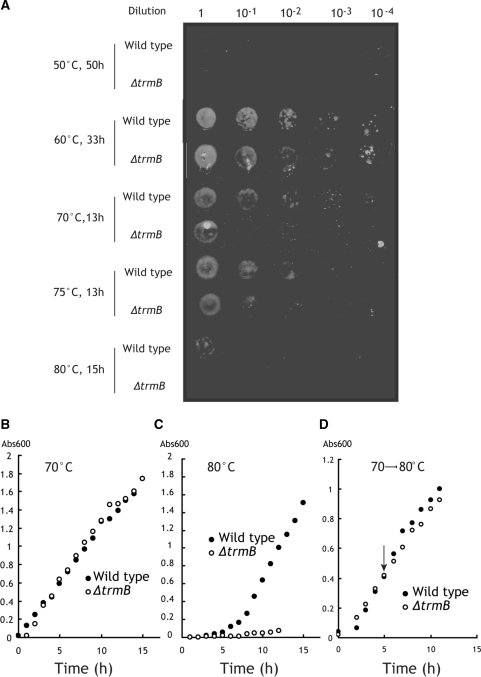
We examined the growth phenotype of the ΔtrmB strain at various temperatures. Figure 3A shows the results of plate cultures at 50, 60, 70, 75 and 80°C. The wild-type strain can live at a wide range of the temperature (50–80°C) although growth at 50°C is very slow (Figure 3A). Below 60°C, the wild-type and ΔtrmB strains exhibited similar growth. However, above 70°C, a growth defect in the ΔtrmB strain was observed. The ΔtrmB strain showed a long lag phase at 70°C as compared with the wild type (Figure 3A) although the growth curve of the ΔtrmB strain in liquid culture was almost the same as that of the wild type (Figure 3B). When the ΔtrmB strain was cultured at 75 or 80°C, the growth defect was more clearly observed. At 80°C in particular, growth on a plate was not observed within 15 h (Figure 3A) and the growth in liquid culture showed a severe growth defect (Figure 3C). In fact, bacteriolysis is often observed during culture at 80°C: Figure 3C shows one example in which cell number began to decrease around the 12 h mark. These experimental results suggested that the m7G46 modification in tRNA is required for viability of T. thermophilus at high temperatures.
Figure 3.
Growth phenotypes of the wild-type and ΔtrmB strains. (A) The wild-type and ΔtrmB strains were serially diluted, spotted onto plates containing rich medium, and incubated at the temperatures indicated. Incubation time is indicated next to temperature. The growth curves of the wild-type and ΔtrmB strains in liquid cultures at 70°C (B), 80°C (C) and 70–80°C (D). The arrow in panel D indicates the shift point of temperature from 70°C to 80°C.
We tested several culture conditions for preparation of tRNA and proteins at high temperatures. As a result, we found that the ΔtrmB strain could survive at 80°C in liquid culture when it was cultured at 70°C until the middle log phase and then the culture temperature could be shifted from 70 to 80°C (Figure 3D). The existence of TrmB seems to be more important in lag and early log phases than after middle log phase. Hereafter in this article, we describe these culture conditions as 70–80°C.
35S-Met incorporation into proteins in the ΔtrmB strain decreased not only at 80°C but also at 70°C
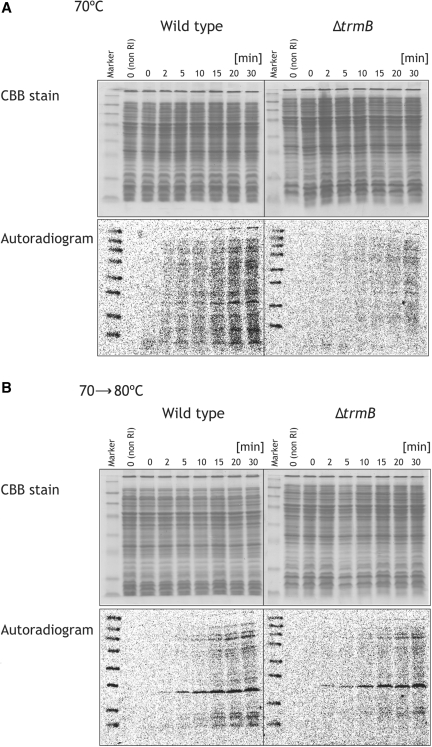
We compared in vivo protein synthesis activities of the wild-type and ΔtrmB strains by monitoring 35S-Met incorporation. When the cell density had reached 0.5A600 nm, 35S-Met was directly added into the culture medium. Figure 4A shows the results at 70°C. Unexpectedly, the speed of 35S-Met incorporation into proteins in the ΔtrmB cells was clearly slowed as compared with that in the wild-type cells. Thus, although the apparent growth curves of both strains in liquid culture are similar at 70°C, protein synthesis activity of the ΔtrmB strain is lower than that of the wild-type strain even at 70°C. At 70°C, protein synthesis is not rate limiting unlike other processes such as DNA replication. In contrast, 35S-Met incorporation of the ΔtrmB cells cultured at 70–80°C is clearly inferior to that of the wild-type cells (Figure 4B), consistent with the growth phenotypes. These experimental results demonstrate that the lack of m7G46 modification in tRNA causes depression of protein synthesis at high temperatures.
Figure 4.
Protein synthesis activities in the wild-type and ΔtrmB strain. The protein synthesis activities of the wild-type (left) and ΔtrmB (right) strains were compared by 35S-Met incorporation at 70°C (A) and 70–80°C (B). 35S-Met was added at the zero points and samples were taken out at 2, 5, 10, 15, 20 and 30 min. Total proteins were analyzed by 15% SDS–PAGE. The gels were stained with Coomassie brilliant blue (upper). 35S-Met incorporation was monitored with a Fuji-photo film imaging analyzer (lower). Non RI means the sample before addition of 35S-Met.
Other modifications are decreased in the class I tRNA fraction from the ΔtrmB cells cultured at 70–80°C
We have observed decreases in some modifications in tRNA from the ΔtrmB strain at 70°C as described above. To clarify whether the lack of the m7G46 modification was having effects on other modifications at high temperatures, we prepared class I tRNA fractions from the wild-type and ΔtrmB cells cultured at 70–80°C. Figure 5 shows results of the nucleosides analyses. As expected, the amounts of various modified nucleosides were decreased in the ΔtrmB sample in addition to the disappearance of m7G (Figure 5, lower). Thus, we confirmed that the content of Ψ, m2G, m5U, m6A and m1G + Gm in the class I tRNA from the ΔtrmB cells cultured at 70–80°C decreased compared with those from the wild-type cells.
Assays with tRNA modification enzymes demonstrate the hypo-modifications in Class I tRNA from the ΔtrmB strain
Hypo-modifications in class I tRNA from the ΔtrmB strain suggested that the lack of the m7G46 modification affects the activities of several tRNA modification enzymes. To confirm this idea, we performed assays using tRNA modification enzymes. The Gm18, m1G37, m5U54 and m1A58 modifications can be produced by TrmH, TrmD, TrmA and TrmI, respectively (Figure 6A). If these modifications decreased in tRNA from the ΔtrmB strain, the tRNA would be a better substrate for these enzymes as compared to the tRNA from the wild-type strain. We prepared five tRNA modification enzymes (T. thermophilus TrmH and TrmI, A. aeolicus TrmB and TrmD, and E. coli TrmA) (Figure 6B). In T. thermophilus, the m5U54 modification is generated by 5,10-methylenetetrahydrofolate-dependent tRNA methyltransferase TrmFO (39), however 5,10-methylenetetrahydrofolate is unstable and the radioisotope labeled compound is not commercially available. Therefore, we prepared the E. coli TrmA protein instead of TrmFO.
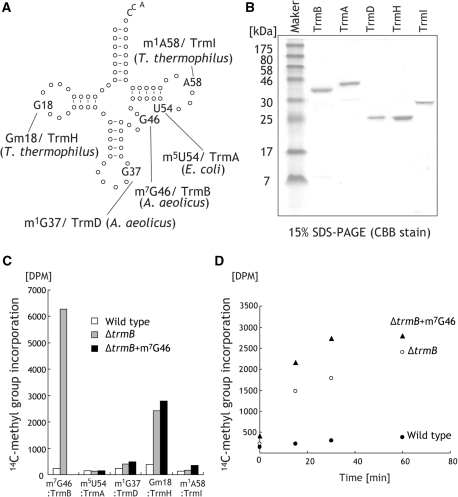
Figure 6.
Methyl-transfer activities of tRNA methyltransferases for the RNAs from the wild-type and ΔtrmB strains. (A) Modified nucleotides are indicated in a tRNA cloverleaf structure together with the responsible modification enzymes (TrmB, TrmA, TrmD, TrmH and TrmI). Sources of the enzymes are indicated in parenthesis. (B) Purified tRNA methyltransferases (TrmB, TrmA, TrmD, TrmH and TrmI) were analyzed by 15% SDS–PAGE and the gel was stained with Coomassie brilliant blue. (C) 14C-methyl group acceptance activity of RNAs from the wild-type (white) and ΔtrmB (grey) strain. The ΔtrmB + m7G46 (black) annotation on the graph means the methyl acceptance activity of the RNA from the ΔtrmB strain methylated with TrmB. The methyl group acceptance activities at a 60 min period are shown. With the exception of the Gm18 modification by TrmH the graphs represent the apparent initial velocities. The methylation velocities except that for the Gm18 modification were relatively slow because the modified tRNA, which is abundant in the RNA, inhibits the modifications. (D) Methyl group incorporation catalyzed by TrmH was monitored in a time-dependent manner.
Total RNAs from the wild-type and ΔtrmB strains cultured at 70°C were prepared. To clarify the role of the m7G46 modification, we modified total RNA from the ΔtrmB strain with A. aeolicus TrmB (we prepared the total RNA from the ΔtrmB strain with m7G46 modification by in vitro enzymatic formation). Methyl group acceptance activities of these RNAs were examined using the tRNA modification enzymes. In the case of E. coli TrmA, the assay was performed at 37°C. Figure 6C shows the 14C-methyl group incorporation at the 60 min period. The total RNA from the ΔtrmB strain is well methylated by A. aeolicus TrmB, while that from the wild-type strain is scarcely methylated, consistent with the lack of the m7G46 modification in the ΔtrmB strain. E. coli TrmA did not methylate total RNA from the wild-type strain as well as that from the ΔtrmB strain, demonstrating that U54 is nearly completely modified to m5U54 or m5s2U54 in both the wild-type and ΔtrmB strains. A. aeolicus TrmD methylated the RNA from the ΔtrmB strain. Thus, a fraction of the G37 in the ΔtrmB RNA was not completely modified in the cells. Furthermore, when G46 was modified to m7G46 by A. aeolicus TrmB, the velocity of the m1G37 modification by TrmD was slightly increased. Although the source of the TrmD was A. aeolicus, the presence of the G46 modification increased m1G37 formation by TrmD. This tendency was more clearly observed with the Gm18 modification. In the case of the Gm18 modification, time-dependent experiments were performed to compare the velocities (Figure 6D). G18 in the RNA from the wild-type strain is near fully modified to Gm18, as confirmed by the fact that the RNA was scarcely modified by TrmH. In contrast, the RNA from the ΔtrmB strain was efficiently methylated by TrmH, suggesting that G18 in the ΔtrmB strain is hypo-modified. Furthermore, in vitro introduction of m7G46 clearly accelerated Gm18 formation. Moreover, this tendency was also observed with the m1A58 modification by TrmI, although A58 is considerably modified to m1A58 in both the wild-type and ΔtrmB cells (Figure 6C).
These experimental results suggest that there is a tRNA modification network, in which the m7G46 modification has a positive effect on other modifications. In T. thermophilus, the m7G46 modification by TrmB enhances the formation velocities of Gm18 by TrmH, m1G37 by TrmD and m1A58 by TrmI. The lack of the m7G46 modification causes hypo-modifications of Gm18 and m1G37 (m1A58 is slightly hypo-modified). In the nucleoside analysis (Figure 5B), the contents of Ψ, m2G, m5U, m6A and m1G + Gm decreased. The Ψ modifications are introduced into various positions by multiple enzymes. The enzymes responsible for the m2G6 and m6A37 modifications have not been identified. Therefore, the effect of the m7G46 modification on these enzymes could not be experimentally verified. However, our experimental results demonstrated that the presence of the m7G46 modification induces nearly full modification of several modified nucleotides such as Gm18 and m1G37.
Aminoacylation of purified tRNAPhe
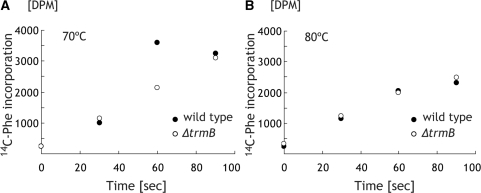
Protein synthesis activity of the ΔtrmB strain is low compared with that of wild-type strain at 70°C as well as at 70–80°C. The proportion of some modified nucleotides in Class I tRNA from the ΔtrmB strain are low compared with those from the wild-type strain. Do these hypo-modifications affect amino acid charging activity of tRNA? We partially purified the Phe-RS fraction and measured the Phe charging activity of tRNAPhe. In these experiments, we purified tRNAPhe from wild-type and ΔtrmB cells cultured at 70 and 70–80°C. Figure 7 shows the aminoacylations at 70°C (left) and 80°C (right) of the purified tRNAPhe from wild-type and ΔtrmB cells cultured at 70°C. As shown in Figure 7, there is no difference between Phe charging velocities of the purified tRNAPhe from wild-type and the ΔtrmB strain at 70 and 80°C. Furthermore, we examined Phe charging activity of the tRNAPhe from the wild-type and the ΔtrmB cells cultured at 70–80°C and confirmed that Phe charging velocities were the same at 70 and 80°C (data not shown). Thus, the lack of the m7G46 and the presence of hypo-modifications do not affect the Phe charging activity of tRNAPhe.
Figure 7.
Phe charging activities of purified tRNAPhe. Phe charging activities at 70°C (A) and 80°C (B) were measured by 14C-Phe incorporation into the purified tRNAPhe from the wild-type (filled circles) and ΔtrmB (open circles) strains. These graphs show one set of data from two independent experiments.
Melting temperatures of the class I tRNA fractions from the ΔtrmB strain decrease
The lack of m7G46 and hypo-modifications of other modified nucleotides does not affect amino acid charging activity (at least in the case of Phe charging activity); however, protein synthesis activity of the ΔtrmB strain is clearly inferior to that of the wild type. To address this problem, we measured melting temperatures of class I tRNA fractions from the wild-type and ΔtrmB strains (Table 1). To clarify the contribution of the m7G46 modification to melting temperature, we prepared yeast tRNAPhe transcript containing m7G46 by in vitro methyl-transfer reaction with A. aeolicus TrmB. As shown in Table 1, the presence of the m7G46 modification increased the melting temperature by only 0.1°C. However, class I tRNA fractions showed a clear difference in melting temperatures. The melting temperature of class I tRNA from the ΔtrmB strain was 76.2°C while that from the wild-type strain was 79.7°C. Thus, the lack of m7G46 and hypo-modifications cause a 3.5°C decrease in the melting temperature. At 80°C, some tRNA species from the ΔtrmB strain may be structurally loosened. Furthermore, we measured melting temperatures of the class I tRNA fractions from the wild-type and ΔtrmB cells cultured at 70–80°C. Unexpectedly, the melting temperature of the class I tRNA fraction of the ΔtrmB strain increased to 78.5°C (Table 1). In spite of enhancement of the hypo-modifications, the melting temperature increased. This experimental result prompted us to investigate the tRNA population.
Table 1.
Melting temperatures of tRNA
| tRNA | Melting temperature (°C) |
|---|---|
| yeast tRNAPhe transcript | 71.6 |
| yeast tRNAPhe transcript containing m7G46 | 71.7 |
| Class I tRNA | |
| 70°C Wild-type | 79.7 |
| 70°C ΔtrmB | 76.2 |
| 70–80°C Wild-type | 79.7 |
| 70–80°C ΔtrmB | 78.5 |
Degradation of some kinds of tRNA species in the ΔtrmB cells cultured at 70–80°C
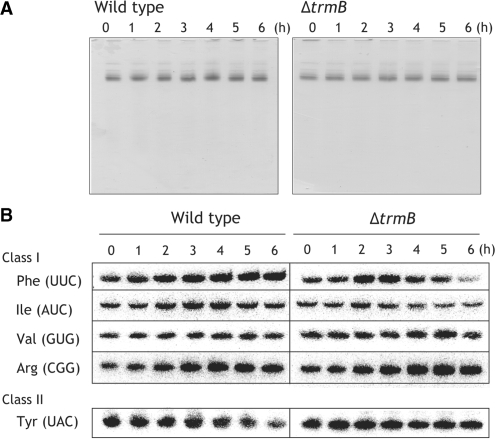
We performed northern hybridization to investigate the tRNA population (Figure 8). As shown in Figure 8A, small RNA (mainly tRNA and 5S rRNA) fractions were prepared from the wild-type and the ΔtrmB cells cultured at 70–80°C. In the experiments, zero time refers to the start point of the temperature shift to 80°C. We prepared four class I tRNA probes and one class II tRNA probe (tRNATyr). These class I tRNA species contain the m7G46 modification in the wild-type strain. As shown in Figure 8B, the class I tRNA species in the wild-type cells did not decrease within 6 h: tRNAPhe and tRNAArg slightly increased and tRNATyr slightly decreased. In contrast, tRNAPhe and tRNAIle in the ΔtrmB cells clearly decreased (Figure 8B). Thus, populations of tRNA species in the ΔtrmB cells are changed through degradation. These experimental results explain the increase in the melting temperature of the class I tRNA from the ΔtrmB cells cultured at 70–80°C. Furthermore, the results may explain the discrepancy in the results from experiments concerning the m5U content. In the nucleoside analysis (Figure 5), the content of the m5U was decreased in the class I tRNA from the ΔtrmB strain. In contrast, the TrmA assay showed that U54 in the ΔtrmB strain is near fully modified to m5U54 or m5s2U54 (Figure 6C). U54 hypo-modified tRNA may be preferentially degraded at high temperatures. If so, the degradation of U54 hypo-modified tRNA raises the melting temperature of the class I tRNA fraction. Based on the results from northern hybridization, we concluded that the lack of the m7G46 and hypo-modifications of other modified nucleotides cause degradation of some tRNA species (tRNAPhe and tRNAIle) and result in depression of protein synthesis at high temperatures.
Figure 8.
Degradation of tRNAPhe (UUC) and tRNAIle (AUC) in the ΔtrmB cells cultured at 70–80°C. (A) Small RNA fractions of the wild-type (left) and ΔtrmB (right) strains were time-dependently prepared from the cells cultured at 70–80°C. The zero periods mean the shift points of the culture temperature from 70°C to 80°C. 0.02 A260 units of RNA samples were analyzed by 10% PAGE (7 M urea). The gels were stained with toluidine blue. (B) The RNA samples in panel A were analyzed by northern hybridization. Sequences of the DNA probes for tRNA detection are described in the ‘Materials and Methods’ section.
DISCUSSION
The m7G46 modification in tRNA is widely found in eubacteria and eukaryotes. Nevertheless, the role of the m7G46 modification in eubacterial tRNA has not been clarified: the E. coli trmB disruptant showed no growth defect (8). In the current study, we demonstrate the importance of the tRNA m7G46 modification in the extreme thermophilic eubacterium T. thermophilus, which grows at 50–83°C. A summary of our study is shown in Figure 9. In the ΔtrmB cultured at high temperatures (above 70°C), several modified nucleotides in tRNA were hypo-modified in addition to the lack of the m7G46 modification. Although these hypo-modifications do not affect the Phe charging activity of tRNAPhe, they cause a decrease in melting temperature and degradation of tRNAPhe and tRNAIle. Protein synthesis of the ΔtrmB strain is clearly depressed above 70°C. At 80°C, the ΔtrmB strain exhibit a severe growth defect and often shows bacteriolysis during culture. Thus, the m7G46 modification is essential for cell viability and it may act through a tRNA modification network, in which the m7G46 modification has a positive effect on other modifications. It was demonstrated that the presence of the m7G46 in tRNA enhanced Gm18 formation activity by TrmH. Similar results were also obtained with m1G37 by TrmD and m1A58 by TrmI, although these modifications were near fully formed in the ΔtrmB cells. Thus, these results suggest that TrmB may be one of the key enzymes in the tRNA modification network. It is noteworthy that our preliminary experiment using gel-filtration column chromatography showed no direct interaction between TrmB and TrmH proteins (data not shown), although the source of TrmB was A. aeolicus. The m7G46 modification and stabilized local structure of tRNA seem to have a positive effect on the Gm18 formation by TrmH, although there is a potential for interaction of TrmB and TrmH on the precursor tRNA. Further study will be necessary to clarify the protein–protein and/or protein–tRNA interactions in the network.
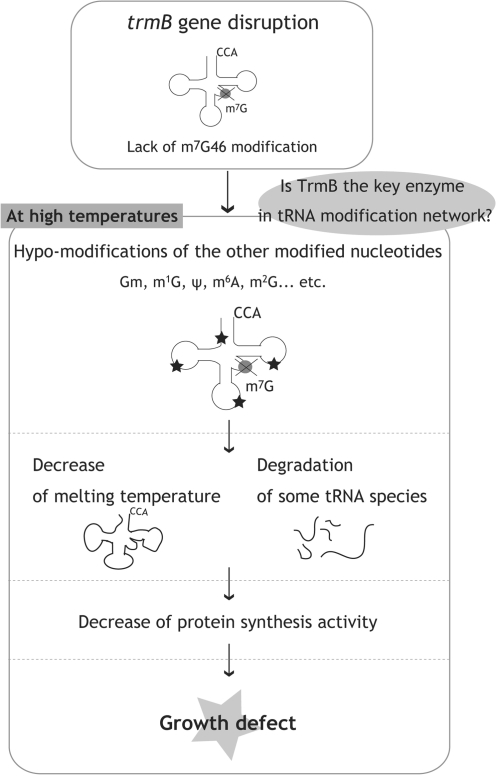
Figure 9.
Summary of this study. Effects of the trmB gene disruption are depicted. The lack of the m7G46 modification causes hypo-modification of other nucleotides in class I tRNA. The melting temperature of the tRNA decreases, and tRNAPhe and tRNAIle are degraded. The degradation of tRNA depresses protein synthesis and the ΔtrmB strain exhibits a severe growth defect at high temperatures. These experimental results suggest the existence of a tRNA modification network, in which the m7G46 modification catalyzed by TrmB may act as one of the key factors.
For more than 50 years, there has been limited information concerning the role of the m7G46 modification in tRNA. In the case of anticodon or anticodon loop modifications, lack of the modification often causes disorder of the codon–anticodon interaction and/or frame shift error (43,44). Therefore, at least, lysidine34 (k2C34) (45), inosine34 (I34) (46) and m1G37 (47) modifications in E. coli are essential for cell viability. In contrast, it is difficult to explain the roles of modified nucleotides in the three-dimensional core of tRNA. In some cases, the disruption of a single three-dimensional (3D) core modification enzyme gene gives no significant apparent phenotype. For example, a null mutant of E. coli truB, which encodes tRNA (Ψ55) synthase, grows normally, although the mutant exhibits a defect in survival upon rapid transfer from 37 to 50°C (48). In the case of the E. coli m5U54 modification which is produced by TrmA, the TrmA protein is essential for viability although the known catalytic activity of TrmA is not necessary (33). Because the TrmA protein exists not only in a tRNA bound form but also a 16S rRNA bound form in E. coli cells, the TrmA protein seems to have multiple functions (49). Thus, studies of gene disruptant mutants of 3D core modification enzymes are not straightforward. In fact, we have constructed several gene disruptant strains of T. thermophilus tRNA modification enzymes and some of them show no apparent change in growth phenotype. For example, a null mutant of T. thermophilus trmH grows normally at 70 and 80°C (Iwashita,C. and Hori,H., unpublished results). In contrast, the ΔtrmB strain showed a severe growth defect at high temperatures as demonstrated in the current study. Therefore, we consider that the m7G46 modification may be one of the key factors in the tRNA modification network. Our findings in the current study may explain the wide distribution of the m7G46 modification in eubacterial tRNA. Because T. thermophilus is an extreme thermophilic eubacterium, we were able to elucidate the importance of the m7G46 modification at high temperatures, in which unmodified tRNA transcripts are melted. It is likely that primitive life was born on Earth in a high-temperature environment. Thus, RNA modifications in the primitive life are likely to be more important than those in modern mesophiles. In T. thermophilus, two modifications, m5s2U54 (50) and m1A58 (31), have been reported to contribute to viability at high temperatures. The m5s2U54 modification raises the melting temperature of tRNA (50) and the presence of m1A58 induces the m5s2U54 modification (51). The m7G46 modification seems to reflect this m5s2U54 and m1A58 modification network because our in vitro experiment showed that the m7G46 modification enhances the speed of m1A58 formation. However, because the m1A58 modification in the ΔtrmB strain is nearly complete, the modification enzyme for the m1A58 (TrmI) would seem to be abundant in the cells. The lack of m7G46 mainly has a positive effect on other modifications such as Gm18 which is catalyzed by TrmH. Although G46 is one of the positive determinants for TrmH as described in our previous report (52), the effect of the G46 modification on TrmH activity has not been elucidated. In E. coli, the Gm18 modification contributes to translational accuracy in conjunction with Ψ55 as demonstrated by the fact that a mutant strain lacking both the Gm18 and Ψ55 modifications shows a growth defect and increased frameshift error frequency (53). In the T. thermophilus ΔtrmB strain, a similar phenomenon may occur because the level of Gm and Ψ modifications in the ΔtrmB cells decreases. In the class I tRNA from ΔtrmB cells, we found that the content of the m1G37 modification is not 100%. The hypo-modification of m1G37 directly perturbs accuracy of protein synthesis, because this modification prevents frameshift errors (43,54).
Although prevention of tRNA degradation by modified nucleotides is predicted from the results of in vitro experiments (55), in vivo degradation of the hypo-modified tRNA in bacterial cells has not been experimentally verified. Our experimental results from the current study clearly reveal the degradation of the hypo-modified tRNA in eubacterial cells. Recently, it has been reported that tRNA modifications in yeast function as a quality control system for tRNA. For example, the m7G46 modification in yeast contributes to the stability of tRNA in conjunction with the other modified nucleotides such as m5C (16). Furthermore, the m1A58 modification in yeast is essential for viability through the stability of initiator tRNAMet (56,57). In the T. thermophilus ΔtrmB strain, degradation of tRNAPhe and tRNAIle were observed at high temperatures. Therefore, a primitive quality control system, in which tRNA modifications are monitored, may exist in eubacterial cells. Further study will be necessary to clarify whether a tRNA quality control system in eubacteria exists or not.
FUNDING
Funding for open access charge: Research Fellowships for Young Scientist (20 4827 to C.T.), a Grant-in-aid (20034041 to H.H.) for Science Research on Priority Areas, and a Grant-in-aid (19350087 to H.H.) for Science Research from the Japan Society for the Promotion of Science (JSPS).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. Yaeta Endo (Ehime University) and Prof. Kazuya Nishikawa (Gifu University) for use of laboratory facilities. They also thank Dr Naoki Shigi (National Institute of Advanced Industrial Science and Technology, Japan) for valuable discussion.
REFERENCES
- 1.Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SH, Sussman JL, Suddath FL, Quigley GJ, McPherson A, Wang AH, Seeman NC, Rich A. The general structure of transfer RNA molecules. Proc. Natl Acad. Sci. USA. 1974;71:4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 6.Garcia GA, Goodenough-Lashhua DM. Mechanisms of RNA-Modifying and -Editing Enzymes. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. Washington, DC: American Society for Microbiology; 1998. pp. 555–560. [Google Scholar]
- 7.Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Bie LG, Roovers M, Oudjama Y, Wattiez R, Tricot C, Stalon V, Droogmans L, Bujnicki JM. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J. Bacteriol. 2003;185:3238–3243. doi: 10.1128/JB.185.10.3238-3243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zegers I, Gigot D, van Vliet F, Tricot C, Aymerich S, Bujnicki JM, Kosinski J, Droogmans L. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 2006;34:1925–1934. doi: 10.1093/nar/gkl116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto H, Watanabe K, Ikeuchi Y, Suzuki T, Endo Y, Hori H. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 2004;279:49151–49159. doi: 10.1074/jbc.M408209200. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11:821–830. doi: 10.1261/rna.2030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartlidge RA, Knebel A, Peggie M, Alexandrov A, Phizicky EM, Cohen P. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J. 2005;24:1696–1705. doi: 10.1038/sj.emboj.7600648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16:52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 14.Tomikawa C, Ochi A, Hori H. The C-terminal region of thermophilic tRNA (m7G46) methyltransferase (TrmB) stabilizes the dimer structure and enhances fidelity of methylation. Proteins. 2008;71:1400–1408. doi: 10.1002/prot.21827. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto K, Toyooka T, Tomikawa C, Ochi A, Takano Y, Takayanagi N, Endo Y, Hori H. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex) FEBS Lett. 2007;581:1599–1604. doi: 10.1016/j.febslet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takano Y, Takayanagi N, Hori H, Ikeuchi Y, Suzuki T, Kimura A, Okuno T. A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol. Microbiol. 2006;60:81–92. doi: 10.1111/j.1365-2958.2006.05080.x. [DOI] [PubMed] [Google Scholar]
- 19.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 1999;126:951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 20.Hoseki J, Okamoto A, Takada N, Suenaga A, Futatsugi N, Konagaya A, Taiji M, Yano T, Kuramitsu S, Kagamiyama H. Increased rigidity of domain structures enhances the stability of a mutant enzyme created by directed evolution. Biochemistry. 2003;42:14469–14475. doi: 10.1021/bi034776z. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Mastuno Y, Kuroda Y, Nishimura Y, et al. Structural genomics projects in Japan. Nature Struct. Biol. 2000;7:943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto Y, Yano T, Kuramitsu S. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 2001;506:231–234. doi: 10.1016/s0014-5793(01)02926-x. [DOI] [PubMed] [Google Scholar]
- 23.Hori H, Suzuki T, Sugawara K, Inoue Y, Shibata T, Kuramitsu S, Yokoyama S, Oshima T, Watanabe K. Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells. 2002;7:259–272. doi: 10.1046/j.1365-2443.2002.00520.x. [DOI] [PubMed] [Google Scholar]
- 24.Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 25.Awai T, Kimura S, Tomikawa C, Ochi A, Ihsanawati, Bessho Y, Yokoyama S, Ohno S, Nishikawa K, Yokogawa T, et al. Aquifex aeolicus tRNA (N2,N2-Guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 2009;284:20467–20478. doi: 10.1074/jbc.M109.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchino Y, Kato M, Sugisaki H, Nishimura S. Nucleotide sequence of starfish initiator tRNA. Nucleic Acids Res. 1979;6:3459–3469. doi: 10.1093/nar/6.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson BC, Jager G, Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 1997;25:4093–4097. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure. 2004;12:593–604. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe K, Nureki O, Fuakai S, Ishii R, Okamoto H, Yokoyama S, Endo Y, Hori H. Roles of conserved amino acid sequence motifs in the SpoU (TrmH) RNA methyltransferase family. J. Biol. Chem. 2005;280:10368–10377. doi: 10.1074/jbc.M411209200. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe K, Nureki O, Fukai S, Endo Y, Hori H. Functional categorization of the conserved basic amino acid residues in TrmH (tRNA (Gm18) methyltransferase) enzymes. J. Biol. Chem. 2006;281:34630–34639. doi: 10.1074/jbc.M606141200. [DOI] [PubMed] [Google Scholar]
- 31.Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ny T, Björk GR. Growth rate-dependent regulation of transfer ribonucleic acid (5-methyluridine) methyltransferase in Escherichia coli B/r. J. Bacteriol. 1980;142:371–379. doi: 10.1128/jb.141.1.67-73.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson BC, Gustafsson C, Berg DE, Björk GR. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc. Natl Acad. Sci., USA. 1992;89:3995–3998. doi: 10.1073/pnas.89.9.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbonavicius J, Jager G, Björk GR. Amino acid residues of the Escherichia coli tRNA(m5U54)methyltransferase (TrmA) critical for stability, covalent binding of tRNA and enzymatic activity. Nucleic Acids Res. 2007;35:3297–3305. doi: 10.1093/nar/gkm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byström AS, Björk GR. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol. Gen. Genet. 1982;188:440–446. doi: 10.1007/BF00330046. [DOI] [PubMed] [Google Scholar]
- 36.Takeda H, Toyooka T, Ikeuchi Y, Yokobori S, Okadome K, Takano F, Oshima T, Suzuki T, Endo Y, Hori H. The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells. 2006;11:1353–1365. doi: 10.1111/j.1365-2443.2006.01022.x. [DOI] [PubMed] [Google Scholar]
- 37.Toyooka T, Awai T, Kanai T, Imanaka T, Hori H. Stabilization of tRNA (m1G37) methyltransferase [TrmD] from Aquifex aeolicus by an intersubunit disulfide bond formation. Genes Cells. 2008;13:807–816. doi: 10.1111/j.1365-2443.2008.01207.x. [DOI] [PubMed] [Google Scholar]
- 38.Pachmann U, Zachau HG. Yeast seryl tRNA synthetase: two sets of substrate sites involved in aminoacylation. Nucleic Acids Res. 1978;5:961–973. doi: 10.1093/nar/5.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria – evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, Hori H, Nureki O. Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:8180–8185. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grawunder U, Schön A, Sprintzl M. Sequence and base modifications of two phenylalanine-tRNAs from Thermus thermophilus HB8. Nucleic Acids Res. 1992;20:137. doi: 10.1093/nar/20.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe K, Shinma M, Oshima T, Nishimura S. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 1976;72:1137–1144. doi: 10.1016/s0006-291x(76)80250-1. [DOI] [PubMed] [Google Scholar]
- 43.Farabaugh PJ, Björk GR. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbonavicius J, Qian Q, Durand JMB, Hagervall TG, Björk GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 46.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinghorn SM, O’Byrne CP, Booth IR, Stansfield I. Physiological analysis of the role of truB in Escherichia coli: a role for tRNA modification in extreme temperature resistance. Microbiology. 2002;148:3511–3520. doi: 10.1099/00221287-148-11-3511. [DOI] [PubMed] [Google Scholar]
- 49.Gustafsson C, Björk GR. The tRNA-(m5U54)-methyltransferase of Escherichia coli is present in two forms in vivo, one of which is present as bound to tRNA and to a 3′-end fragment of 16S rRNA. J. Biol. Chem. 1993;268:1326–1331. [PubMed] [Google Scholar]
- 50.Shigi N, Suzuki T, Terada T, Shirouzu M, Yokoyama S, Watanabe K. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 2006;281:2104–2113. doi: 10.1074/jbc.M510771200. [DOI] [PubMed] [Google Scholar]
- 51.Shigi N, Sakaguchi Y, Suzuki T, Watanabe K. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 2006;281:14286–14306. doi: 10.1074/jbc.M511675200. [DOI] [PubMed] [Google Scholar]
- 52.Hori H, Yamazaki N, Matsumoto T, Watanabe Y, Ueda T, Nishikawa K, Kumagai I, Watanabe K. Substrate recognition of tRNA (Guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 1998;273:25721–25727. doi: 10.1074/jbc.273.40.25721. [DOI] [PubMed] [Google Scholar]
- 53.Urbonavicius J, Durand JM, Björk GR. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J. Bacteriol. 2002;184:5348–5357. doi: 10.1128/JB.184.19.5348-5357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Björk GR, Wikström PM, Byström AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 55.Kumagai I, Watanabe K, Oshima T. A thermostable tRNA (guanosine-2′)-methyltransferase from Thermus thermophilus HB27 and the effect of ribose methylation on the conformational stability of tRNA. J. Biol. Chem. 1982;257:7388–7395. [PubMed] [Google Scholar]
- 56.Anderson J, Phan L, Cuesta R, Carison BA, Pak M, Asano K, Björk GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3652. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA (1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]