Abstract
Mono-ubiquitylation of a transactivator is known to promote transcriptional activation of certain transactivator proteins. For the Sacchromyces cerevisiae transactivator, GAL4, attachment of mono-ubiquitin prevents destabilization of the DNA–transactivator complex by the ATPases of the 26S proteasome. This inhibition of destabilization depends on the arrangement of ubiquitin; a chain of ubiquitin tetramers linked through lysine 48 did not display the same protective effect as mono-ubiquitin. This led to an investigation into the properties of ubiquitin that may be responsible for this difference in activity between the different forms. We demonstrate the ubiquitin tetramers linked through lysine 63 do protect from proteasomal-mediated destabilization. In addition, we show that the mutating the isoleucine residue at position 44 interferes with proteasomal interaction in vitro and will abolish the protective activity in vivo. Together, these data implicate the hydrophobic patch of ubiquitin as required to protect transactivators from destabilization by the proteasomal ATPases.
INTRODUCTION
Many short-lived and damaged proteins are degraded by the ubiquitin-proteasome system (UPS) (1). The UPS is comprised of the 26S proteasome, the small protein ubiquitin, and the protein machinery used to attach ubiquitin to target proteins. Most proteins are targeted for degradation by attachment of a poly-ubiquitin chain containing multiple ubiquitin monomers linked together through lysine 48. Poly-ubiquitin chains containing four or more monomers of ubiquitin are efficiently recognized by proteins in the regulatory particle (RP) of proteasome. The RP then removes the ubiquitin chain and the six proteasomal ATPases (Rpt 1–6) assist in translocation of the target protein into the interior of the barrel shaped core particle (CP) where the proteolytic active sites are located. The CP can be capped on either end by the RP.
The proteolytic activity of the UPS is intimately involved in RNA polymerase II transcription at many levels. It has long been known that the UPS can negatively regulate transcription by proteolysis of activators, thus keeping their level too low to drive gene transcription (2–4). On the other hand, proteasome-mediated proteolysis has been found to have a stimulatory effect on the transcription of many genes, for example, through the degradation of repressor proteins such as IκB (5). It has also been shown that proteasome-mediated turnover of activators, coactivators and other promoter-bound transcription factors are essential for the expression of some genes, though the mechanistic basis of this phenomenon is not clear. Finally, the proteasome is involved in the efficient termination of transcription and clearance of the RNAP II from sites of DNA damage (6).
The UPS also affects transcription through non-proteolytic mechanisms. Chromatin immunoprecipitation followed by microarrays (ChIP-chip protocol) have revealed that proteasomal proteins are associated with DNA throughout the yeast genome (7,8). This suggests that the proteasomal proteins played a role in nucleic acid metabolism and, in agreement with this view, several roles of the proteasome have been found at different stages in transcriptional regulation. These roles include chromatin modification (9,10) and transcriptional elongation (11–14), both of which occur independent of proteolytic activity. Studies of the GAL and heat shock genes in yeast have shown that the proteasomal ATPases, but not the 20S CP, are required for efficient elongation in vitro and in vivo (11,12,15). It was shown that the transactivator Gal4 binds directly to two of the Rpt proteins (Rpt4 and Rpt6) and acts to recruit a fragment of the proteasome that includes the six ATPases (Rpts 1–6), Rpn1 and Rpn2, and perhaps other proteins, but excludes the 20CP as well as the 19S RP lid sub-complex to GAL promoters in vivo (16,17). The mechanism by which this sub-complex of the 19S RP stimulates elongation is unclear, but it has been speculated to be involved in the remodeling of initiation complexes into elongation complexes and in the partial disassembly of nucleosomes in the pathway of the elongating polymerase.
More recently, a second non-proteolytic activity of the proteasomal ATPase complex was discovered, which is the ATP-dependent destabilization of activator-promoter complexes (18). This destabilization activity requires physical contact between the activation domain (AD) of the Gal4 transactivator and two of the proteasomal proteins, Rpt4 and Rpt6, and probably involves the unfolding of the activator by the proteasomal ATPases, though this has not been shown conclusively (19). This potent activity can strongly repress GAL transcription by preventing stable association of the activator with the promoter in vivo.
Interestingly, however, this activity is manifest only in the context of certain Gal4 mutants, whereas the wild-type protein is immune to this activity in vivo. Recent investigations have revealed that the mutations that render Gal4 sensitive to this ‘stripping’ activity also prevent it from being mono-ubiquitylated within the DNA-binding domain, suggesting that mono-ubiquitylation of the activator serves to protect it from the proteasomal APTase complex (20). This provides a potential explanation for the stimulatory effect that mono-ubiquitylation has been shown to have on some activators [also see ref. (20) for another possible mechanism].
In agreement with this idea, it was found that high levels of soluble Ub blocks the destabilization of Gal4–DNA complexes in vitro, arguing that Ub contacts with either the activator or proteasomal ATPases down-regulate the stripping reaction and that these interactions can be driven in trans by high levels of Ub (19). There are several known ubiquitin receptors present in the proteasome. Rpn10 is a known poly-ubiquitin chain receptor, but deletion of the protein doesn’t grossly inhibit degradation of poly-ubiquitylated substrates in vivo (21,22). The ATPase Rpt5 is known to interact with lysine-48-linked tetra-ubiquitin chains using a cross-linking strategy, but monomeric ubiquitin did not demonstrate any detectable interaction by the same cross-linking methodology (23). Finally, Rpn13 has recently been demonstrated to bind to both monomeric and lysine 48-linked ubiquitin chains (24,25). To determine the identity of the ubiquitin receptor in the context of the Gal4 system, a novel chemical cross-linking and label transfer strategy was used (26). Ubiquitin was found to bind directly to Rpn1 and Rpt1 in the proteasomal ATPase complex and that these interactions disrupt the AD-Rpt6/Rpt4 interactions, causing the dissociation of the Gal4-proteasomal ATPase complex and terminating the stripping reaction.
One of the interesting observations from the previous study was that while mono-ubiquitylation added in trans would prevent destabilization of the transactivators from DNA, lysine-48-linked tetra-ubiquitin did not display a protective effect even at much higher concentrations. This difference between monomeric and chain forms of ubiquitin was also seen for interaction of ubiquitin to the ATPase Rpt5 shown by Pickart and co-workers (23). The explanation for this difference between forms of ubiquitin was not apparent in either of the prior studies.
In the current study, we set out to determine what surfaces of ubiquitin were important for its protective function. Proteasomal-mediated destabilization assays were used as a tool to isolate surfaces of ubiquitin for further study. These studies reveal that ‘exposed’ versions of ubiquitin, but not versions that form higher order packed quantanary structures, would effectively inhibit destabilization of Gal4/DNA complexes. Linkages of poly-ubiquitin chains that bury a hydrophobic patch, centered around isoleucine 44, do not effectively prevent destabilization. Further, mutation of the hydrophobic patch abrogates the ability of ubiquitin to interact with the proteasomal subunit Rpt1. In vivo, ubiquitin with the I44A mutation is no longer able to prevent proteasomal-mediated destabilization when fused to a mutant form of Gal4. We conclude that the hydrophobic patch of ubiquitin is important for the inhibition of destabilization of activator–DNA complexes.
MATERIALS AND METHODS
General methods and strains used
For chromatin immunoprecipitation (ChIP) experiments, yeast strain Sc726 (SUG1 gal4::HIS3) was used (27). These strains were transformed with single copy plasmids (derived from pSB32) expressing either wild type Gal4 or a mutant form of the protein expressed from the native Gal4 promoter. In each case the encoded proteins were tagged at their N-termini with three tandem copies of the T7 epitope tag (Novagen). Genetic fusion of ubiquitin to T7 tagged Gal4D in the pSB32 vector was done by removing Ub from GST-Ub-Gap71-VP16 (18) using a NcoI digest and inserting Ub into the NcoI site at the start codon of the T7 tag.
The steady-state levels of the Ub-Gal4D fusions were monitored using expression from a multi-copy plasmid with the native GAL4 promoter. These proteins were extracted from the pSB32 vector with the restriction enzymes BamHI and EcoRI and inserted into the YEp351 multi-copy vector. The constructs were then transformed into Sc726. α-Galactosidase assays were done using Sc244 (strain 21) (a gal4-2, Gal80, ura3-52, leu2-3 112, ade1, MEL1) transformed with the pSB32 plasmids mentioned above.
26S proteasomes were purified using a FLAG affinity tag as described (28) with modifications (11). The hexahistidine tagged Rpt1 protein in the pET-28a vector was kindly provided by George DeMartino (UTSW). Hexahistidine tagged ubiquitin and CCPGCC-tagged ubiquitin have been described (19). Mutations in hexahistidine tagged ubiquitin were made using QuikChange mutagenesis (Strategene). Hexahistidine tagged proteins were purified using standard IMAC procedures (Qiagen). Ubiquitin chains were purchased from Boston Biochem.
Destabilization of activator–DNA complexes by the proteasome
The destabilization assay has been described previously (18) with changes (19). Destabilization in the presence of mono-Ub or tetra-Ub chains was done by adding protein to the reaction mix immediately before addition of the activator–DNA complex.
Competition of ubiquitin/Rpt1 interaction using ubiquitin mutants
Cross-linking and label transfer reactions between ubiquitin and Rpt1 have been described previously (19). Competition reactions using ubiquitin mutants were performed by adding a 3-fold excess of purified ubiquitin to the Rpt1 protein lysate 15 min before adding the cross-linkable form of ubiquitin and then performing the reaction as described.
Expression level of the ubiquitin-Gal4D proteins
A ΔGal4 strain (Sc726) was transformed with YEp351 multi-copy plasmid containing the indicated Ub-Gal4D fusion protein. A 500 ml culture of these strains was grown to mid-log phase in complete media lacking leucine with raffinose as the carbon source. Galactose was added for 2 h and the cells were collected by centrifugation. A 1 ml aliquot was saved to measure the cell density at an OD of 600 nm and an equal amount of cells were resuspended in 50 ml of water and 1 × SDS loading buffer. The cells were subjected to three cycles of freeze/thaw and cell debris was spun down at 14 K for 10 min. The lysate was loaded on gel and subject to SDS–PAGE and western blotting with an anti-Gal4 N-terminal antibody.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to the protocol described (17). Cells were treated grown in raffinose containing medium, with galactose added to 2% to induce the GAL genes. Induction was carried out for 30 min prior to addition of formaldehyde. Immunoprecipitations were carried out using anti-Gal4 N-terminal antibody as previously described (17).
Quantitative PCR of precipitated chromatin was performed using an iCycler Thermal Cycler and the IQ SYBR Green Supermix, 2 × mix containing 100 mM KCl, 40 mM Tris–HCl, pH 8.4, 0.4 mM each dNTP, 50 U/ml iTaq DNA polymerase, 6 mM MgCl2, SYBR Green I, 20 nM fluorescein, stabilizers (Biorad, Hercules, CA). Relative fold enrichment of specific DNA was calculated by comparing Ct values derived from primers against the GAL7 promoter from samples precipitated with the specific antibody compared to an unspecific control antibody relative Ct values from to the total, unprecipitated DNA from each sample. Primers used for analysis have been described (20). In the graphed figures the Gal4D sample was arbitrarily set as 1.
Quantification of mRNA transcripts
Total RNA was isolated from 10 ml of cells OD600 0.6–0.8 after addition of galactose. Cells were centrifuged for 5 min at 3000g in a Sorvall RT7 centrifuge with a RTH-750 swing bucket rotor. Cells were washed with PBS and centrifuged as before. Cell pellets were frozen in liquid nitrogen and stored at −80°C. Cell pellets were resuspended in 400 µl water and 400 µl water-saturated phenol was added and vortexed 1 min. The mixture was incubated at 65°C for 45 min. The aqueous layer was removed and extracted with water-saturated phenol followed by chloroform. RNA was treated with RQ1 DNase (Promega corp., Madison, WI) for 1 h. RNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform. The RNA was precipitated by adding 40 µl 3 M NaOAc pH 5.3 and 1 ml 95% EtOH. RNA quantity was measured by measuring OD260. Total RNA of 1 µg was used to make cDNA using the Stratascript first strand cDNA synthesis kit (Stratagene, La Jolla, CA) and oligo dT. CDNA was measured by quantitative PCR as above using GAL1 and ACTI primers. The Ct value from each sample was used to calculate the ratio of GAL1/ACT1. Replicated from three samples were averaged and graphed. The primers used for analysis have been described (20).
α-Galactosidase assays
α-Galactosidase assays have been described (29). Briefly, Sc244 with the pSB32 vectors containing Gal4 and Gal4 mutants were grown in synthetic complete lacking leucine with raffinose as the carbon source. At an OD 600 nm of 0.6 galactose was added to 2% final. After a 45 min induction, the cells were pelleted and lysed by bead disruption. Total protein of 50 µg was used for each assay and the nM min−1 µg−1 of p-Nitrophenyl produced was determined. The mean and standard error of the mean was graphed with the activity from Gal4 set as 100%.
RESULTS
Identification of Ub residues required for inhibition of destabilization
We had previously reported an assay to monitor the ability of the proteasomal ATPases to destabilize transactivator–DNA complexes in a non-proteolytic manner (see Supplementary Figure 1) (18). Briefly, a small, biotinylated piece of DNA containing 5 Gal4-binding sites (UASG) is tethered to a StreptAvidin-conjugated bead and the binding sites are saturated with transactivator. After washing, the transactivator–DNA complex is exposed to purified 26S or purified 19S proteasome in the presence of ATP and an excess of non-biotinylated DNA containing the UASG sites, which is required to see the full stripping activity (18). As the transactivator is removed from the DNA by the proteasome it is ‘trapped’ by the excess of non-biotinylated DNA. Western blots are used to monitor the amount of the transactivator remaining on the tethered DNA in the presence of the proteasome. Using this assay, the ability of the proteasome to destabilize transactivator–DNA complexes can be monitored. Addition of monomeric ubiquitin would inhibit the ability of the proteasome to destabilize transactivator–DNA interactions, but addition of lysine-48-linked tetra ubiquitin chains failed to inhibit the destabilization activity of the proteasome (19).
A hypothesis consistent with these observations is that a surface of Ub exposed in the monomer, but hidden in lysine-48-linked polymers, might be critical for the protective activity. Structural studies have suggested that part of surface of the lysine-48-linked ubiquitin is buried at the interface of tetramer in the ‘closed’ confirmation (30,31). This ‘closed’ confirmation might result in the necessary proteasomal interaction surface of ubiquitin being buried and prevent the chains from inhibiting destabilization. Thus the lysine-48-linked tetra ubiquitin would not inhibit destabilization when added to the assay in trans. Indeed, the buried surface patch of ubiquitin in the lysine 48-linked chains includes the ‘hydrophobic patch’ of ubiquitin, implicated as important in many interactions and functions of ubiquitin (32,33). The failure of the small-ubiquitin modifier, SUMO, to inhibit destabilization also supports this hypothesis. Although SUMO is structurally similar to ubiquitin, it lacks a clear hydrophobic patch that is present on ubiquitin monomers (34,35).
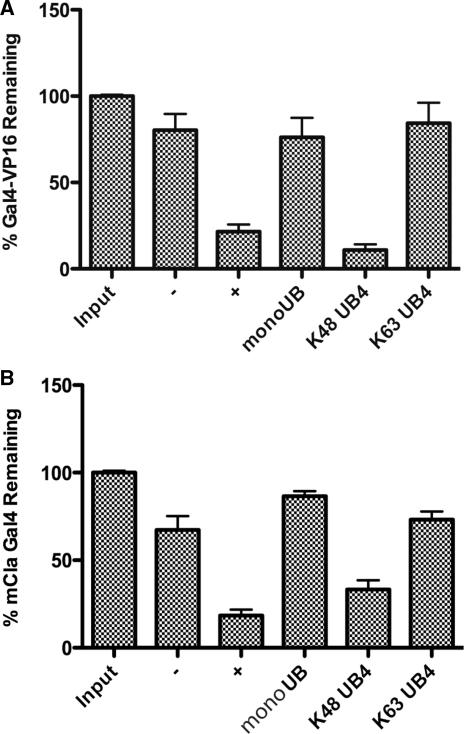
To test the importance of the hydrophobic patch, lysine-63-linked tetra ubiquitin was included in the assay. Ubiquitin chains that contain the lysine-63-linkage display a more open structure and the hydrophobic patch is solvent exposed and not buried (36). This allowed us to use the biochemical assay as a tool to quickly isolate a region of the ubiquitin that is required for inhibition of destabilization. The lysine-63-linked ubiquitin was just as effective at preventing destabilization of the Gal4-VP16 protein as monomeric ubiquitin (Figure 1A). As seen previously, the lysine 48-linked ubiquitin had no activity in preventing proteasomal-mediated destabilization of transactivator–DNA complexes. This result was also true when the assay was repeated with the mCla version of Gal4 (Figure 1B). The mCla Gal4 protein contains both the native DNA binding domain and AD of Gal4 but with the middle region removed to make a protein more soluble for in vitro studies. The mCla Gal4 protein induces transcription of the GAL genes and responds to the all of the endogenous signals in the same manner as full-length Gal4 (37). The ability of mono-ubiquitin to prevent destabilization of activator–DNA complexes depends on a solvent exposed hydrophobic patch.
Figure 1.
Ability of ubiquitin forms to inhibit proteasomal-mediated destabilization. (A) Destabilization of transactivator–DNA complexes by 26S proteasome in the presence of ubiquitin forms. Controls without (−) or with (+) proteasome are indicated. Reactions performed in the in the presence of 10 µM mono-Ub (lane 4), 10 µM K48 linked Ub4 (lane 5), or 10 µM K63 linked Ub4 (lane 6) are also indicated. The average amount and SEM of Gal4-VP16 retained by DNA was measured in three experiments by western blot and normalized with total protein input (lane 1) being set as 100%. (B) Same as (A), but the Gal4-VP16 protein was replaced with mCla Gal4 protein.
Mutation of the hydrophobic patch of ubiquitin reduces Rpt1 interaction
The ability of mono-ubiquitin or lysine-63-linked tetra-ubiquitin to inhibit destabilization but the inability of SUMO or lysine-48-linked tetra-ubiquitin suggests that the hydrophobic patch of ubiquitin is required. Mono-ubiquitin is known to interact with the Rpt1 and Rpn1 subunits in a manner that prevents the interaction of Gal4 with the Rpt4/Rpt6 subunits (19). These mutually exclusive proteasomal interactions are thought to be the reason behind the ability of mono-ubiquitin to inhibit proteasomal-mediated destabilization. If ubiquitin–proteasome interaction is inhibited by mutation of the hydrophobic patch this would strengthen the claim that the hydrophobic patch is required to inhibit the destabilization of transactivators by the proteasome.
To test this idea, a point mutation was made to disrupt the hydrophobic patch of ubiquitin at isoleucine-44. This mutation (or the control mutation, D58A) allowed for a competition experiment to test if the mutated form of ubiquitin would affect the ubiquitin/Rpt1 interaction. Previously, our lab had described a novel cross-linking and label transfer assay (26). This assay relies on a small tag on the protein of interest (in this case ubiquitin), which serves as a docking point for the cross-linking reagent containing a biotin tag. After cross-linking and label transfer, only direct-binding partners of the protein of interest become biotinylated. This assay was used to demonstrate that mono-ubiquitin interacted with the Rpt1 and Rpn1 subunits in the context of the 26S proteasome and was also able to interact with the bacterially expressed Rpt1 subunit (19).
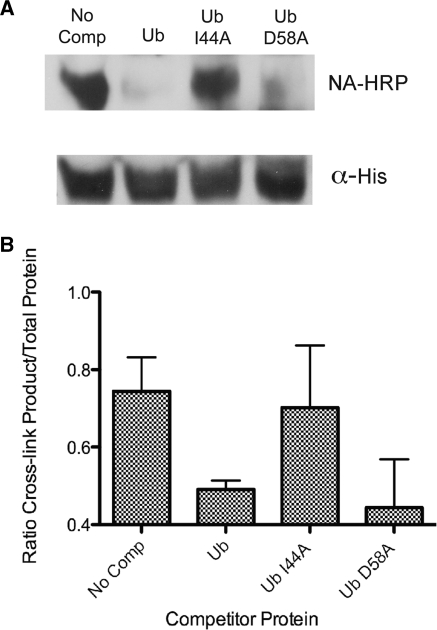
To test the mutated ubiquitin forms, the cross-linkable form of ubiquitin (CCPGCC-Ub) was mixed with bacterial lysate containing a hexahistidine tagged Rpt1 protein. After cross-linking and label transfer the samples were subject to IMAC to enrich for the Rpt1 protein and biotinylation was probed by blotting (Figure 2). Without addition of any competitor, the biotinylation of the Rpt1 protein was easily detected (Figure 2A, No Comp, graphed in Figure 2B) demonstrating the interaction between Rpt1 and monomeric ubiquitin is easily detected. Not surprisingly, addition of an excess of wild-type ubiquitin reduced the biotinylation of Rpt1 to almost undetectable levels. This effect was also seen when the competition cross-linking experiment was performed with Ub D58A. Both wild-type and the D58A versions of ubiquitin are able to out compete a tagged version of ubiquitin when added in excess. However, competition using Ub I44A did not decrease the interaction between CCPGCC-UB and Rpt1, as Rpt1 was strongly biotinylated. Analysis of multiple assays demonstrated that adding UB I44A was the same as having no competitor form of ubiquitin (Figure 2B). Ub I44A does not prevent the interaction of Rpt1 and wild-type, tagged ubiquitin most likely because its ability to interact with Rpt1 is disrupted by the mutation. This demonstrates the importance of the hydrophobic path of ubiquitin for Rpt1 interaction and further demonstrates the importance of this patch for ability of ubiquitin to inhibit proteasomal-mediated destabilization.
Figure 2.
Mutation of the hydrophobic patch of ubiquitin reduces Rpt1 interaction. (A) Biotinylated Rpt1 product (NA-HRP) present after cross-linking and label transfer between CCPGCC-ubiquitin and Rpt1 is shown. Competitor ubiquitin is indicated above each lane. Total His6-Rpt1 in the assay is shown by the α-His blot. (B) The average ratio and SEM of three experiments of cross-linked Rpt1 to total Rpt1 is graphed for each condition.
Mutation of I44 of ubiquitin abolishes the protective effect in vivo
The biochemical assays above demonstrate the importance of the hydrophobic path for interaction with the proteasomal subunit Rpt1 and in inhibition of proteasomal-mediated destabilization of transactivators. These biochemical assays served as a useful tool to implicate this region of ubiquitin. Further confirmation of the functional effects of these mutations requires in vivo experiments. To test the role of the hydrophobic patch of ubiquitin in a cellular environment, the same two point mutations were made in ubiquitin in the context of the ubiquitin–Gal4D fusion proteins. The Gal4D version of Gal4 is missing a small portion of the AD. The Gal4D protein is not effectively mono-ubiquitylated and this leads to a defect in DNA occupancy due to susceptibility to proteasomal-mediated destabilization (20). However, the portion of the AD required for transcriptional activation remains. This partial truncation of Gal4 provides a useful system of to study the effect of mono-ubiquitylation. Fusion of ubiquitin to Gal4D was previously shown to partially restore activity and DNA occupancy of the Gal4D protein. Would the hydrophobic patch mutation prevent the rescue of Gal4D by ubiquitin?
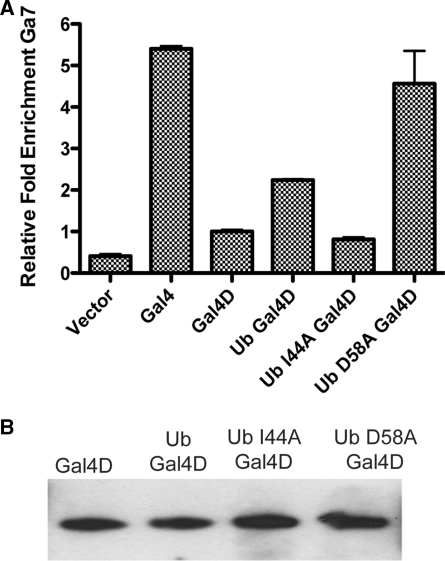
DNA occupancy of the fusion protein was checked by ChIP using antibodies directed against the N-terminus of the Gal4 protein. Occupancy of the fusion protein on the promoter of the Gal4 responsive genes GAL1-10 (Supplementary Figure S2) and GAL7 (Figure 3A) were measured. On both promoters, the isoleucine-44 to alanine mutation resulted in a loss of the ability of ubiquitin to promote occupancy of Gal4D, as measured by qPCR following ChIP. Mutation of another region on the surface of ubiquitin (D58A) did not cause a decrease in occupancy and resulted in slightly higher occupancy. The hydrophobic patch of ubiquitin is required for the rescue of DNA occupancy of a partial truncation of Gal4.
Figure 3.
Mutation of the hydrophobic patch of ubiquitin prevents rescue of Gal4D. (A) A ChIP assay was used to monitor the DNA occupancy of the different Gal4 constructs. The relative fold enrichment of GAL7 promoter DNA is indicated for the Gal4D construct indicated below each bar with the Gal4D construct set as 1. (B) Expression levels of Gal4D constructs. The steady-state level of the indicated protein was measured by western blot with an antibody raised against Gal4.
The expression level of Gal4D is known to alter the activity of the construct. Massive over expression in the protein will result in a partial recovery of the activity of Gal4D presumably due to forced occupancy of the promoter driven by the over expression (38). To be certain that the changes seen in DNA occupancy were not due to expression level, the steady-state level of the ubiqutin–Gal4D fusions were compared to Gal4D (Figure 3B). There was little difference in protein levels seen in the cellular lysate of the different constructs. Thus, the difference in occupancy measured in panel A cannot be solely due to differences in expression levels and must be a result of the ubiquitin mutations.
Mutation of I44 of ubiquitin reduces the activity of ubiquitin–Gal4D
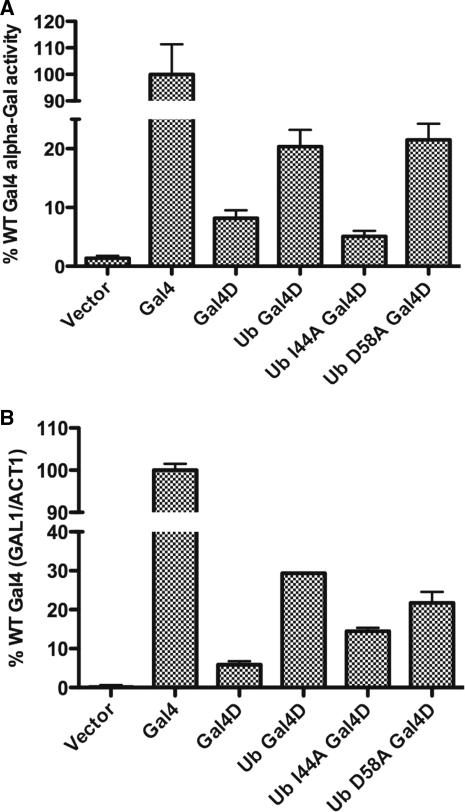
The activity of the ubiquitin–Gal4D mutant constructs was monitored to determine if the transcriptional output correlated with the occupancy levels seen above. If true, then the mutations of ubiquitin are most likely only affecting DNA occupancy because of changes in inhibition of proteasomal-mediated destabilization and not causing changes in transcription activity to the Gal4 protein. An enzyme assay was used to monitor the production α-galactosidase, the gene product of the Gal4 responsive MEL1 gene (Figure 4A). Adding ubiquitin to the N-terminus of Gal4D (Ub Gal4D) increased enzymatic activity by more than 2-fold compared with Gal4D. Mutation of the hydrophobic patch (I44A) decreased activity levels of the Ub-Gal4D constructs to levels similar to Gal4D lacking the ubiquitin fusion. Mutation of other residues (D58A) did not decrease the activity of the Ub-Gal4D constructs.
Figure 4.
The I44A mutation abolishes the ability of ubiquitin to promote Gal4D activity. (A) An α-galactosidase assay measuring the MEL1 gene product produced from the indicated constructs. The average and SEM of three experiments are shown normalized to the levels produced with wild-type Gal4. (B) The relative ratio of GAL1 and ACT1 mRNA is graphed based on qPCR quantization of changes in Ct values from strains expressing the indicated Gal4 form. The average and SEM of three experiments are shown normalized to the levels of GAL1 produced with wild-type Gal4.
As a second measure of activity, the mRNA produced from the GAL1 gene was monitored by quantitative PCR (Figure 4B and Supplementary Figure S3). The ability of ubiquitin to promote activity of the Gal4D protein was decreased by mutation of the hydrophobic patch of ubiquitin (I44A). Mutation of other residues of ubiquitin had little effect on the activity of the Ub-Gal4D fusion protein (D58A). Both assays used to measure activity demonstrated that the hydrophobic patch of ubiquitin is required to promote transcriptional activation of galactose responsive genes.
DISCUSSION
In this study we set out to find a possible explanation for the differences between mono-ubiquitin and lysine-48-linked poly-ubiquitin chains to inhibit the destabilization activity of the proteasome. Monomeric or completely solvent exposed forms of ubiquitin chains (lysine-63-linked) protected activator–DNA complexes from destabilization, but tightly packed or ‘closed’ forms (lysine-48-linked) did not (Figure 1). This biochemical assay suggested that the hydrophobic patch, buried in the closed form, might be important for the ubiquitin/proteasome interaction. We then used mutational studies of the hydrophobic patch of mono-ubiquitin in vitro and in vivo as a more physiological relevant look at the importance of the hydrophobic patch.
Mutation of the hydrophobic patch produced a form of ubiquitin that was no longer able to bind to the Rpt1 protein (Figure 2). In vivo, mutation of the hydrophobic patch of ubiquitin prevented partial rescue of the occupancy of the Gal4D protein on the promoters of galactose responsive genes (Figure 3), which correlated with the inability of a mutated hydrophobic patch to restore activity to the protein (Figure 4). Mutation of other regions on the surface of ubiquitin did not cause any decrease in the activity of ubiquitin. We conclude that the hydrophobic patch of ubiquitin is required to inhibit the destabilization activity of the proteasomal ATPases on transactivator-DNA complexes.
The hydrophobic patch, made of up L8, I44 and V70 residues, has long been known to be an important region of the surface to ubiquitin. Mutation of these residues is known to disrupt proteolysis and endocytosis of target proteins (32,33). There are now several structural studies that suggest that many of the ubiquitin binding proteins interact directly with the hydrophobic patch of ubiquitin (39,40). The ability of point mutations to the hydrophobic patch to alter the activity of ubiquitin suggests that these structural studies are physiological relevant and the hydrophobic patch is an interaction region for ubiquitin binding proteins.
Mutation of this patch resulted in a form of ubiquitin that cannot compete for Rpt1 binding in vitro. In addition, mutation of the hydrophobic patch, but not other regions on the surface of ubiquitin, would also disrupt the protective effect of a monomeric ubiquitin in vivo. The fact that mutation would disrupt the activity of monomeric forms of ubiquitin suggests that the effect requires direct interaction with an intact hydrophobic patch.
However, a final verdict on the role of the hydrophobic patch of ubiquitin in proteasomal-mediated destabilization will require more in-depth structural studies. Our previous cross-linking strategies can provide information about potential interacting partners within the RP of the proteasome and implicated Rpn1 and Rpt1 as binding partners. The cross-linking studies provided no information about potential surfaces or residues on the surface of ubiquitin that were important for this interaction. The comparisons between different forms of ubiquitin chains in vitro and mutational data from in vivo studies seen here clearly demonstrate that the hydrophobic patch of ubiquitin is important for the inhibition of destabilization.
This study also further defines the connection between two non-proteolytic roles of the proteasomal-ATPases, proteasomal-mediated destabilization of transactivators (18,20) and promoting efficient transition into an elongation complex (11,12). Both activities required an exposed AD to recruit the proteasomal ATPases. If the transactivator is not protected by mono-ubiquitylation or, as this study shows lacks a hydrophobic patch, then the activator is stripped from the DNA. However, the presence of the hydrophobic patch of mono-ubiquitin allows interaction with the proteasome through Rpt1 and Rpn1. This interaction protects the transactivator and allows the proteasomal ATPases to transition into their role of promoting transcription elongation. The exposed hydrophobic patch acts as an important physical feature determining which of those two non-proteolytic activities of the proteasomal ATPase will dominate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM 087283]. Funding for open access charge: National Institutes of Health [GM 087283].
Conflicts of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Prof George DeMartino (UTSW) for kindly providing the bacterially expressed Rpt1 constructs.
REFERENCES
- 1.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 2.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 3.Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 6.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl Acad. Sci. USA. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Sikder D, Johnston SA, Kodadek T. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J. Biol. Chem. 2006;281:27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- 9.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 11.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatroy particle of the proteasome is required for efficient transcriiption elongation by RNZ Polymerase II. Mol. Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 12.Ferdous A, Kodadek T, Johnston SA. A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA Polymerase II. Biochemistry. 2002;41:12798–12805. doi: 10.1021/bi020425t. [DOI] [PubMed] [Google Scholar]
- 13.Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Bhat KP, Turner JD, Myers SE, Cape AD, Ting JP, Greer SF. The 19S proteasome ATPase Sug1 plays a critical role in regulating MHC class II transcription. Mol. Immunol. 2008;45:2214–2224. doi: 10.1016/j.molimm.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Sulahian R, Sikder D, Johnston SA, Kodadek T. The proteasomal ATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 2006;34:1351–1357. doi: 10.1093/nar/gkl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archer CT, Burdine L, Kodadek T. Identification of Gal4 activation domain-binding proteins in the 26S proteasome by periodate-triggered cross-linking. Mol. Biosyst. 2005;1:366–372. doi: 10.1039/b510019d. [DOI] [PubMed] [Google Scholar]
- 17.Fernando Gonzalez AD, Thomas K, Stephen AJ. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 18.Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, Johnston SA. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 2007;21:112–123. doi: 10.1101/gad.1493207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archer CT, Burdine L, Liu B, Ferdous A, Johnston SA, Kodadek T. Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J. Biol. Chem. 2008;283:21789–21798. doi: 10.1074/jbc.M803075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archer CT, Delahodde A, Gonzalez F, Johnston SA, Kodadek T. Activation domain-dependent monoubiquitylation of Gal4 protein is essential for promoter binding in vivo. J. Biol. Chem. 2008;283:12614–12623. doi: 10.1074/jbc.M801050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M. Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26 S protease subunit S5a. J. Biol. Chem. 1995;270:29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- 22.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 24.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Archer CT, Burdine L, Gillette TG, Kodadek T. Label transfer chemistry for the characterization of protein-protein interactions. J. Amer. Chem. Soc. 2007;129:12348–12349. doi: 10.1021/ja072904r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell SJ, Johnston SA. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J. Biol. Chem. 2001;276:9825–9831. doi: 10.1074/jbc.M010889200. [DOI] [PubMed] [Google Scholar]
- 28.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Kew OM, Douglas HC. Genetic co-regulation of galactose and melibiose utilization in Saccharomyces. J. Bacteriol. 1976;125:33–41. doi: 10.1128/jb.125.1.33-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J. Mol. Biol. 1994;236:601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 31.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 32.Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- 33.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 34.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 35.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 36.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 37.Ding WV, Johnston SA. The DNA binding and activation domains of Gal4p are sufficient for conveying its regulatory signals. Mol. Cell. Biol. 1997;17:2538–2549. doi: 10.1128/mcb.17.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto K, Adachi Y, Toh-e A, Oshima Y. Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein. J. Bacteriol. 1980;141:508–527. doi: 10.1128/jb.141.2.508-527.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nature Struct. Mol. Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.