Abstract
The regulation of DNA repair enzymes is crucial for cancer prevention, initiation, and therapy. We have studied the effect of ultraviolet B (UVB) radiation on the expression of the two nucleotide excision repair factors (XPC and XPD) in human keratinocytes. We show that hypoxia-inducible factor-1α (HIF-1α) is involved in the regulation of XPC and XPD. Early UVB-induced downregulation of HIF-1α increased XPC mRNA expression due to competition between HIF-1α and Sp1 for their overlapping binding sites. Late UVB-induced enhanced phosphorylation of HIF-1α protein upregulated XPC mRNA expression by direct binding to a separate hypoxia response element (HRE) in the XPC promoter region. HIF-1α also regulated XPD expression by binding to a region of seven overlapping HREs in its promoter. Quantitative chromatin immunoprecipitation assays further revealed putative HREs in the genes encoding other DNA repair proteins (XPB, XPG, CSA and CSB), suggesting that HIF-1α is a key regulator of the DNA repair machinery. Analysis of the repair kinetics of 6-4 photoproducts and cyclobutane pyrimidine dimers also revealed that HIF-1α downregulation led to an increased rate of immediate removal of both photolesions but attenuated their late removal following UVB irradiation, indicating the functional effects of HIF-1α in the repair of UVB-induced DNA damage.
INTRODUCTION
Solar ultraviolet B (UVB) radiation is the primary environmental risk factor responsible for induction of non-melanoma skin cancer which includes basal cell carcinomas and squamous cell carcinomas, the most common types of human malignancy worldwide. A major deleterious effect of UVB is the induction of well-defined structural alterations in DNA (1). UVB-induced DNA damage sets in motion a highly complex well-coordinated series of responses whereby DNA damage and stalled replication forks can be detected. This in turn can trigger DNA repair, cell cycle delay or apoptosis (2). The ultimate fate of cells with damaged DNA is dependent on the type and extent of damage, DNA repair capacity and UVB-induced apoptotic signaling pathways (3,4). Understanding the interplay between various factors involved in the regulation of cellular responses to UVB could enhance current knowledge regarding cancer prevention, initiation, and therapy.
Hypoxia-inducible factor-1 (HIF-1) is the key transcription factor induced by hypoxic conditions and additionally responds to other cellular stresses under normoxic conditions (5,6). HIF-1 is a heterodimeric protein consisting of two α and β subunits (7). In normoxia, HIF-1α is rapidly targeted for ubiquitination and proteasomal degradation after its hydroxylation by prolyl-hydroxylases (PHDs) (8–11). PHD activity decreases under hypoxic conditions, resulting in the stabilization and accumulation of HIF-1α. When stabilized, HIF-1α translocates to the nucleus, binds to the hypoxia response element (HRE) of target genes and participates in the regulation of numerous genes involved in angiogenesis, glycolysis, apoptosis, migration and metastasis (8,12,13). We and others have shown that HIF-1α expression is modulated after UVB exposure and plays an important role in the regulation of cellular responses to this type of genotoxic stress (14–16). UVB induces reactive oxygen species (ROS), which in turn have a biphasic effect on HIF-1α expression. Whereas rapidly produced cytoplasmic ROS downregulate HIF-1α expression, delayed mitochondrial ROS result in its upregulation (14). Furthermore, we have shown that the pro-apoptotic effect of HIF-1α in irradiated cells is accompanied by p53 modulation (14), suggesting a functional link between UVB-induced ROS production, HIF-1α variation and DNA repair.
Among DNA repair systems, nucleotide excision repair (NER) is the major pathway for repairing numerous types of damage induced by UVB irradiation, including cyclobutane pyrimidine dimers (CPD), 6-4 photoproducts (6-4PP) (17,18), and some ROS-induced damage (18). NER proceeds through two distinct yet overlapping pathways: transcription-coupled repair, which specifically removes lesions from the transcribed strand of active genes, and global genome repair (GGR), which removes lesions throughout the genome (19). In GGR, XPC recognizes structural DNA abnormalities associated with damaged bases. Then, TFIIH (including XPB, XPD and several other subunits), most likely together with XPG, XPA and RPA, unwinds the DNA helix through its DNA helicase activity. After incisions have been made on both sides of the lesion by XPF-ERCC1 and XPG, the oligonucleotide containing the damaged base(s) is released. Finally, the gapped DNA region is restored by a DNA polymerase and DNA ligase (17,20). Defective NER is associated with several human diseases, such as xeroderma pigmentosum, which has a high incidence of skin cancers, Cockayne syndrome and trichothiodystrophy (21). Furthermore, genetic studies reveal a strong association between NER gene polymorphisms and the occurrence of many types of cancer (22).
Despite the functional and clinical importance of NER in maintaining genetic stability, little is known about the regulatory mechanisms underlying its expression. As mentioned above, our previous data suggest a link between UV exposure, HIF-1α and DNA repair (14). This raises an important question: how does modulation of HIF-1α regulate DNA repair? Several clues point towards a possible answer. First, UV irradiation can induce NER factors in cells (23–26). Second, UV irradiation modulates the expression of HIF-1α, a transcription factor (14–16). Third, software analysis reveals that there are multiple potential HREs in the promoter regions of these two important NER enzymes, XPC and XPD. Here, we tested the hypothesis that HIF-1α plays a critical role in the transcriptional regulation of XPC and XPD expression during UV-induced DNA repair by binding directly to the HREs in their promoter regions.
MATERIALS AND METHODS
Source of keratinocytes and irradiation procedure
Keratinocytes were isolated from normal human skin in patients undergoing plastic surgery, grown in MCDB153 medium, and irradiated at a dose of 200 mJ/cm2 using a Biotronic device (Vilber Lourmat, Marne la Vallée, France) equipped with a dosimeter. The UVB lamp emitted a continuous spectrum between 280 and 380 nm (major peak at 312 nm) (27).
Lentiviral vector constructs and keratinocyte transduction
The lentivector used for transduction (THshHIF1W) has been previously described (14). Briefly, TPW plasmid was used as the backbone for the construction of the THshHIFW by replacing hPGK with a cassette containing the H1 promoter followed by a shRNA sequence (5′-GATGTTAGCTCCCTATATCCC-3′) that targeted the HIF-1α mRNA. Production of lentiviral particles, determination of the titer of viral supernatant and keratinocyte transduction were performed as previously described (28).
Western blotting procedure
Western blotting was performed as previously described (29). Briefly, equal amounts of total protein were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrophoretically transferred to PVDF membranes. Membranes were then incubated overnight at 4°C with a 1:1000 dilution of the anti-XPD (GeneTex, Euromedex, Mundolsheim, France), anti-XPC (GeneTex) or anti-HIF-1α (BD Transduction Laboratories, Le Pont de Claix, France) antibodies. After additional incubation with 1:10,000 dilution of an anti-immunoglobin horseradish peroxidase-linked antibody (Vector Laboratories, Biovalley S.A, Marne la Vallée, France) for 1 h, blots were developed using the chemiluminescence ECL reagent (Amersham Bioscience, Saclay, France).
Quantitative real-time polymerase chain reaction
Total cellular RNA was extracted using TRIzol® (Invitrogen, Cergy-Pontoise, France) according to the manufacturer’s instructions. Total cellular RNA (2 µg) was reverse transcribed at 42°C for 60 min using the 1st Strand cDNA synthesis kit (Roche, Meylan, France). Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out for XPC, XPD and Tubulin using the SYBR Green method with an ABI instrument. Primer sequences are listed in Supplementary Table S1. The reactions were cycled 40 times after initial polymerase activation (50°C, 2 min) and initial denaturation (95°C, 15 min) using the following parameters: denaturation at 95°C for 15 s; annealing and extension at 60°C for 1 min. A final fusion cycle (95°C, 30 s; 60°C, 30 s; 95°C, 30 s) terminated these reactions. The standard curve demonstrated a linear relationship between the cycle threshold (Ct) values and the cDNA concentration. The relative expression of each gene was assessed by considering the Ct and efficiency values, and normalized according to tubulin expression level.
Nuclear extraction preparation and gel shift assay
Nuclear proteins were prepared using the NE-PER® nuclear and cytoplasmic extraction kit (Pierce Biotechnology, Rockford, IL, USA) in accordance with the manufacturer’s instructions. Complementary oligonucleotides were annealed to make double-stranded probes, which were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase (Promega, Madison, WI, USA). The radiolabeled probes were incubated with or without nuclear extract at 22°C for 20 min, and the samples were then electrophoresed on 8% non-denaturing polyacrylamide gel. The gel was subjected to autoradiography at –80°C. For the competition assay, nuclear extract was incubated with the anti-HIF-1α (Novus Biologicals, Littleton, CO, USA) or anti-Sp1 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) or 3.5 pmol (100 times excess) of unlabeled competition oligonucleotides for 30 min prior to adding the [32P]-labeled probe.
Chromatin immunoprecipitation assay
The assay was performed using the EZ ChIPTM Chromatin Immunoprecipitation Kit (Upstate Biotech, Waltham, MA, USA) in accordance with the manufacturer’s instructions. Briefly, cells were fixed with 1% formaldehyde for 10 min at 37°C, and nuclear extracts were obtained and sonicated to produce DNA of ∼500 bp. Protein–DNA complexes were immunoprecipitated with anti-HIF-1α antibody (Novus NB 100-105) or normal Rabbit IgG (negative control). The immunoprecipitated DNA was then purified and eluted with 50 μl of elution buffer. PCR amplification was done using 2 μl of DNA sample with different sets of primers (Supplementary Table S2). Amplification of soluble chromatin prior to immunoprecipitation was used as an input positive control. qRT-PCR was carried out on eluted DNA with the same primer sets, using Super Array RT2 SYBR Green (Biomol, TEBU, Le Perray en Yvelines, France) according to the manufacturer’s instructions with the AB biosystem machine.
Plasmid constructs and luciferase assays
The regions of XPC and XPD promoter containing HREs were amplified from human genomic DNA by PCR using primers (Supplementary Table S3) and cloned into pGEM-7 (Promega), in which an SV40 minimal promoters with the following sequence had already been inserted: 5′-ctatgcatagagggtatataatggaagctcgacttccag-3′. The integrity of the corresponding sequences was checked by sequencing and the cassette (HRE/SV40 minimal promoter) was further cloned into pGL4.10-Basic (Promega), which encodes firefly luciferase. The C-HRE3 containing the basic promoter region of XPC promoter was inserted into pGL4.10 without SV40 minimal promoter. The mutations in C-HRE3 were generated in pGL4.10 containing wild-type C-HRE3, using a site-directed mutagenesis kit (Promega) according to the manufacturer’s instructions. The primer used to mutate the HRE site was as follows: 5′-caacgaaggggcgGggccaagc-3′, in which the HRE is marked in bold italics and the mutated base is marked in bold uppercase italics.
Keratinocytes were transduced by shHIF-1α 24 h after seeding on 96-well plates (2 × 104 cells per well). One day after transduction, the keratinocytes were co-transfected with 32 ng of promoter-pGL4 reporter plasmid and 16 ng of control Renilla plasmid using Fugene-6 reagent (Roche). The latter plasmid was used to normalize transfection efficiency. Keratinocytes were exposed to UVB 48 h later and then harvested at different time intervals by adding 20 µl of luciferase lysis buffer (Promega). The cell lysates were kept at –80°C and then subjected to luciferase assays using the Dual-GloTM Luciferase Assay System (Promega) in accordance with the manufacturer’s instructions. The firefly luciferase activity expressed in each well was normalized by the Renilla luciferase activity. For all transfection assays, at least three independent experiments were performed in triplicate.
Determination of 6-4PP and CPD repair kinetics
Exponentially growing asynchronous keratinocytes were exposed to UVB. At different time intervals after irradiation, cells were fixed with 4% formaldehyde for 10 min at room temperature and then permeabilized overnight in cold 70% ethanol. Cells were then resuspended in either 0.5% Triton-X-100/2N HCl (for CPD detection) or 0.5% Triton-X-100/0.2N HCl (for 6-4PP detection) for 10 min at room temperature. After washing with Tris-base 1M (pH 10) and then with PBS, keratinocytes were incubated overnight at 4°C with 100 µl PBS-TF (4% FBS/0.25% Tween-20/PBS) containing a 1:100 dilution of either the anti-6-4PP (Kamiya Biomedical, Seattle,WA, USA) or anti-CPD (Kamiya Biomedical) antibodies. After washing twice with PBS, cells were resuspended in 100 µl PBS-TF containing Alexa- Fluor 488-coupled secondary antibody (1 : 200) for 1 h at room temperature. Repair kinetics was then monitored using flow cytometry (FACSalibur, Becton Dickinson) to quantify the change in geometric mean fluorescence over time.
Statistics
Student’s t-test was applied for statistical evaluation and a P-value < 0.05 was considered significant. Results are presented as means ± SD.
Results
XPC expression in human keratinocytes is sensitive to HIF-1α modulation
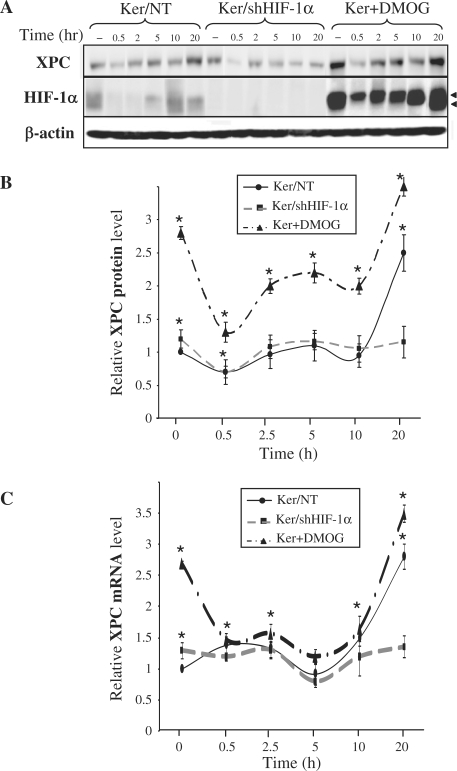
We have previously demonstrated that exposure of keratinocytes to UVB induces a biphasic effect on HIF-1α expression, including a dramatic decrease in HIF-1α protein immediately after irradiation, followed by a gradual increase of 2.5–10 h post-irradiation [(14), Figure 1A]. We wanted to determine whether, as a transcription factor, HIF-1α influences the kinetics of XPC expression following irradiation. To this end, we investigated XPC expression in keratinocytes in which HIF-1α is either downregulated using shRNA lentiviral transfer or conversely stabilized by incubation with dimethyloxaloylglycine (DMOG), a specific inhibitor of PHDs which blocks hydroxylation, and therefore subsequent degradation, of HIF-1α under normoxic conditions. Following exposure of non-transduced keratinocytes to UVB irradiation, there was an immediate decrease in XPC protein level followed by a gradual increase which peaked at 20 h (Figure 1A and B). Interestingly, the late upsurge could be due to an increased transcription of the XPC messenger, whereas the earlier protein drop occurred despite increased mRNA transcription (Figure 1C). Compared to non-transduced cells, the baseline XPC protein expression was higher in the cells silenced for HIF-1α (1.2-fold) and in the cells incubated with DMOG (2.8-fold) (Figure 1A and B). This observation at the protein level correlated with an increase in XPC mRNA (Figure 1C). Furthermore, XPC expression following UVB irradiation varied depending on whether HIF-1α was present or absent. Indeed, in HIF-1α-downregulated keratinocytes, the primary decrease in XPC protein expression was still noticeable, whereas the late XPC increase observed in non-transduced cells was abrogated (Figure 1B). This observation correlated with the absence of late increased XPC mRNA (Figure 1C). In DMOG-treated keratinocytes, the accumulated HIF-1α was associated with enhanced late XPC protein and mRNA expression levels, although there was certainly a saturation effect as observed by the small difference with the non-irradiated cells (Figure 1B and C). Taken together, our results show that HIF-1α downregulation led to increased XPC expression prior to irradiation, but inhibited its late increase of XPC following irradiation. On the contrary, HIF-1α upregulation resulted in a significant increase in XPC expression levels. These data strongly suggest that HIF-1α is actively involved in XPC regulation.
Figure 1.
Effect of UVB on XPC expression. Keratinocytes were harvested at the indicated time points after irradiation. (A) Total protein extracts were assessed for the presence of the XPC and HIF-1α protein by western blotting. β-actin was used as a loading control. Arrowheads indicate the two HIF-1α forms [see also ref. (14)]. (B) The band intensities of XPC proteins were quantified densitometrically. (C) The relative level of XPC mRNA was determined by qRT-PCR. The results are expressed as the mean ± SD of three independent experiments. –, no UVB exposure; Ker/NT, non-transduced keratinocytes; Ker/shHIF-1α, keratinocytes transduced with shHIF-1α; Ker + DMOG, keratinocytes treated with dimethyloxaloylglycine. *P < 0.05 for cells at the indicated time point versus non-transduced cells prior to irradiation.
Study of the XPC promoter reveals hypoxia response elements with multiple functions
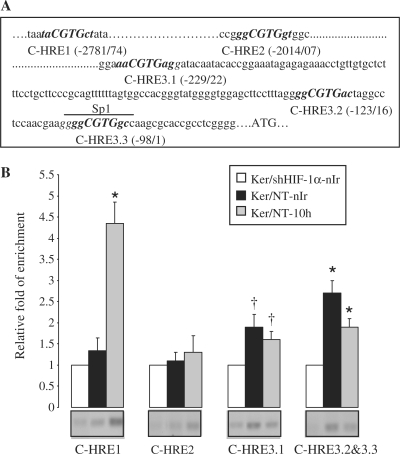
HREs are composite regulatory elements comprising the conserved HIF-binding site with an (A/G)CGTG core sequence and highly variable flanking sequences (30). To determine whether HIF-1α can bind directly to the promoter region of XPC, we searched for the consensus HRE in the human XPC promoter. Five candidate HREs were found within 3 kb upstream from the ATG translation initiation codon (Figure 2A). To analyze binding of HIF-1α to these HREs, we performed ChIP experiments. As shown in Figure 2B, HREs complexed with HIF-1α were immunoprecipitated with anti-HIF-1α antibody and detected by PCR amplification with the primers spanning C-HRE1, C-HRE3.1 and C-HRE3.2/3.3. Interestingly, while the regions containing C-HRE3.1 and C-HRE3.2/3.3 immunoprecipitated in both non-irradiated and irradiated cells, immunoprecipitation of C-HRE1 was substantially increased in the UVB-irradiated cells (Figure 2B).
Figure 2.
Differential binding of HIF-1α on XPC promoter in non-irradiated and irradiated keratinocytes. (A) Five-nucleotides sequences matching the consensus HRE [(A/G)CGTG, marked in bold] are present in the 3 kb upstream region of human XPC gene, here referred to as C-HRE1, -HRE2, -HRE3.1, -HRE3.2 and -HRE3.3. The nucleotide sequences were numbered in relation to the translational start codon, ATG. The upperlined sequence represents the Sp1 binding site sharing a sequence in common with C-HRE3.3. (B) Ten hours after UVB-exposure, irradiated and non-irradiated keratinocytes were subjected to ChIP assay using an anti-HIF-1α antibody. Bands indicate PCR products using primers that span the indicated HREs. The sets of primers used are indicated in Supplementary Table S2. The relative levels of corresponding precipitated HRE fragments following ChIP were quantified by qRT-PCR. PCR amplification in shHIF-1α-transduced cells detected a minimal level of HREs which was normalized to one for each experiment. Ker/NT-10 h, non-transduced keratinocytes harvested 10 h after UVB exposure; nIr, non-irradiated keratinocytes. The results are expressed as the mean ± SD of three independent experiments. *P < 0.01 and †P < 0.05 for each HRE versus shHIF-1α−transduced keratinocytes.
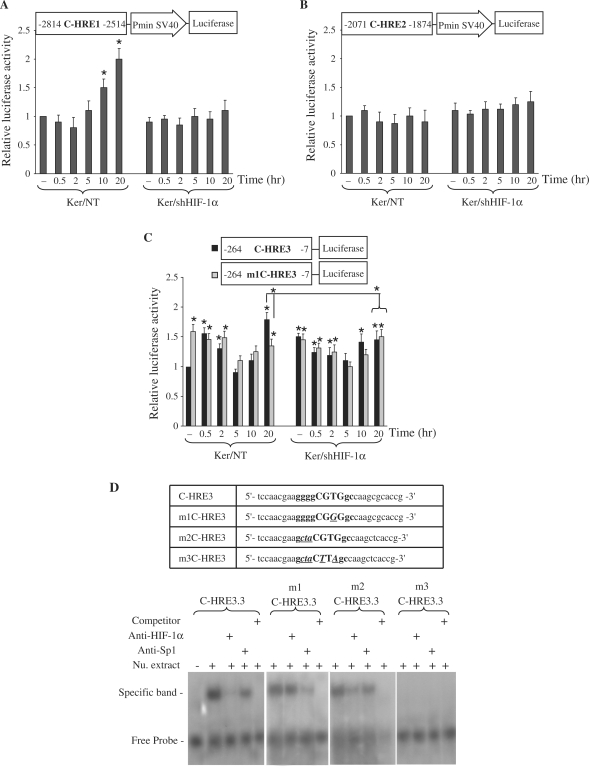
To further characterize the role of HREs in the regulation of XPC expression following UVB-irradiation, luciferase reporter plasmid was used. Sequences spanning HRE have been shown to play a pivotal role in forming a fully functional HRE, modulating the transcriptional response, and conferring tissue specificity (30). Therefore, fragments containing each HRE and its flanking region were cloned into a luciferase reporter plasmid. These constructs were transiently transfected into non-transduced or shHIF-1α-transduced cells, and their luciferase activities determined at different time intervals following irradiation (Figure 3). Cells transfected by C-HRE1-luciferase revealed an increase in luciferase activities 10 h after UVB-irradiation, compared with non-irradiated cells. This was not observed in the shHIF-1α-transduced keratinocytes transfected with C-HRE1-luciferase (Figure 3A). Results from cells transfected by C-HRE2-luciferase indicated that luciferase activities were not affected by UVB-irradiation or by down regulation of HIF-1α (Figure 3B). Assessment of luciferase activities in cells transfected by C-HRE3-luciferase showed that downregulation of HIF-1α resulted in increased basal levels of luciferase activity (Figure 3C). Furthermore, following irradiation of non-transduced cells transfected by C-HRE3-luciferase, there was an immediate moderate increase in luciferase activity levels, followed by a gradual decrease and a 2-fold increase 20 h post-irradiation. Interestingly, both immediate and late increases were abrogated in cells transduced by shHIF-1α (Figure 3C). Taken together, these results suggest that the C-HRE1 sequence exclusively participates in the late UVB response, whereas the C-HRE3 one involved in a more complex regulation of XPC prior to and after irradiation.
Figure 3.
Functional analysis of putative HREs present in the XPC promoter. Luciferase reporter gene constructs containing C-HRE1 (A), C-HRE2 (B) or C-HRE3 (C) fragments were transfected into non-transduced or shHIF-1α-transduced keratinocytes. Cells were harvested at indicated time points after UVB irradiation. Firefly luciferase activity was normalized to Renilla luciferase activity. The luciferase assays were done in triplicate. For each reporter, the mean ± SD luciferase activity is shown (n = 3) relative to the activity in non-transduced cells prior to irradiation. The schematic representation of the plasmid construct used in each experiment is shown at the top of each diagram. *P < 0.05 for cells at the indicated time point versus NT cells prior to irradiation. (D) EMSA shows that HIF-1α- and Sp1-binding sites on C-HRE3 in XPC promoter are functional. The oligonucleotide sequences used in EMSA are shown in the top panel. C-HRE3 corresponds to the wild-type probe, m1C-HRE3 contains a mutation in the HIF-1α binding site, m2C-HRE3 is a probe mutated in the Sp1-binding site, and m3C-HRE3 contains mutations in both sites. Uppercase letters indicate the core of the HIF-1α binding site, and bold letters indicate the Sp1 binding site that shares a common sequence with C-HRE3.3. Mutated bases are indicated with bold, italics and underlining. Nu. extract, nuclear extract.
HIF-1α competes with Sp1 for C-HRE3 in XPC promoter
An increase in both XPC mRNA and protein levels was observed in cells knocked down for HIF-1α (Figure 1A–C). In addition, XPC mRNA and luciferase activity levels were increased immediately after UVB-irradiation (0.5 and 2 h) in non-transduced cells, while a drop in HIF-1α expression was observed at these time points (Figures 1C and 3C). Therefore, we wondered whether HIF-1α could compete with other transcription factors in the region containing C-HRE3. Analysis of the C-HRE3 region demonstrated that the HIF-1α-binding site (5′-ggCGTGgc-3′) and the Sp1 binding site (5′-ggggCGTGgc-3′) share overlapping bases (Figure 2A). The consensus sequence of Sp1-binding site is [5′-(G/T)(G/A)GGCG(G/T)(G/A)(G/A)(C/T)-3′]. To assess the binding of HIF-1α and Sp1, ESMA was performed using wild and mutated oligonucleotides (Figure 3D). Incubation of a 32P-labeled wild-type probe (C-HRE3) with nuclear extracts of non-irradiated keratinocytes resulted in a shifted protein–DNA complex band (Figure 3D, lane 2). The addition of anti-HIF-1α and, to a lesser extent, anti-Sp1 antibody decreased the amount of shifted band (Figure 3D, lanes 3 and 4, respectively), while addition of excessive unlabeled probes abolished the shifted nuclear protein–DNA band (Figure 3D, lane 5), indicating the existence of HIF-1α and Sp1 binding sites on this oligonucleotide. Incubation of nuclear extracts with a labeled probe containing a mutation in the HIF-1α binding site (m1C-HRE3) formed a shifted protein–DNA complex, which was reduced by the addition of anti-Sp1 but not by anti-HIF-1α antibody. On the other hand, the addition of anti-HIF-1α antibody to a mixture of nuclear extracts and labeled oligonucleotides containing the mutation in Sp1 binding site (m2C-HRE3) led to decreased band shifting. Finally, incubation of an oligonucleotide with mutations in both HIF-1α and Sp1 binding sites (m3C-HRE3) with nuclear extracts failed to form a protein–DNA band, supporting the idea that C-HRE3 contains HIF-1α and Sp1 binding sites.
To investigate the role of Sp1 in XPC expression, a second C-HRE3-luciferase construct was made by substituting the T nucleotide in the HIF-1α binding site for a G nucleotide designated m1C-HRE3. In non-transduced cells transfected by this new construct, the basal level of luciferase activity (non-irradiated cells) was 1.5 times higher compared with the level measured in cells transfected by normal C-HRE3-luciferase plasmid (Figure 3C). The luciferase activity levels were maintained at the same level and did not increase at 20 h, as did normal C-HRE3-luciferase-transfected cells. These results suggest that HIF-1α binding to C-HRE3 in non-irradiated cells reduces the attachment of Sp1 to the XPC promoter. The immediate downregulation of HIF-1α after irradiation (0.5 h) allows Sp1 binding to the XPC promoter, leading to the initial increase in XPC mRNA expression.
HIF-1α positively regulates XPD expression
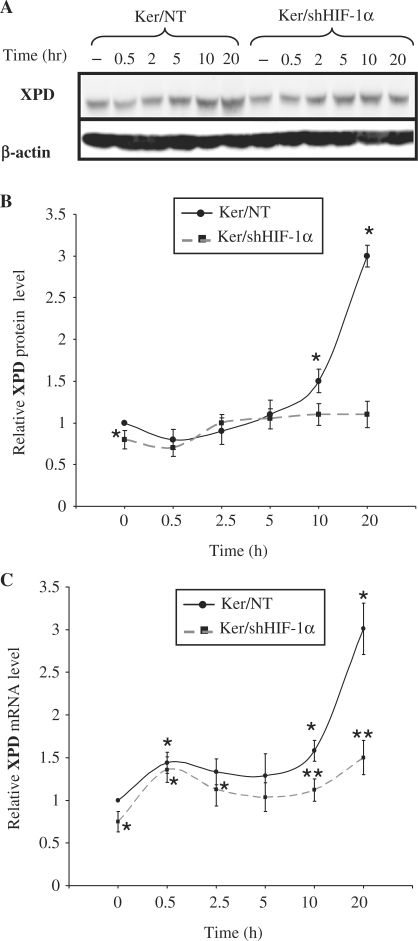
As multiple proteins are involved in removing DNA lesions after UVB irradiation, we wondered whether HIF-1α activity is limited to XPC regulation or modulates the expression of other proteins. Therefore, we investigated the effect of HIF-1α on the expression of XPD, which participates not only in the NER repair system but also plays a fundamental role in transcription. An analysis of XPD expression in UVB irradiated non-transduced keratinocytes showed it to be upregulated, following a slight reduction immediately after irradiation (Figure 4A and B). The basal expression of XPD protein in cells transduced by shHIF-1α showed a moderate but significant decrease before irradiation (Figure 4A and B). Furthermore, downregulation of HIF-1α abrogated the late increase in XPD protein level following UVB-irradiation (Figure 4A and B). Quantifying of XPD mRNA expression levels in non-transduced cells after irradiation indicated a rapid increase, followed by a late, strong upregulation (Figure 4C). Downregulation of HIF-1α had no effect on the immediate increase in XPD mRNA expression following UVB, but abrogated its late increase (Figure 4C). These results suggest that HIF-1α plays an important role in XPD mRNA expression in both the non-irradiated and irradiated conditions.
Figure 4.
Effect of UVB on XPD expression. Keratinocytes were harvested at the indicated time points after irradiation. (A) Total protein extracts were assessed for the presence of the XPD protein by western blot. β-actin was used as a loading control. (B) The protein bands corresponding to XPD were quantified densitometrically. (C) the relative levels of XPD mRNA were quantified by qRT-PCR. The results are expressed as the mean±SD of three independent experiments. *P < 0.05 for cells at the indicated time point versus non-transduced cells prior to irradiation. **P < 0.05 for transduced cells versus corresponding non-transduced cells.
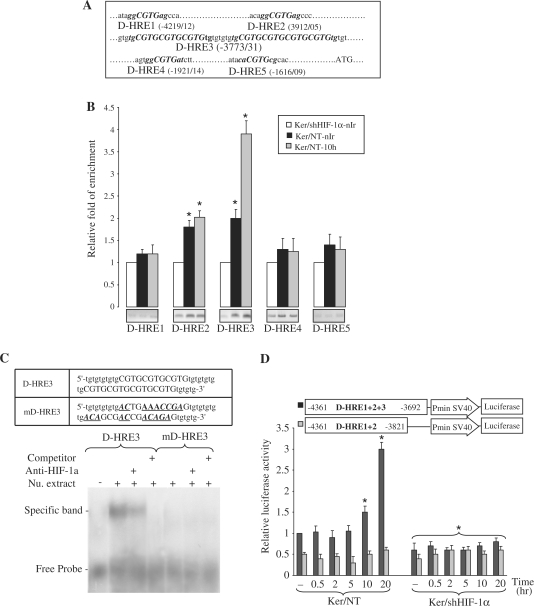
To identify putative HIF-1α binding sites in the XPD promoter, we explored the 4.5 kb region upstream of the ATG translational initiation codon. Eleven candidate HREs, hereafter termed D-HREs, were found (Figure 5A) and subjected to ChIP assays. As shown in Figure 5B, PCR amplification of immunoprecipitated DNA was increased with the primers spanning D-HRE2 as well as D-HRE3 (comprising 7 HRE repeats) in both non-irradiated and irradiated cells. To further confirm the binding of HIF-1α to these sequences, we carried out ESMA analyses using olignucleotides containing wild-type (D-HRE3) and mutated (mD-HRE3) sequences (Figure 5C). Incubation of D-HRE3 oligonucleotide with nuclear extracts led to delayed protein–DNA band migration. Adding anti-HIF-1α antibody to the mixture reduced the amount of shifted bands, whereas adding excess unlabeled probe abolished the formation of this band. Subjecting the mutated oligonucleotide mD-HRE3 to electrophoretic mobility shift assay (EMSA) analysis failed to form this protein–DNA shifted band, suggesting the existence of HIF-1α binding sites on this oligonucleotide (Figure 5C).
Figure 5.
Binding of HIF-1α to the XPD promoter in non-irradiated and irradiated keratinocytes. (A) Eleven-nucleotides sequences matching the consensus HRE (marked in bold) are present in the 4 kb upstream region of the human XPD gene, here referred to as D-HRE1 to 5, in which D-HRE3 includes seven overlapping HREs. (B) Ten hours after UVB-exposure, irradiated and non-irradiated keratinocytes were subjected to ChIP analysis using an anti-HIF-1α antibody. Bands indicate PCR products amplified using primers that span the indicated HREs. The relative levels of corresponding precipitated HRE fragments following ChIP were quantified by qRT-PCR. The results are expressed as the mean±SD of three independent experiments. *P < 0.01 for each HRE versus shHIF-1α-transduced keratinocytes. (C) EMSA analysis of the XPD promoter using HRE mutant probe and anti-HIF-1α antibody. The sequences of the oligonucleotide probes are presented in the top panel. D-HRE3 is the wild-type probe and mD-HRE3 is the probe mutated in the HIF-1α-binding sites. Mutated bases are indicated with bold, italics and underlining. (D) Relative luciferase activities were measured in NT or shHIF-1α-transduced keratinocytes transfected with luciferase reporter constructs containing either the D-HRE1 + 2 + 3 or the D-HRE1 + 2 at the indicated time points after UVB irradiation. For each reporter, the mean luciferase activity is shown (±SD, n = 3) relative to the activity in the NT cells prior to irradiation. *P < 0.05 for cells at the indicated time point versus NT cells prior to irradiation.
To investigate the function of D-HREs found in the XPD promoter, the fragment containing D-HRE1 + 2 + 3 was amplified by PCR and inserted in a luciferase reporter plasmid (Figure 5D). Analysis of cells transfected with D-HRE1 + 2 + 3 plasmid showed that the promoter activity was induced 10–20 h after irradiation. HIF-1α downregulation led to a significant decrease in basal activity of this promoter and totally abrogated the late induction of the luciferase activity. These results, which are consistent with the XPD mRNA expression and ChIP assay, indicate that this fragment plays a role in the regulation of XPD expression. To better define the individual role of each D-HRE elements enclosed in the D-HRE1 + 2 + 3, a plasmid containing only the D-HRE1 + 2 was constructed. Analysis of luciferase activity in cells transfected by this construct did not show any significant variation following irradiation (Figure 5D). Moreover, the basal activity of this promoter was lower than that of D-HRE1 + 2 + 3, indicating the important role of D-HRE3 in the modulation of XPD expression. To rule out the role of D-HRE4 and D-HRE5 in XPD expression, the constructs containing these D-HREs were transfected into keratinocytes. Luciferase activity assays revealed that neither D-HRE4 nor D-HRE5 had a significant effect on XPD expression (Supplementary Figure S1). Overall, our results show that D-HRE3 has a positive role both in basal expression of XPD and in its late overexpression following UVB irradiation.
HIF-1α downregulation increases immediate repair of UVB-induced DNA damage but attenuates late repair
To investigate whether the effects of HIF-1α on XPC and XPD expression might affect NER, we evaluated the repair kinetics of 6-4PPs and CPDs, the most frequent types of photolesions removed primarily by NER (31) with different kinetics (32).
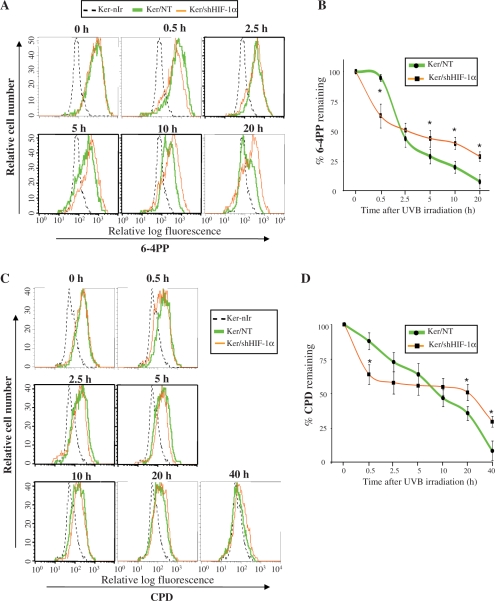
After exposure of non-transduced keratinocytes to UVB, there was a slight repair (5% ± 3) within the first 0.5 h, followed by a rapid drop of 6-4PPs resulting in 56 ± 7% removal of 6-4PPs 2.5 h post-irradiation. The gradual repair of 6-4PPs in the next hours led to 92 ± 6% removal by 20 h (Figure 6A and B). Downregulation of HIF-1α resulted in an increased rate of immediate repair of 6-4PPs but attenuated their late removal, i.e. 40 ± 10% of 6-4PPs was removed within 0.5 h after irradiation while removal by 20 h post-irradiation was 71 ± 4% (Figure 6A and B).
Figure 6.
Effect of HIF-1α downregulation on repair kinetics of 6-4PPs and CPDs. Keratinocytes were harvested at different times after UVB-irradiation and levels of 6-4PPs and CPD were analyzed by flow cytometry following fluorescent immunostaining with the corresponding specific antibody. (A) Representative histogram illustrating the 6-4PPs level at different time points after irradiation in non-transduced or shHIF-1α-transduced keratinocytes. (B) The percentage of 6-4PPs remaining at various time points after irradiation was evaluated by comparison with the initial levels. The results are expressed as the mean ± SD of three independent experiments. (C) Representative histogram illustrating levels of CPDs at different time points after irradiation. (D) The percentage of CPDs remaining at different time points after irradiation was evaluated and the results are expressed as the mean ± SD of three independent experiments. *P < 0.05 for shHIF-1α-transduced cells at the indicated time points versus corresponding non-transduced cells.
Kinetics of CPDs repair in non-transduced cells revealed that CPDs are gradually removed following UVB irradiation, i.e. 12 ± 6% removal at 0.5 h after irradiation reaching slowly 91 ± 7% at 40 h (Figure 6C and D). In keratinocytes silenced for HIF-1α, the immediate CPDs removal was higher (36% ± 7 by 0.5 h) compared to non-transduced cells, while the rate of late CPDs repair was much more slower removing 70 ± 4% of CPD by 40 h post-irradiation (Figure 6C and D).
Taken together, these results indicate that HIF-1α could regulate the repair kinetics of 6-4PPs as well as that of CPDs through the regulation of XPC and XPD expression.
DISCUSSION
Besides being already recognized as the main transcription factor induced under hypoxic conditions, HIF-1α has recently been shown to respond to other stress factors, including UVB irradiation. Consistent with this notion, we highlighted for the first time the functional roles of two HREs (C-HRE1 and C-HRE3.3) and a cassette of seven overlapping HREs (D-HRE3) in the UVB-induced modulation of XPC and XPD expression, respectively.
Effect of HIF-1α on XPC mRNA expression
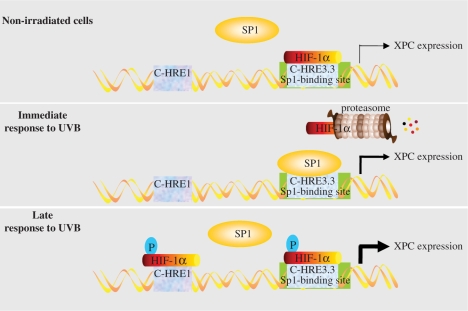
Studies of the XPC promoter revealed that the region from –175 to –1 of the XPC start codon contains the most cis-elements involved in regulation of XPC expression in non-irradiated cells (33). Two putative Sp1 and AP1 sites are predicted to be in this region. Furthermore, it has been suggested that hypermethylation of 17 CpG islands (especially at Sp1 and AP1 sites) modulates XPC expression negatively (33). Consistently, we found that C-HRE3.3, which is located in this area and shares the overlapping base with the Sp1 binding site, participates actively in the regulation of XPC expression in non-irradiated and irradiated conditions. Furthermore, we found another HRE (C-HRE1) in XPC promoter involved exclusively in the regulation of XPC expression after irradiation. Based on our previous results on the effects of UVB irradiation on HIF-1α expression (14) and our present data about the role of HREs in regulation of XPC expression, the following model provides a mechanistic explanation for the effect of HIF-1α and Sp1 on XPC expression (Figure 7). Regarding HIF-1α expression, we have already shown that UVB induces a biphasic variation in HIF-1α protein expression, including a rapid downregulation of this transcription factor, followed by a delayed gradual upregulation. Our previous results also showed that phosphorylation of HIF-1α through UVB-induced activation of p38 MAPK and JNK1 is required for its late stabilization and accumulation (14). It is likely that the occupation of C-HRE3.3 by the weak, non-phosphorylated HIF-1α transcription factor in non-irradiated cells impairs the binding of Sp1 to its DNA-binding site. Rapid degradation of HIF-1α upon UVB exposure allows Sp1 binding, leading to the moderate primary upregulation of XPC. After some delay, UVB irradiation induces phosphorylated HIF-1α (14), which binds to C-HRE1 and C-HRE3.3, and promotes the secondary marked increase in XPC expression. Data from the experimental decrease and increase in HIF-1α levels further support this model. In shHIF-1α-transduced keratinocytes, the reduced level of HIF-1α leaves Sp1 unhindered to bind to the XPC promoter. Sp1 binding leads to stronger induction of XPC than non-phosphorylated HIF-1α, resulting in moderately increased XPC expression at baseline. Furthermore, the late post-UVB surge of phosphorylated HIF-1α is absent in these cells, which creates a relatively stable level of XPC. In DMOG-treated cells, accumulated HIF-1α in both its non-phosphorylated and phosphorylated forms (Figure 1A) binds to C-HRE1 and C-HRE3.3 in either non-irradiated or irradiated cells, leading to XPC upregulation.
Figure 7.
Schematic representation of the proposed model for the effect of UVB-induced HIF-1α modulation on XPC expression.
The interaction between HIF-1α and Sp1 has been described in the regulation of another DNA repair pathway, namely mismatch repair (MMR). Studies investigating the mechanism underlying hypoxia-promoted MMR suppression suggest that HIF-1α competes with Myc for binding to Sp1, which in turn directly binds to the MSH2 promoter (34). These authors proposed that Sp1 binds to the MSH2 promoter as a molecular switch by recruiting Myc in normoxia but HIF-1α in hypoxia, and that this replacement leads to the downregulation of MSH2. In support of this hypothesis, there are no HRE sites on the MSH2 promoter (35). Sp1, but not HIF-1α, is directly bound to the MSH2 promoter under hypoxic conditions (34). In contrast, our study specifically shows that both HIF-1α and Sp1 can bind directly to the same overlapping sequences (C-HRE3.3) on the XPC promoter, and that converting the HRE to the Sp1 binding site abolishes binding of HIF-1α and vice versa.
It has been shown that UV-induced XPC expression is p53-dependent in some cell lines (23). A putative p53 response element located at −2458/−2407 in the XPC promoter has been suggested to be the regulatory element mediating XPC upregulation (23). Increased luciferase activity in keratinocytes transfected with C-HRE1 and C-HRE3, which lack this putative p53 response element, suggests that the direct binding of p53 to the XPC promoter is not necessary for XPC upregulation following UVB irradiation. It may be that p53 participates in the regulation of XPC through interaction with HIF-1α. Such an interaction has been proposed as one mechanism whereby HIF-1α influences hypoxia-induced apoptosis (36) as well as UVB-induced apoptosis (14). Overall, our results establish the importance of HIF-1α in XPC expression.
Effect of HIF-1α on repair kinetics of 6-4PPs and CPDs
Aspects of DNA repair specific to UVB-induced damage appear to correlate with the varying levels of XPC protein produced through HIF-1α regulation. Our data reveal a biphasic variation in XPC protein expression following UVB irradiation of human keratinocytes with an immediate downregulation, followed by a gradual increase. The direct binding of 26S proteasome to XPC following UV irradiation is thought to be responsible for its early degradation (37). Subsequent de novo synthesis of XPC can overwhelm its degradation and lead to the late increase that we observed. It has been proposed that post-translational modifications such as sumoylation protect the newly synthesized protein from degradation (37,38). Our results indicate that HIF-1α downregulation leads to increased XPC protein levels in non-irradiated cells while it abolishes the late increase in XPC expression after UVB irradiation. This effect of HIF-1α on XPC expression is of particular interest with regard to the repair kinetics of 6-4PPs and CPDs. In fact, it has been shown that 6-4PPs are quickly removed after irradiation, whereas removal of CPDs occurs more gradually, i.e. while 80–100% of 6-4PPs are typically removed within 6–8 h after irradiation, 30–60% of CPDs are repaired by 24 h post-irradiation (31,39,40). In agreement with these data, we found that 70% of 6-4PPs and only 36% of CPDs were removed within 5 h following UVB irradiation in non-transduced keratinocytes. Concerning the effect of HIF-1α on the repair kinetics of these photolesions, our results reveal that their immediate removal by 0.5 h post-irradiation is more rapid in HIF-1α-knockdown keratinocytes than in non-trancduced cells, suggesting that the basal level of XPC is critical for immediate damage recognition and removal. In contrary, the late removal of both 6-4PPs and CPDs is much slower in HIF-1α-downregulated keratinocytes than in non-transduced cells, indicating the importance of newly synthesized XPC following UVB irradiation in NER repair efficiency.
Effects of HIF-1α on XPD mRNA expression and on other NER factors
Since HIF-1α is involved in NER through XPC expression and contributes to MMR regulation, we investigated a potential regulatory role for other DNA repair proteins, specifically other NER proteins. We suspected positive regulation of XPD by HIF-1α since HIF-1α-downregulated keratinocytes exhibited both an attenuation of basal XPD expression and an abrogation of UVB-induced XPD expression. Upregulation of XPD expression after pretreatment of keratinocytes by DMOG supported this idea (data not shown). Our hypothesis was confirmed by the increase in luciferase expression driven by the cassette containing the 7 overlapping HREs of the XPD promoter (Figure 5D). Consistent with our results, it has also been documented that tandem-arrayed HREs can sometimes form a functional HRE. Two or three adjacent HREs were found in the genes encoding transferrin (41), several glycolytic enzymes (42,43), and glucose transporter 1 (44). Regarding other NER factors, analysis of the human XPB, XPG, CSA and CSB gene promoters revealed the presence of a putative HIF-1α response element able to mediate sequence-specific DNA binding to HIF-1α in vitro (ChIP assays, Table 1), which strongly suggests that HIF-1α plays a key role in NER regulation.
Table 1.
ChIP assay on putative HREs in the promoter of five NER factors
| Genes | Sequence 5′→3′ | Location | Relative fold enrichmenta |
|
|---|---|---|---|---|
| Ker/NT-nIr | Ker/NT-10 h | |||
| XPB | ggCGTGag | −3960/–3953 | 1.1 ± 0.3 | 2.9 ± 0.4 |
| caCGTGgt | −3104/–3097 | 1.5 ± 0.4 | 0.9 ± 0.5 | |
| ggCGTGat | −1647/–1640 | 0.8 ± 0.6 | 1.1 ± 0.4 | |
| ggCGTGag | −1431/–1424 | 1.2 ± 0.5 | 1.1 ± 0.3 | |
| ggCGTGag | −379/–372 | 3.8 ± 0.6 | 4.5 ± 0.7 | |
| XPF | ggCGTGag | −2201/–2194 | 1.1 ± 0.3 | 1.3 ± 0.2 |
| gaCGTGaa | −380/–373 | 1.9 ± 0.5 | 1.5 ± 0.4 | |
| XPG | ggCGTGag | −3314/–3037 | 1.1 ± 0.6 | 2.2 ± 0.6 |
| ggCGTGgt | −1898/–1891 | 0.9 ± 0.5 | 1.2 ± 0.4 | |
| taCGTGct | −832/–825 | 4.0 ± 0.3 | 4.5 ± 0.6 | |
| CSA | cgCGTGcc | −2590/–2583 | 1.1 ± 0.4 | 1.0 ± 0.5 |
| ggCGTGgc | −642/–635 | 0.8 ± 0.3 | 1.1 ± 0.4 | |
| caCGTGct | −276/–269 | 3.0 ± 0.5 | 5.0 ± 0.7 | |
| CSB | ggCGTGgt | −1267/1260 | 0.9 ± 0.2 | 1.6 ± 0.3 |
| taCGTGga | −270/–263 | 1.2 ± 0.3 | 1.1 ± 0.4 | |
| caCGTGgt | −104/–97 | 5.0 ± 0.6 | 4.0 ± 0.8 | |
aTen hours after UVB-exposure, irradiated and non-irradiated keratinocytes were subjected to ChIP assay using an anti-HIF-1α antibody. The relative levels of corresponding precipitated HRE fragments following ChIP were quantified by qRT-PCR using primers that span the indicated HREs. The PCR amplification in shHIF-1α-transduced cells detected a minimal level of HREs which was normalized to one for each experiment. The results are expressed as the mean ± SD of three independent experiments.
In conclusion, expression of NER proteins following UVB exposure is influenced by HIF-1α modulation. Our results further support the concept that HIF-1α is central to many pathways triggered by UVB irradiation. Considering the effects of HIF-1α in the regulation of NER, MMR (34), and perhaps other repair systems (45), its pivotal role may reasonably be anticipated in photocarcinogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Union GENESKIN Coordination Action LSH-2003-2.1.1-7 (to A.T.); NIH Grants K01-AR048582-03 and R03 CA125855 (to A.L.K.); R01 CA105136 (to D.R.B.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yucui Zhu, M.D, Muriel Cario-Andre, Ph.D, and Catherine Pain for technical assistance.
REFERENCES
- 1.Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 2.Latonen L, Laiho M. Cellular UV damage responses – functions of tumor suppressor p53. Biochim. Biophys. Acta. 2005;1755:71–89. doi: 10.1016/j.bbcan.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Assefa Z, Van Laethem A, Garmyn M, Agostinis P. Ultraviolet radiation-induced apoptosis in keratinocytes: on the role of cytosolic factors. Biochim. Biophys. Acta. 2005;1755:90–106. doi: 10.1016/j.bbcan.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Kulms D, Schwarz T. Independent contribution of three different pathways to ultraviolet-B-induced apoptosis. Biochem. Pharmacol. 2002;64:837–841. doi: 10.1016/s0006-2952(02)01146-2. [DOI] [PubMed] [Google Scholar]
- 5.Haddad JJ, Land SC. A non-hypoxic, ROS-sensitive pathway mediates TNF-alpha-dependent regulation of HIF-1alpha. FEBS Lett. 2001;505:269–274. doi: 10.1016/s0014-5793(01)02833-2. [DOI] [PubMed] [Google Scholar]
- 6.Kietzmann T, Gorlach A. Reactive oxygen species in the control of hypoxia-inducible factor-mediated gene expression. Semin. Cell Dev. Biol. 2005;16:474–486. doi: 10.1016/j.semcdb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Bardos JI, Chau NM, Ashcroft M. Growth factor-mediated induction of HDM2 positively regulates hypoxia-inducible factor 1alpha expression. Mol. Cell. Biol. 2004;24:2905–2914. doi: 10.1128/MCB.24.7.2905-2914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fandrey J, Gorr TA, Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc. Res. 2006;71:642–651. doi: 10.1016/j.cardiores.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl Acad. Sci. USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 14.Rezvani HR, Dedieu S, North S, Belloc F, Rossignol R, Letellier T, de Verneuil H, Taieb A, Mazurier F. Hypoxia-inducible factor-1alpha, a key factor in the keratinocyte response to UVB exposure. J. Biol. Chem. 2007;282:16413–16422. doi: 10.1074/jbc.M611397200. [DOI] [PubMed] [Google Scholar]
- 15.Turchi L, Aberdam E, Mazure N, Pouyssegur J, Deckert M, Kitajima S, Aberdam D, Virolle T. Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ. 2008;15:1472–1480. doi: 10.1038/cdd.2008.74. [DOI] [PubMed] [Google Scholar]
- 16.Wunderlich L, Paragh G, Wikonkal NM, Banhegyi G, Karpati S, Mandl J. UVB induces a biphasic response of HIF-1alpha in cultured human keratinocytes. Exp. Dermatol. 2008;17:335–342. doi: 10.1111/j.1600-0625.2007.00640.x. [DOI] [PubMed] [Google Scholar]
- 17.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 18.D'E;rrico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 20.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol. Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 23.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl Acad. Sci. USA. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dazard JE, Gal H, Amariglio N, Rechavi G, Domany E, Givol D. Genome-wide comparison of human keratinocyte and squamous cell carcinoma responses to UVB irradiation: implications for skin and epithelial cancer. Oncogene. 2003;22:2993–3006. doi: 10.1038/sj.onc.1206537. [DOI] [PubMed] [Google Scholar]
- 25.Fitch ME, Cross IV, Turner SJ, Adimoolam S, Lin CX, Williams KG, Ford JM. The DDB2 nucleotide excision repair gene product p48 enhances global genomic repair in p53 deficient human fibroblasts. DNA Repair (Amst) 2003;2:819–826. doi: 10.1016/s1568-7864(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 26.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezvani HR, Mazurier F, Cario-Andre M, Pain C, Ged C, Taieb A, de Verneuil H. Protective effects of catalase overexpression on UVB-induced apoptosis in normal human keratinocytes. J. Biol. Chem. 2006;281:17999–18007. doi: 10.1074/jbc.M600536200. [DOI] [PubMed] [Google Scholar]
- 28.Rezvani HR, Cario-Andre M, Pain C, Ged C, deVerneuil H, Taieb A. Protection of normal human reconstructed epidermis from UV by catalase overexpression. Cancer Gene Ther. 2007;14:174–186. doi: 10.1038/sj.cgt.7701000. [DOI] [PubMed] [Google Scholar]
- 29.Rezvani HR, Ged C, Bouadjar B, de Verneuil H, Taieb A. Catalase overexpression reduces UVB-induced apoptosis in a human xeroderma pigmentosum reconstructed epidermis. Cancer Gene Ther. 2008;15:241–251. doi: 10.1038/sj.cgt.7701102. [DOI] [PubMed] [Google Scholar]
- 30.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci. STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell DL. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 32.Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, van Zeeland AA, Mullenders LH. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 2005;4:571–582. doi: 10.1016/j.dnarep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Wu YH, Tsai Chang JH, Cheng YW, Wu TC, Chen CY, Lee H. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene. 2007;26:4761–4773. doi: 10.1038/sj.onc.1210284. [DOI] [PubMed] [Google Scholar]
- 34.Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol. Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability–a calculated mechanism underlying tumor progression. J. Mol. Med. 2007;85:139–148. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- 36.Greijer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 2004;57:1009–1014. doi: 10.1136/jcp.2003.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang QE, Praetorius-Ibba M, Zhu Q, El-Mahdy MA, Wani G, Zhao Q, Qin S, Patnaik S, Wani AA. Ubiquitylation-independent degradation of Xeroderma pigmentosum group C protein is required for efficient nucleotide excision repair. Nucleic Acids Res. 2007;35:5338–5350. doi: 10.1093/nar/gkm550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Auclair Y, Rouget R, Affar el B, Drobetsky EA. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc. Natl Acad. Sci. USA. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell DL, Haipek CA, Clarkson JM. (6-4)Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat. Res. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 41.Rolfs A, Kvietikova I, Gassmann M, Wenger RH. Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J. Biol. Chem. 1997;272:20055–20062. doi: 10.1074/jbc.272.32.20055. [DOI] [PubMed] [Google Scholar]
- 42.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 43.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 44.Okino ST, Chichester CH, Whitlock JP., Jr Hypoxia-inducible mammalian gene expression analyzed in vivo at a TATA-driven promoter and at an initiator-driven promoter. J. Biol. Chem. 1998;273:23837–23843. doi: 10.1074/jbc.273.37.23837. [DOI] [PubMed] [Google Scholar]
- 45.Um JH, Kang CD, Bae JH, Shin GG, Kim DW, Kim DW, Chung BS, Kim SH. Association of DNA-dependent protein kinase with hypoxia inducible factor-1 and its implication in resistance to anticancer drugs in hypoxic tumor cells. Exp. Mol. Med. 2004;36:233–242. doi: 10.1038/emm.2004.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.