Abstract
Specific detection of mRNA cleavage by 5′RACE is the only method to confirm the knockdown of mRNA by RNA interference, but is rarely reported for in vivo studies. We have combined 5′-RNA-linker-mediated RACE (5′-RLM-RACE) with real-time PCR using a molecular beacon to develop a rapid and specific method termed MBRACE, which we have used to detect small-interfering RNA (siRNA)-induced cleavage of ApoB, RRM1 and YBX1 transcripts in vitro, and ApoB in vivo. When RNA from siRNA-transfected cells was used for 5′-RLM-RACE and a cleavage site-specific molecular beacon probe was included in subsequent real-time PCR analysis, the specific mRNA cleavage product was detected. Detection of siRNA-mediated cleavage was also observed when RNA from mouse liver following administration of ApoB-specific siRNA was analysed, even in cases where ApoB knockdown measured by real-time PCR was <10%. With its sensitivity and specificity, this variation on the 5′RACE method should prove a useful tool to detect mRNA cleavage and corroborate knockdown studies following siRNA use in vivo.
INTRODUCTION
The inhibition of gene expression by RNAi has great therapeutic potential but remains hampered by inefficient delivery of siRNA and potential off-target effects. Although many avenues of delivery are being investigated, including the use of localised delivery by direct injection and topical administration, and systemic delivery with intravenous administration (1–3) very few studies have included data to confirm that the effects of siRNA observed in vivo are due to an RNAi-mediated mRNA cleavage mechanism rather than non-specific events. siRNAs have the potential to trigger an innate immune response through the activation of Toll-like receptors (TLR 3, 7 and 8) and also by binding to proteins such as retinoic acid inducible gene 1, and this itself may cause a modulation in gene expression which could account for the observed effects attributed to siRNA-mediated RNAi (4,5).
The importance of confirming that mRNA knockdown following siRNA administration has occurred via an RNAi-mediated event is highlighted by two recent studies reporting the considerable contribution of the innate immune system to apparent in vivo knockdown, suggesting that many of the reports of in vivo efficacy of siRNAs can be explained by a general down-regulation of transcription that is stimulated by the double stranded RNA structure of siRNA without involving RNAi. Kleinman et al. (6) demonstrated that the intravitreal injection of siRNA could lead to down regulation of vascular endothelial growth factor a (Vegfa) and its receptor (Vegfr1) in a sequence-independent manner, as siRNAs with no homology to Vegfa or Vegfr1 were as effective as specific siRNAs (6). More recently, it was demonstrated that siRNAs able to inhibit viral replication in vivo do so via stimulation of the innate immune system rather than through an RNAi-mediated event (7), and that the negative control used in these studies had an unusually low immunostimulatory activity, and had also been a component of several other in vivo studies assessing immunostimulatory siRNA.
Confirmation that an observed mRNA knockdown has occured via an RNAi-based cleavage event can be demonstrated using the well-established technique of Rapid Amplification of cDNA Ends (RACE), as siRNAs cleave their target sequence following a canonical pattern 10 bp from the 5′-end of the antisense strand (8,9). The slicing action of the RISC-associated Argonaute 2 protein at this position cleaves the target mRNA into two distinct fragments: a 5′ fragment with a 3′ hydroxyl group, and a 3′ fragment with a 5′ phosphate. Either of these fragments can be amplified using RACE in a modified version of the RLM-RACE protocol (10,11). The amplified fragment must then be cloned and sequenced to ascertain the junction between the RNA linker sequence and the target RNA sequence which, if mRNA cleavage has occurred via RNAi, should correlate with the position as predicted by the siRNA sequence (12–17). Although RACE-based detection has been utilised to assess the action of antisense-based inhibition of expression (18), it has only rarely been used in the corroboration of knockdown data observed in an ever increasing range of studies using RNAi in vivo (12–17). This may be because RLM-RACE appears to be an inefficient process for detecting mRNA cleavage products, despite the inherent specificity of using target and linker-specific primers. In standard 5′-RLM-RACE, a dephosphorylation step is used to remove any RNAs that have a 5′phosphate (including nicked transcripts), effectively enriching for the capped full-length mRNA, which can then be de-capped and ligated. To detect siRNA-cleaved transcripts, which are also phosphorylated RNAs, this enrichment step cannot be performed; hence any nicked RNAs are also amplified thereby decreasing the efficiency and sensitivity of the reaction. Sequence analysis of this heterogenous population of amplicons determines if a correct product has been derived from the siRNA-mediated cleavage of the target transcript.
In order to overcome these problems, we have modified the second PCR of 5′-RLM-RACE amplification to be a real-time PCR using a molecular beacon hybridisation probe (19) specific to the junction between the RNA linker and mRNA cleavage site. The specificity of molecular beacon probes has been well proven to distinguish between sequence alterations of one nucleotide (20), enabling their use in an expanding field of hybridisation-based applications (21). This high degree of specificity means the junction region between the RNA linker sequence and the cleaved siRNA site can be detected in real-time, eliminating the need to run further nested PCR reactions, clone the candidate amplicons and then confirm their identity via sequencing. We have used this method to detect siRNA-mediated cleavage of ApoB, RRM1 and YBX1 transcripts in vitro, and also of ApoB in vivo.
MATERIALS AND METHODS
Primers, siRNAs and molecular beacon probes
The sequences of the primers, siRNAs and molecular beacons used are detailed in Table 1. RRM1 and YBX1 siRNAs and all primers for qPCR were from Invitrogen Corporation (Carlsbad, CA, USA). The ApoB-specific siRNA ApoB-1 and a mismatched control siRNA were of identical sequence to those used by Alnylam (12), and were synthesised by TriLink Biotechnologies (Carlsbad). The primers and probes for the MBRACE assays were designed using Beacon DesignerTM from PREMIER Biosoft International (Palo Alto, CA, USA). MBRACE primers were purchased from Invitrogen and the molecular beacon probes were synthesised by Integrated DNA Technologies, Inc. (Coralville, IA, USA).
Table 1.
Sequences of siRNAs, primers and probes used in this study
| siRNAs | ||
| Name | Passenger strand | Guide strand |
| ApoB-1 | GUCAUCACACUGAAUACCAAU | AUUGGUAUUCAGUGUGAUGAmC*mA*C |
| ApoB1 mm control | GUGAUCAGACUCAAUACGAAU | AUUCGUAUUGAGUCUGAUCAmC*mA*C |
| RRM1-2 | CCCAGUUACUGAAUAAGCAGAUCUU | AAGAUCUGCUUAUUCAGUAACUGGGCU |
| RRM1-3 | GCAAACUCACUAGUAUGCACUUCUA | UAGAAGUGCAUACUAGUGAGUUUGCCU |
| RRM1-15 | GAUUGUAAAUCCUCACUUAdTdT | UAAGUGAGGAUUUACAAUCdTdT |
| YBX1-9 | CCAGUUCAAGGCAGUAAAUdTdT | AUUUACUGCCUUGAACUGGdTdT |
| qPCR Primers | ||
| Target | Forward | Reverse |
| ApoB | GGCACTGTGGGTCTGGAT | TTCTTCTCTGGAGGGGACTG |
| RRM1 | GGCAAACTCACTAGTATGCACTTC | AAATAATACATCCCAGTCTTCAAACC |
| YBX1 | GGAGTTTGATGTTGTTGAAGGA | AACTGGAACACCACCAGGAC |
| Polr2a | TTACTCCCCTGCATGGTCTC | TGGGAGACATAGCACCACCT |
| LMNA | TGAGGCCAAGAAGCAACTTCA | CTCATGACGGCGCTTGGT |
| Gene-specific primers for first-strand cDNA synthesis | ||
| ApoB | AGAACCCGTGATTCAACCTG | |
| RRM1 | AAGCAGTGCTAAAGGGGTGA | |
| YBX1 | CCGGATGATGGTAGAGATGG | |
| First-round RACE gene-specific primers | ||
| ApoB | GCTCCCATGTGGTGTAGATGCGTTGGA | |
| RRM1 | TGCTGCATTTGATGGTTCCCAGGTTCTG | |
| YBX1 | TCTGGGCGTCTGCGTCGGTAATTGA | |
| MBRACE primers | ||
| Target site | Forward | Reverse |
| ApoB-1 | ACTGGAGCACGAGGACACTG | GGAAGAAAGGAAATGGGCAACG |
| RRM1-2 | CGACTGGAGCACGAGGAC | AGCCCTCATAGGTTTCGTATGG |
| RRM1-3 | CGACTGGAGCACGAGGAC | GATTAGCCGCTGGTCTTGTC |
| RRM1-15 | ACTGGAGCACGAGGACAC | AGGAATTTCTGGTATGCTCTG |
| YBX1-9 | CGACTGGAGCACGAGGACAC | TGCTGGTAATTGCGTGGAGGAC |
| Molecular beacon probes | ||
| ApoB-1 | F-CGCGATCCCAGCATTGGTATTCTTTCTACTCCTTCAGATCGCG-B | |
| RRM1-2 | F-CGCGATCAAAGATCTGCTTTTCTACTCCTTCAGTCCGATCGCG-B | |
| RRM1-3 | F-CGCGATCCCGTAGAAGTGCATTTCTACTCCTTCAGATCGCG-B | |
| RRM1-15 | F-CGCGATCAGATCTTTCAATAAGTGAGGATTTCTACTCCGATCGCG-B | |
| YBX1-9 | F-CGCGATCTGCATATTTACTGCCTTTCTACTCCTTCAGATCGCG-B | |
All sequences are given in 5′-3′ orientation. The siRNA sequences are RNA except dTdT deoxynucleotides ends. The Stealth duplexes (RRM1-2 and RRM1-3) are dsRNA with proprietary modifications of the passenger strand; m, 2′-O-Methyl ribose-modified nucleotides; *phosphorothioate linkage; F, FAM dye label; B, Black Hole Quencher.
Cell lines and transfection
The Hepa1-6 and A549 cell lines used in this study were obtained from ATCC. Hepa1-6 cells were grown in DMEM and A549 cells in RPMI-1640 medium, both supplemented with 10% heat-inactivated fetal bovine serum (FBS) (all from Invitrogen), at 37°C in humidified air with 10% CO2. To measure siRNA-mediated knockdown, cells were transfected with specific or control siRNAs using LipofectamineTM RNAiMAX (Invitrogen) as per the manufacturer’s instructions with modifications (22). Briefly, siRNAs and LipofectamineTM RNAiMax were diluted in DMEM or RPMI 1640 without serum, and incubated for 5–10 min at room temperature. The diluted RNAiMAX was added drop-wise to the siRNA, and lipoplex formation was allowed to proceed for 20–30 min at room temperature. Lipoplexes were then transferred to multi-well tissue culture plates and overlaid with 5 × 103 cells per cm2. Following overnight incubation, cells were isolated and mRNA knockdown was measured as described below.
Isolation of RNA
For in vitro samples, RNA was isolated using the PureLink™ 96 RNA Purification system as per the manufacturer’s protocol (Invitrogen). For in vivo samples, tissue was homogenised in 2 ml Trizol (Invitrogen), debris removed by centrifugation and samples divided into 2 × 1 ml aliquots. To each aliquot, 0.2 ml chloroform was added followed by centrifugation. The aqueous phase was retained and an equal volume of 70 % ethanol was added and mixed. This was applied to the PureLink™ 96 RNA Purification system as above. Purified RNA quality and concentration was assessed with a NanoDrop 1000 spectrophotometer.
Real-time RT–qPCR
First-strand cDNA was synthesised from ∼200 ng RNA from cultured cells or 1 µg RNA from liver samples as follows: RNA was treated with DNase I for 15 min at room temperature, followed by incubation for 10 min at 65°C in the presence of 0.6 mM EDTA. After the addition of random primers (1.5 µg) the reaction was incubated for a further 10 min at 65°C. After cooling on ice for 1 min, Superscript® III polymerase (Invitrogen) was added in the presence of 5 mM DTT and 1 mM dNTPs, and the reaction incubated at 25°C for 5 min, followed by 1 h at 55°C. For real-time PCR, the cDNA was diluted 1 : 4 in 10 mM Tris pH 7.0, and reactions carried out on a LightCycler® 480 (Roche) using LightCycler® 480 SybrGreen I Master mix (Roche) and gene-specific primers at 180 nM. Levels of ApoB, RRM1 or YBX1 mRNA were normalised to the reference gene RNA polymerase IIa (Polr2a) for mouse cell lines and in vivo samples, and lamin A/C (LMNA) for human cell lines, and relative change in mRNA following treatment with specific or control siRNA was calculated from triplicate technical replicates of each using the 2 ΔΔCt method (23). Standard deviations were also calculated, based on the Ct standard deviation of the three technical replicates and converted to fold change.
5′-RLM-RACE
5′-RLM-RACE was performed using the GeneRacerTM kit (Invitrogen) with the manufacturer’s instructions modified as follows. Briefly, 100 ng total RNA was directly ligated to the RNA linker without prior treatment. After phenol/chloroform extraction and precipitation, first-strand cDNA was synthesised using a gene-specific primer (Table 1). From this reaction, 1 µl was used in first-round 5′RACE reactions using the GeneRacer 5′ primer and target gene-specific primer with cycling as described in the GeneRacerTM kit manual: 1 cycle of 94°C for 2 min, then 5 cycles of 94°C for 30 s and 72°C for 1 min, then 5 cycles of 94°C for 30 s and 70°C for 1 min, then 20 cycles of 94°C for 30 s and 68°C for 1 min. The second-round 5′RACE reaction used 1 µl of the first-round reaction and internal primers (GeneRacerTM Nested and gene-specific nested) with cycling as above but with an extension time of 15 s. From this reaction, 10 µl was analysed on a 3% agarose gel. To confirm the site of mRNA cleavage, a region corresponding to the size of the predicted 5′RACE amplicon was excised. The DNA was cloned using the TA Cloning® kit (Life Technologies) and sequenced using an ABI3730 sequencer, with BigDyeTM Terminator 3.1 Ready Reaction Cycling kit (Applied Biosystems, Foster City, CA, USA).
MBRACE
The first round 5′RACE reaction product described above was used as a template for the MBRACE reaction. This contained 1 µl from the first-round RACE reaction and the TaqMan® Probe Master (Roche). For the final optimised method, primers and probes were used at the following concentrations: 180 nM MB-R, 3.6 µM MB-F and 250 nM molecular beacon probe (all specific for the target gene). Reactions were run on a LightCycler® 480 with the following cycling conditions: 95°C for 10 min, then 45–55 cycles of 95°C for 10 s, 62°C for 30 s and 72°C for 8 s, followed by 40°C for 30 s.
Hydrodynamic tail-vein injection
Female CD-1 nude mice, 5–6 weeks of age and weighing 20–23 g were assigned to treatment groups such that the mean body weights were similar for each group. The animals were placed in a restraining device and injected without anaesthesia. Fifty microgram of ApoB-1 siRNA or the control siRNA in 1.8 ml Ringer’s Solution (147 mM NaCl, 4 mM KCl, 1.13 mM CaCl2) was rapidly (within 5–7 s) injected into one of the tail side veins through a 27-gauge needle. The injection was repeated 8 and 24 h later. Twenty-four hours after the last injection, animals were humanely euthanised by CO2 asphyxiation and livers collected. Liver tissues were snap-frozen in liquid nitrogen and stored at −80°C prior to RNA isolation. Animal experiments were performed with animal care ethics approval by the Genesis Research and Development Animal Ethics Committee, under Code of Ethical Conduct approved by the National Animal Ethics Advisory Committee and Ministry of Agriculture and Forestry, in accordance with the New Zealand Animal Welfare Act 1999.
RESULTS
Molecular beacon RACE detects siRNA-mediated mRNA cleavage
Standard 5′RLM-RACE can be used to confirm that mRNA knockdown as measured by real-time RT–qPCR is the result of specific siRNA-directed cleavage of mRNA directed by siRNAs, but it is time consuming and rarely carried out. To improve the speed and sensitivity of the 5′-RLM-RACE assay, we modified the method to allow sequence-specific fluorescence-based detection of the cleaved target mRNA. This involved addition of an RNA linker and performing 5′RACE as standard, but instead of performing two rounds of PCR to detect a product on a gel, the final round was performed as real-time PCR incorporating a molecular beacon probe spanning the junction between the RNA linker sequence and the 5′-end of the cleaved mRNA, and termed this method MBRACE (Figure 1). Molecular beacon probes give a fluorescent signal only when hybridised to an exact complement, so even if multiple products were generated by PCR, only the cDNA template from cleaved target mRNA ligated to the RNA linker should be detected. The method was tested first on two ApoB clones of known sequence generated from a prior 5′-RLM-RACE experiment using a siRNA specific for ApoB called ApoB-1 (12): one clone was the expected product with the RNA linker attached to the ApoB-1-directed siRNA cleavage site; the second clone contained a 4-nucleotide insertion between the linker and the cleavage site (Figure 2A). MBRACE reactions were performed and as shown in Figure 2B, only the specific product (cloned positive) gave a fluorescent signal whereas the clone with the 4 bp insertion (cloned negative) showed only the background fluorescence.
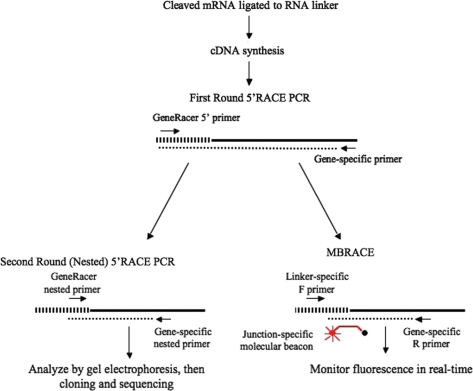
Figure 1.
Comparison of 5′-RLM-RACE and MBRACE methods. The initial steps are the same in both methods, in which an RNA linker (striped bar) is attached to siRNA-cleaved mRNA (solid line). After cDNA synthesis by reverse transcription, a first round PCR reaction is set up using one primer specific for the linker sequence and a second primer specific to the gene target and product generated (dotted line). For standard 5′RACE (left), a second round PCR is performed using internal primers (Nested). Products are then analyzed by agarose gel electrophoresis, and amplicons of the predicted size are excised, cloned and sequenced to confirm that the product is derived from siRNA-mediated cleavage. For MBRACE, the first round PCR products are also used as template for a second round with the same nested primers, but the addition of a molecular beacon spanning the junction between the linker and cleaved mRNA enables amplification to be detected in real-time using a LightCycler® 480.
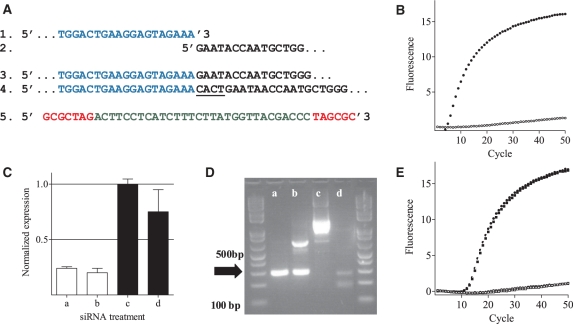
Figure 2.
Specific detection of mRNA cleavage products with MBRACE in vitro. (A) Schematic representation of sequences of: (1) the RNA linker (blue); (2) ApoB cleavage site; (3) positive (RNA linker ligated to cleavage product) and (4) negative (4 bp insertion between linker and mRNA fragment underlined) clones; and (5) the molecular beacon probe used to detect linker ligation to the cleaved mRNA fragment (red text represents the 7-bp stem). (B) Amplification curves showing fluorescent signal generated from positive (filled square) and negative (open square) cDNA clones. (C) RT–qPCR analysis of ApoB mRNA in Hepa1-6 cells transfected with either 1 (a) or 10nM (b) ApoB-1 siRNA, or 1 (c) or 10 nM (d) mismatch control siRNA. (D) The same samples were used as template in standard 5′-RLM-RACE analysis, and analyzed by electrophoresis. The correct product indicated by the arrow has a size of 290 bp. (E) Amplification curves from MBRACE reactions using the template from siRNA-treated cells as in (C) filled symbols represent samples transfected with ApoB-1 siRNA, and open symbols are samples transfected with mismatch control siRNA at 10nM (open square) or 1nM (open triangle). All MBRACE experiments were performed in duplicate, but for clarity only a single, representative replicate is shown.
The MBRACE assay is specific to the siRNA target gene and target site
The method was further tested on a number of in vitro samples. RNA prepared from Hepa1-6 cells transfected with ApoB-1 or a control siRNA was initially analysed by RT–qPCR to confirm knockdown of mRNA (Figure 2C) then tested in the standard 5′-RLM-RACE method, with two rounds of PCR prior to analysis by agarose gel electrophoresis (Figure 2D). RT–qPCR showed clear knockdown of ApoB transcript with ApoB-1 siRNA (a and b, Figure 2C) compared to the control siRNA (c and d, Figure 2C). In 5′-RLM-RACE analysis, the predicted product of 290 bp was clearly evident in cells treated with ApoB-1 siRNA (lanes a and b, Figure 2D). A band of the correct size was also observed in the sample from cells treated with 1 nM control siRNA (lane d, Figure 2D). This underlies the drawback of analysing 5′-RLM-RACE products based on size alone, the identity of which must be confirmed by cloning and sequencing. To test the MBRACE method, the products from the first round 5′-RLM-RACE PCRs were then used as template in molecular beacon-containing real-time PCR reactions (Figure 2E). Only the samples transfected with ApoB-1 siRNA showed a fluorescent signal, whereas there was no amplification with the control siRNA samples. These data demonstrate that the true product of ApoB-1 siRNA-mediated cleavage was only present in cells transfected with the ApoB-1 siRNA and that the other products observed by gel electrophoresis following standard 5′-RLM-RACE were artefacts not derived from siRNA-cleaved ApoB.
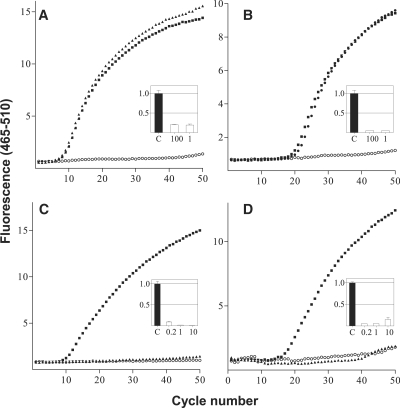
The specificity of the MBRACE protocol was next tested for the ability to detect specific cleavage of two further mRNA species each targeted with two different siRNAs. A549 cells were transfected with siRNA specific for either of three independent sites in the RRM1 (RRM1-2, RRM1-3 and RRM1-15) and one site in the YBX1 transcript (YBX1-9). RT–qPCR experiments demonstrated that all four siRNAs were highly effective in inducing knockdown of their target mRNA (Figure 3A–D insets). When real-time PCR was performed using molecular beacons specific to each of the siRNA cleavage sites, a fluorescent signal was detected only with the molecular beacon specific for that siRNA cleavage site (Figure 3A–D). Confirming the specificity of the MBRACE reaction, each molecular beacon produced a fluorescent signal only when applied to a template derived from cells treated with the matching siRNA. This is clearly evident in Figure 3C and D, where the addition of a molecular beacon specific for the RRM1-15-mediated cleavage resulted in a fluorescent signal only when the template was derived from A549 cells treated with RRM1-15 siRNA, and was not detected following treatment with YBX1-9 siRNA. Likewise, the molecular beacon complementary to the cleavage site induced by YBX1-9 siRNA only produced a fluorescent signal when used with template from YBX1-9 siRNA-treated cells.
Figure 3.
Application of MBRACE assay to other targets in vitro. MBRACE assays of RNA extracted from A549 cells transfected with (A) 1 nM and 100 nM of RRM1-2, (B) 1 nM and 100 nM of RRM1-3, (C) 10 nM YBX1-9 or (D) 10 nM RRM1-15. Filled symbols are samples transfected with RRM1 or YBX1 siRNAs; open symbols are from cells only control samples. YBX1 and RRM1 templates in (C) and (D) were tested with specific molecular beacon primers and probes (filled square) and also tested with molecular beacon primers and probes specific for RRM1-15 (filled triangle) and YBX1-9, (filled triangle), respectively. Inset graphs show siRNA-mediated mRNA knockdown using the same siRNAs as measured by RT–qPCR.
Real-time PCR and 5′-RLM-RACE are limited in their effectiveness for detection of cleaved mRNA in vivo
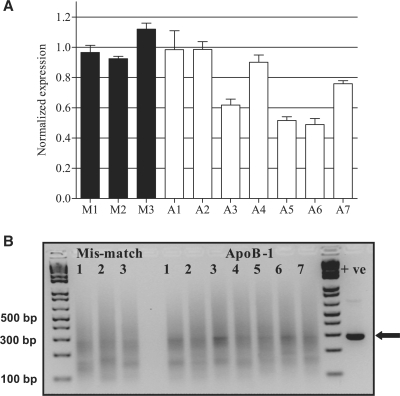
To test the ability of MBRACE to confirm siRNA-mediated knockdown in vivo, hydrodynamic tail-vein injection (HTVI) of ApoB-1 siRNA was carried out. Twenty-four hours after delivery of siRNA for ApoB or mis-matched control, mice livers were harvested and RNA prepared. These were processed for analysis by real-time RT–qPCR to measure ApoB mRNA knockdown, and by 5′-RLM-RACE to detect the presence of the specifically cleaved mRNA fragments. In four mice receiving the ApoB-1 siRNA, RT–qPCR analysis showed that knockdown of ApoB was minimal (Figure 4A, A1, A2, A4 and A7), whilst the remaining three mice in this group had ApoB mRNA levels that were reduced by about 30–50%. To detect the presence of cleaved ApoB transcript, standard 5′-RLM-RACE was performed. Agarose gel electrophoresis of the 5′-RLM-RACE products showed a correct sized band (290 bp) within a smear (Figure 4B). Interestingly, two of the three mice showing the greatest mRNA knockdown also had a clearly detectable band of around 290 bp, which is the size predicted following cleavage by ApoB-1 siRNA (Figure 4A and 4B, A3 and A6). A number of other ApoB-1 samples appeared to show increased staining in that region of the gel, but so too did the control samples treated with mismatched siRNA.
Figure 4.
ApoB knockdown and detection of mRNA cleavage products in mouse liver. (A) qRT–PCR of ApoB mRNA levels from the livers of mice treated with ApoB-1 siRNA or a mis-matched control siRNA by the HTVI method. Levels of ApoB mRNA were normalised to the average of the mis-match siRNA samples. (B) Standard 5′-RLM-RACE of the same samples, analyzed by electrophoresis on a 3% TAE-agarose gel using 1 kb Plus ladder (Invitrogen). An in vitro positive control (+ve) was prepared and analyzed in parallel; the band of correct size (290 bp) is indicated with an arrow.
MBRACE detects ApoB-1 siRNA activity in vivo
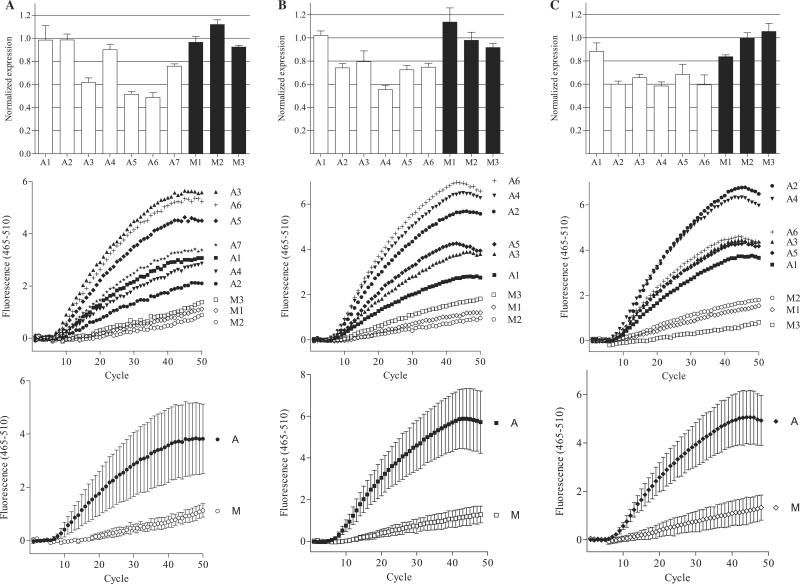
The in vivo samples from Figure 4 were tested in a MBRACE assay, using the same 5′–RLM-RACE first-round PCR samples as template. These results showed that RNA isolated from mice receiving the ApoB-1 siRNA gave a positive fluorescent signal, higher than that observed with the mismatch control siRNA samples (Figure 5A), indicating detection of the specifically cleaved ApoB product. Samples from ApoB-1-treated mice that showed no detectable knockdown by real-time PCR or gel electrophoresis gave a fluorescent signal, confirming that some specific cleavage of the ApoB transcript had occurred (compare Figure 4 with Figure 5A). This was confirmed by analysing two additional HTVI experiments by MBRACE, where all samples derived from mice that were treated with the ApoB-1 siRNA showed specifically cleaved ApoB mRNA detected by the molecular beacon probe (Figure 5B and C). This was particularly clear in one experiment, where even though from RT–qPCR analysis ApoB mRNA was reduced by only 10% in one sample (Figure 5C, A1), the MBRACE method clearly detected the ApoB-1 siRNA-cleaved product above the mismatch control samples.
Figure 5.
Detection of ApoB mRNA cleavage in vivo with MBRACE. Detection of ApoB knockdown in vivo using the MBRACE method was tested in three independent HTVI experiments, (A–C). Top panels show RT–qPCR analysis of ApoB expression in animals treated with the ApoB-specific siRNA ApoB-1 (A1–A7 in experiment A and A1–A6 in experiments B and C) or mismatch control siRNA (M1–M3). Amplification curves from the same samples were used for MBRACE reactions and fluorescence from the individual templates is shown in the middle panels, with average fluorescence from all specific (A) or mismatch control (M) siRNA-treated animals shown in the bottom panels. All individual MBRACE experiments were performed in duplicate but for clarity only single replicates are shown.
DISCUSSION
The use of siRNA in vivo remains hampered by off-target effects and difficulties associated with confirming that the observed mRNA knockdown is the result of an RNAi-mediated event. When tested in vitro, knockdown of target mRNA by siRNA is generally effective and can be detected easily with real-time PCR, as the majority of cells are transfected and mRNA levels are often decreased by over 90%. Similarly, the specificity of siRNA-mediated knockdown observed in vitro can be confirmed with 5′-RLM-RACE amplification and sequencing to determine target-site cleavage, a simple albeit time-consuming and labour-intensive process. We have used molecular beacon probes as a tool to aid detection of siRNA-mediated knockdown in both in vitro and in vivo studies. The stem-loop-based hybridisation specificity of molecular beacons has seen their application in a variety of fields, for example utilizing their ability to give real-time results for the provision of genetic identification of single nucleotide differences (19,20), as well as their use in real-time monitoring of gene expression in isothermic amplification strategies (24). We found that using molecular beacon probes to detect specific cleavage of mRNA in vitro dramatically increased the speed at which we were able to confirm siRNA-mediated cleavage of the target mRNA.
The MBRACE method was applied to five siRNAs across three genes and in each case specifically detected the siRNA-mediated cleavage of the mRNA. This confirmed the initial results obtained with the cloned ApoB RACE products and demonstrates the broad applicability of this method to rapid detection of siRNA-mediated mRNA cleavage events. Using the Premier Biosoft molecular beacon probe designer for our cleavage sites of interest generated probes with moderate to high predicted activity in each instance. Although this may vary for other sites depending on the sequence of the target site, in such cases, the method could be further optimised by altering the sequence of the RNA linker. Interestingly, the efficiency of cleavage site detection by the molecular beacon was not always equivalent. The molecular beacon specific for the cleavage of RRM1 mRNA induced by siRNA RRM1-3 was notable in that the log-linear phase of amplification appeared much later (>20 cycles) compared with the other target sites (all around 10 cycles). Differences in Cp between different targets even within the same gene are to be expected at this point. Although the knockdown of mRNA for both RRM1-2 and RRM1-3 siRNAs is very similar, the sensitivity of each cleaved mRNA to subsequent degradation may differ subtly leading ultimately, after two rounds of PCR, to the difference in amplification seen in the MBRACE assay; also the primer combinations after reverse transcriptase treatment are specific for each target region, and may not be optimal with the general reaction conditions used. Similarly, a dose response was observed in knockdown with the YBX1-9 siRNA, but this was not reflected in the Cp and total fluorescence of the MBRACE assay, which was similar at all siRNA concentrations for each site. This is likely due to the template for MBRACE being derived from the first round of RACE PCR, an end-point assay. Therefore, subsequent real-time PCR amplifications will likely begin with templates of similar concentration, and will not reflect the original differences in mRNA concentration.
After optimizing the MBRACE reaction in vitro, we applied this to the in vivo knockdown of ApoB mRNA in mouse livers using a previously published siRNA sequence (12). To overcome the need for cholesterol conjugation or a delivery vehicle to introduce siRNA into the liver, we used the HTVI, one of the few methods for introducing foreign nucleic acids in vivo without the requirement of a delivery vehicle (25). HTVI involves the injection of a large bolus volume (9% of body weight) containing the nucleic acid molecule into the tail vein over a short space of time; although injected nucleic acid can be subsequently found in many organs, the bulk is transferred to the liver (25–27) due to this organ having more access to the vena cava, which takes up the injected volume, and a non-selective entry into hepatocytes (26). HTVI has frequently been used in siRNA in vivo studies (28,29). However, the large volumes used have been shown to cause some damage to cell structure, and the loss of cell viability within the liver (30), suggesting that a decrease in gene expression following HTVI-based delivery of siRNA may be due to non-specific degradation of RNA following cell death, rather than siRNA-induced RNAi. This further underlines the importance of confirming that mRNA knockdown results from an siRNA-mediated cleavage event.
The ApoB-1 siRNA sequence used in these studies has been used successfully in a number of previous studies. Soutschek et al. (12) used cholesterol-conjugated siRNA and achieved 50% knockdown of ApoB in mice, confirmed by 5′-RLM-RACE. Cleavage products were also detected when cholesterol-conjugated siRNA were bound to high density or low density lipoprotein that were subsequently perfused through mouse livers (13); in this case the subsequent 5′-RLM-RACE PCRs were radio-labelled to increase sensitivity. Both studies showed that decreases in liver ApoB mRNA levels of ∼50%, were followed by significant decreases in serum levels of apolipoprotein B. This indicates that the level of knockdown we observed using HTVI is comparable to that seen with the different delivery methods employed in these studies. In comparison, when the same ApoB-1 siRNA was delivered with stable nucleic acid lipid particles (SNALPs), expression was decreased to 10% at 2.5 mg/kg both in mice and in non-human primates (16). Again, this knockdown was confirmed by 5′-RLM-RACE (15).
Apart from the studies listed above, confirmation of the cleavage of specific mRNA sequences directed by the siRNA is not commonly reported. This may be due to the difficulty of amplifying a clean RACE product for latter sequencing, or the difficulty in delivering to organs other than the liver. The imaging of the RACE product can be improved with the use of radiolabelled (10,13) or digoxygenin-labelled primers (18), or by using radiolabelled probes specific to the amplified product (11), whereas two rounds of nested RACE reactions were sufficient to generate RACE products when knockdown was more effective (16). SNALP used as a delivery vehicle was also highly effective with in vivo studies on polo-like kinase 1 (Plk1) and kinase spindle protein (Ksp) in mouse hepatic tumours, achieving 45% knockdown in expression and being confirmed with one round of 5′-RLM-RACE (17). The results of these in vivo studies suggests that more effective knockdown leads to more efficient amplification of a positive 5′-RLM-RACE product. Nevertheless, when MBRACE assays were performed following the delivery of the ApoB-1 siRNA to mouse livers using HTVI, evidence of specific cleavage was observed even in cases where knockdown as measured by real-time PCR was negligible. In three independent studies real-time RT–qPCR analysis consistently showed at best 30–50% knockdown of ApoB mRNA, and even with this level of knockdown clear 5′-RLM-RACE results were not obtained. However, MBRACE results for the samples showed significantly more fluorescence than seen in negative control samples indicating the presence of the specifically-cleaved ApoB transcript. These results confirmed not only the successful delivery of the ApoB-1 siRNA but also that RNAi-mediated cleavage of the target had indeed occurred. These results further demonstrate the sensitivity, combined with the inherent specificity, of the MBRACE method and its utility in the analysis of in vivo experiments.
The development of siRNAs for clinical use is being pursued for an ever widening range of targets, making it all the more surprising, with the precedent set by the studies described earlier, that it is still rare to see confirmation that an siRNA-mediated effect observed in vivo has indeed occurred via RNAi. Two recent reports included RACE data at the in vitro stage of development to validate the activity of the siRNA used. A study targeting the neurotensin receptor 2 (NTS2) mRNA with a 27-mer dicer-substrate siRNA used 5′RACE both to ensure that the 27-mer employed was diced to the active 21-mer siRNA, as well as to confirm that the excised siRNA cleaved the target NTS2 mRNA at the predicted site (31). Similarly, 5′RACE was used to show that β1,3-d-glucan-encapsulated siRNA particles mediated knockdown of the targeted mRNA of mitogen activated kinase kinase kinase kinase 4 (Map4k4) (32). Neither study, however, applied 5′RACE methodology to subsequent in vivo experiments, and successful 5′RACE detection from in vitro samples does not necessarily mean that the subsequent knockdown of the target mRNA observed in vivo also results from an RNAi-mediated cleavage event.
A more surprising trend in recently published reports is the absence of RACE data altogether. Instead, many reports describing the successful application of siRNA in in vivo studies still present real-time RT–qPCR results without confirmatory RACE data (33–38). Some do not measure the decrease in the target mRNA at all, and instead use measurements of just the protein target, the disease phenotype or tumour size (39–41). While all of these studies show activity of siRNA in vivo, they lack the demonstration of a biological link between the observed phenotypic effects and the activation of the RNAi machinery by the delivered siRNA. In light of reports demonstrating that the immune stimulatory properties of siRNAs can lead to some of the effects observed when siRNAs are administered in vivo (6,7), we suggest that incorporation of 5′RACE-based methodologies become a requirement for the publication of studies applying siRNA in vivo. Our results demonstrate that the incorporation of a molecular beacon specific to the siRNA-mediated cleavage site during the second round of RACE in a real-time PCR setting enables specific, rapid and sensitive detection of cleaved mRNA in vivo. Employing this methodology to future studies of RNAi in vivo will help to clarify the relative importance of specific and non-specific effects caused by administration of siRNAs in animal models of metabolic disease, cancer and viral infection without recourse to time-consuming cloning and sequencing steps.
FUNDING
Funding for open access charge: Genesis Research & Development Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dykxhoorn DM, Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- 2.Behlke MA. Progress towards in vivo use of siRNAs. Mol. Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 5.Rossi JJ. Innate immunity confounds the clinical efficacy of small interfering RNAs (siRNAs) Gene Ther. 2009;16:579–580. doi: 10.1038/gt.2009.26. [DOI] [PubMed] [Google Scholar]
- 6.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L, McClintock K, Maclachlan I. Misinterpreting the therapeutic effects of siRNA caused by immune stimulation. Hum. Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 10.Fromont-Racine M, Bertrand E, Pictet R, Grange T. A highly sensitive method for mapping the 5′termini of mRNAs. Nucleic Acids Res. 1993;21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volloch V, Schweitzer B, Rits S. Ligation-mediated amplification of RNA from murine erythroid cells reveals a novel class of beta-globin mRNA with an extended 5′-untranslated regions. Nucleic Acids Res. 1994;22:2507–2511. doi: 10.1093/nar/22.13.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 13.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 14.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl Acad. Sci. USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querbes W, Ge P, Zhang W, Fan Y, Costigan J, Charisse K, Maier M, Nechev L, Manoharan M, Kotelianski V, et al. Direct CNS delivery of siRNA mediates robust silencing in oligodendrocytes. Oligonucleotides. 2009;19:23–30. doi: 10.1089/oli.2008.0165. [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 17.Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, Ambegia E, McClintock K, MacLachlan I. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J. Clin. Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giles RV, Spiller DG, Tidd DM. Detection of ribonuclease H-generated mRNA fragments in human leukemia cells following reversible membrane permeabilization in the presence of antisense oligodeoxynucleotides. Antisense Res. Dev. 1995;5:23–31. doi: 10.1089/ard.1995.5.23. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 20.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 21.Marras SA, Tyagi S, Kramer FR. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Reid G, Coppieters ‘t Wallant N, Patel R, Antonic A, Saxon-Aliifaalogo F, Cao H, Webster G, Watson JD. Potent subunit-specific effects on cell growth and drug sensitivity from optimised siRNA-mediated silencing of ribonucleotide reductase. J. RNAi Gene Silencing. 2009;5:321–330. [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Leone G, van Schijndel H, van Gemen B, Kramer FR, Schoen CD. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson SD, Jackson LN, Chen LA, Rychahou PG, Evers BM. Effectiveness of siRNA uptake in target tissues by various delivery methods. Surgery. 2007;142:262–269. doi: 10.1016/j.surg.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- 27.Suda T, Gao X, Stolz DB, Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]
- 28.De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, He Y, Caguyong MJ, Bloomer S, Herweijer H, et al. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 2006;34:4486–4494. doi: 10.1093/nar/gkl609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebestyén MG, Budker VG, Budker T, Subbotin VM, Zhang G, Monahan SD, Lewis DL, Wong SC, Hagstrom JE, Wolff JA. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J. Gene Med. 2006;8:852–873. doi: 10.1002/jgm.921. [DOI] [PubMed] [Google Scholar]
- 31.Doré-Savard L, Roussy G, Dansereau MA, Collingwood MA, Lennox KA, Rose SD, Beaudet N, Behlke MA, Sarret P. Central delivery of Dicer-substrate siRNA: a direct application for pain research. Mol. Ther. 2008;16:1331–1339. doi: 10.1038/mt.2008.98. [DOI] [PubMed] [Google Scholar]
- 32.Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, Ostroff GR, Czech MP. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Navarro F, Lal A, Basar E, Pandey RK, Manoharan M, Feng Y, Lee SJ, Lieberman J, Palliser D. Durable protection from Herpes Simplex Virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe. 2009;22:84–94. doi: 10.1016/j.chom.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shim J, Byun HO, Lee YD, Lee ES, Sohn S. Interleukin-6 small interfering RNA improved the herpes simplex virus-induced systemic inflammation in vivo Behcet's; disease-like mouse model. Gene Ther. 2009;16:415–425. doi: 10.1038/gt.2008.180. [DOI] [PubMed] [Google Scholar]
- 35.Villares GJ, Zigler M, Wang H, Melnikova VO, Wu H, Friedman R, Leslie MC, Vivas-Mejia PE, Lopez-Berestein G, Sood AK, et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, Röder N, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- 37.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmona S, Jorgensen MR, Kolli S, Crowther C, Salazar FH, Marion PL, Fujino M, Natori Y, Thanou M, Arbuthnot P, et al. Controlling HBV replication in vivo by intravenous administration of triggered PEGylated siRNA-nanoparticles. Mol. Pharm. 2009;6:706–717. doi: 10.1021/mp800157x. [DOI] [PubMed] [Google Scholar]
- 39.Sonoke S, Ueda T, Fujiwara K, Sato Y, Takagaki K, Hirabayashi K, Ohgi T, Yano J. Tumor regression in mice by delivery of Bcl-2 small interfering RNA with pegylated cationic liposomes. Cancer Res. 2008;68:8843–8851. doi: 10.1158/0008-5472.CAN-08-0127. [DOI] [PubMed] [Google Scholar]
- 40.Mangala LS, Zuzel V, Schmandt R, Leshane ES, Halder JB, Armaiz-Pena GN, Spannuth WA, Tanaka T, Shahzad MM, Lin YG, et al. Therapeutic targeting of ATP7B in ovarian carcinoma. Clin. Cancer Res. 2009;15:3770–3780. doi: 10.1158/1078-0432.CCR-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran MA, Gowda R, Sharma A, Park EJ, Adair J, Kester M, Smith NB, Robertson GP. Targeting V600EB-Raf and Akt3 using nanoliposomal-small interfering RNA inhibits cutaneous melanocytic lesion development. Cancer Res. 2008;68:7638–7649. doi: 10.1158/0008-5472.CAN-07-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]